bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_0907_orf1
Length=269
Score E
Sequences producing significant alignments: (Bits) Value
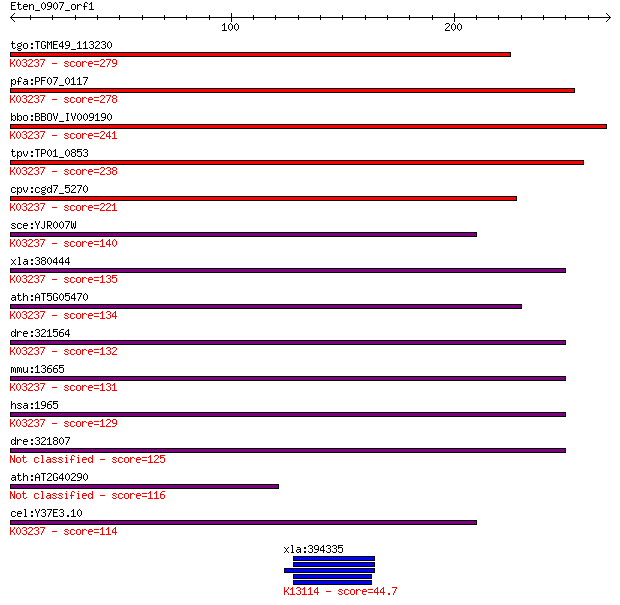
tgo:TGME49_113230 eukaryotic translation initiation factor 2 a... 279 6e-75
pfa:PF07_0117 eukaryotic translation initiation factor 2 alpha... 278 1e-74
bbo:BBOV_IV009190 23.m05857; translation initiation factor 2 a... 241 3e-63
tpv:TP01_0853 eukaryotic translation initiation factor 2 subun... 238 2e-62
cpv:cgd7_5270 Translation initiation factor 2, alpha subunit(e... 221 3e-57
sce:YJR007W SUI2; Alpha subunit of the translation initiation ... 140 4e-33
xla:380444 eif2s1, MGC53429, eif2a; eukaryotic translation ini... 135 1e-31
ath:AT5G05470 EIF2 ALPHA; RNA binding / translation initiation... 134 4e-31
dre:321564 eif2s1, MGC56347, eif2, fb19d01, wu:fb19d01; eukary... 132 1e-30
mmu:13665 Eif2s1, 0910001O23Rik, 2410026C18Rik, 35kDa, Eif2a, ... 131 2e-30
hsa:1965 EIF2S1, EIF-2, EIF-2A, EIF-2alpha, EIF2, EIF2A; eukar... 129 2e-29
dre:321807 eif2s1l, Eif2s1, fb36a08, wu:fb36a08, zgc:56510; eu... 125 1e-28
ath:AT2G40290 eukaryotic translation initiation factor 2 subun... 116 1e-25
cel:Y37E3.10 hypothetical protein; K03237 translation initiati... 114 3e-25
xla:394335 np-a; nuclear protein; K13114 pinin 44.7 4e-04
> tgo:TGME49_113230 eukaryotic translation initiation factor 2
alpha subunit, putative (EC:2.7.7.8); K03237 translation initiation
factor 2 subunit 1
Length=347
Score = 279 bits (714), Expect = 6e-75, Method: Compositional matrix adjust.
Identities = 143/224 (63%), Positives = 163/224 (72%), Gaps = 31/224 (13%)
Query 1 IDLSKSRVSPEDIVKCEERYSKGKKVHQTVRHVALKHNLGVDELNAKVIWPLYKKYGHAL 60
IDLSK RVSPEDIVKCEE++SK KKVHQTVRH A KH + VD+LN VIWPLY+KYGHAL
Sbjct 102 IDLSKRRVSPEDIVKCEEKFSKSKKVHQTVRHAAQKHGMKVDDLNRSVIWPLYRKYGHAL 161
Query 61 DALKAAALDPEEVFGGLDVSEEVKKSLIQDIQLRLAPQALKLRARIDVWCFGKDGIDAVK 120
DALK AA+ P+EVF GL+V EEV+KSLIQDIQLRLAPQALKLRAR+DVWCFGK GIDAVK
Sbjct 162 DALKEAAMRPDEVFAGLEVDEEVRKSLIQDIQLRLAPQALKLRARVDVWCFGKQGIDAVK 221
Query 121 KALLAAKEKAAETPEKAAETPEKAAETPEKAAETPERAVDTPQTQDIAVKLIAPPQYVVV 180
AL A +E D T I +KLIAPPQYVVV
Sbjct 222 AALQAGQEVG-----------------------------DDEVT--INIKLIAPPQYVVV 250
Query 181 TSSFDKESGMQKIQQAISRISQTIKSYRGGDFKQQGEIMVLGSE 224
TS +DKE GM+KI+QA+ IS IKS+ GGDFKQQGEI+V+G +
Sbjct 251 TSCYDKELGMRKIEQAMKAISDKIKSFSGGDFKQQGEIVVMGGD 294
> pfa:PF07_0117 eukaryotic translation initiation factor 2 alpha
subunit, putative; K03237 translation initiation factor 2
subunit 1
Length=329
Score = 278 bits (712), Expect = 1e-74, Method: Compositional matrix adjust.
Identities = 141/255 (55%), Positives = 179/255 (70%), Gaps = 33/255 (12%)
Query 1 IDLSKSRVSPEDIVKCEERYSKGKKVHQTVRHVALKHNLGVDELNAKVIWPLYKKYGHAL 60
IDLSK RVSP+DI+KCEE++SK KKVHQTVRHVA +H + V+ELN K IWPLY++YGHAL
Sbjct 90 IDLSKRRVSPKDIIKCEEKFSKSKKVHQTVRHVAKEHGITVEELNEKAIWPLYERYGHAL 149
Query 61 DALKAAALDPEEVFGGLDVSEEVKKSLIQDIQLRLAPQALKLRARIDVWCFGKDGIDAVK 120
DALK A ++PE VF GLD+SEE+K SL++DI+LRL PQALKLR RIDVWCFG +GIDAVK
Sbjct 150 DALKEATMNPENVFKGLDISEEIKNSLLKDIKLRLTPQALKLRGRIDVWCFGYEGIDAVK 209
Query 121 KALLAAKEKAAETPEKAAETPEKAAETPEKAAETPERAVDTPQTQDIAVKLIAPPQYVVV 180
+AL KE + + I +KLIAPPQYV+V
Sbjct 210 EALKKGKEISNDKVS-------------------------------INIKLIAPPQYVIV 238
Query 181 TSSFDKESGMQKIQQAISRISQTIKSYRGGDFKQQGEIMVLGSEEDRRLEELL--LEVSE 238
TS DK+ GM KIQ+A+ IS IK Y+GGDFKQQGEI+V+G +E++RLEELL +
Sbjct 239 TSCHDKDLGMAKIQEAMKVISDKIKEYKGGDFKQQGEILVIGGDEEKRLEELLDKHDGIS 298
Query 239 SEEEEEEEEEEEDEG 253
S++++ +E+DE
Sbjct 299 SDDDDYNTSDEDDEN 313
> bbo:BBOV_IV009190 23.m05857; translation initiation factor 2
alpha subunit; K03237 translation initiation factor 2 subunit
1
Length=332
Score = 241 bits (614), Expect = 3e-63, Method: Compositional matrix adjust.
Identities = 137/272 (50%), Positives = 173/272 (63%), Gaps = 38/272 (13%)
Query 1 IDLSKSRVSPEDIVKCEERYSKGKKVHQTVRHVALKHNLGVDELNAKVIWPLYKKYGHAL 60
IDLSK RVSPEDIVKCEE++SK KKVHQTVRH+A KH + V+ELN IWPLY++Y +AL
Sbjct 88 IDLSKRRVSPEDIVKCEEKFSKAKKVHQTVRHIAQKHGMSVEELNRICIWPLYQRYPNAL 147
Query 61 DALKAAALDPEEVFGGLDVSEEVKKSLIQDIQLRLAPQALKLRARIDVWCFGKDGIDAVK 120
DALK AA++ +F L ++ EV SLI DIQLRL PQALKLR IDVWCFG DGI+AVK
Sbjct 148 DALKEAAINKTNIFKDLPIAPEVIDSLIADIQLRLTPQALKLRCMIDVWCFGPDGIEAVK 207
Query 121 KALLAAKEKAAETPEKAAETPEKAAETPEKAAETPERAVDTPQTQDIAVKLIAPPQYVVV 180
AL+ AK + +I VKLIAPPQY ++
Sbjct 208 SALMLAKAN---------------------------------EEHNIQVKLIAPPQYEIM 234
Query 181 TSSFDKESGMQKIQQAISRISQTIKSYRGGDFKQQGEIMVLGSEEDRRLEELL----LEV 236
T+ DK+ GM I A+ IS I+SY GG+FKQ+G+I+++G E++R L LL
Sbjct 235 TTCHDKDKGMDIITTALEEISNKIRSYAGGEFKQRGDIIIVGDEDERHLYSLLEGQESSE 294
Query 237 SESEEEEEEEEEEEDEGMGRV-QAGVPEDEVE 267
E + +E EEEDEGMGRV ++ +P D VE
Sbjct 295 EEEDSSSSDESEEEDEGMGRVDESMLPPDAVE 326
> tpv:TP01_0853 eukaryotic translation initiation factor 2 subunit
alpha; K03237 translation initiation factor 2 subunit 1
Length=342
Score = 238 bits (607), Expect = 2e-62, Method: Compositional matrix adjust.
Identities = 136/258 (52%), Positives = 177/258 (68%), Gaps = 26/258 (10%)
Query 1 IDLSKSRVSPEDIVKCEERYSKGKKVHQTVRHVALKHNLGVDELNAKVIWPLYKKYGHAL 60
IDLSK RV+PEDI+KCEE++SK KKVHQTVRH+A KH + V+ELN + IWPLYK Y HAL
Sbjct 89 IDLSKRRVTPEDIIKCEEKFSKSKKVHQTVRHIAQKHGISVEELNRECIWPLYKAYPHAL 148
Query 61 DALKAAALDPEEVFGGLDVSEEVKKSLIQDIQLRLAPQALKLRARIDVWCFGKDGIDAVK 120
DALK AA + E VF + +S+EV SL+QDIQLRL PQALKL+ +DVWCFG +GI+AVK
Sbjct 149 DALKEAASNKENVFKNISISQEVIDSLLQDIQLRLVPQALKLKCSVDVWCFGPEGINAVK 208
Query 121 KALLAAKEKAAETPEKAAETPEKAAETPEKAAETPERAVDTPQTQDIAVKLIAPPQYVVV 180
+L AK P+ T + I+++LIAPPQY ++
Sbjct 209 MSLGKAK------------------------LLDPQVIHTTHLYECISIRLIAPPQYEIL 244
Query 181 TSSFDKESGMQKIQQAISRISQTIKSYRGGDFKQQGEIMVLGSEEDRRLEELL-LEVSES 239
TS FDKE+G+ + Q + I + IKS+ GGDFKQ+ E++V+G +E + LE+LL L +
Sbjct 245 TSCFDKEAGLALMNQTLEVIEKNIKSFAGGDFKQKCEVVVIGDDE-KHLEDLLELHETTD 303
Query 240 EEEEEEEEEEEDEGMGRV 257
EEE +E+EEEEDEGMGRV
Sbjct 304 EEEGDEDEEEEDEGMGRV 321
> cpv:cgd7_5270 Translation initiation factor 2, alpha subunit(eIF2-alpha);
S1-like RNA binding domain ; K03237 translation
initiation factor 2 subunit 1
Length=355
Score = 221 bits (562), Expect = 3e-57, Method: Compositional matrix adjust.
Identities = 111/227 (48%), Positives = 150/227 (66%), Gaps = 30/227 (13%)
Query 1 IDLSKSRVSPEDIVKCEERYSKGKKVHQTVRHVALKHNLGVDELNAKVIWPLYKKYGHAL 60
IDLSK RV+PED+ +CEE+YSK KKV+QTVRH+A K + V+EL+ K+IWPL++KYGHAL
Sbjct 108 IDLSKRRVNPEDVERCEEKYSKMKKVYQTVRHIAQKEGISVEELSEKLIWPLHRKYGHAL 167
Query 61 DALKAAALDPEEVFGGLDVSEEVKKSLIQDIQLRLAPQALKLRARIDVWCFGKDGIDAVK 120
DALK AA++PE V G D+ VK++LIQDI+ RL+PQ LKLRARI V C G DGIDAV+
Sbjct 168 DALKEAAVNPEAVLGEFDLPASVKEALIQDIKFRLSPQQLKLRARIHVCCCGYDGIDAVR 227
Query 121 KALLAAKEKAAETPEKAAETPEKAAETPEKAAETPERAVDTPQTQDIAVKLIAPPQYVVV 180
A++A +E T +I VK IAPPQY+V
Sbjct 228 HAIIAGQETV------------------------------TKDDVEIQVKFIAPPQYLVT 257
Query 181 TSSFDKESGMQKIQQAISRISQTIKSYRGGDFKQQGEIMVLGSEEDR 227
S+ K +G++ I++A+ I I Y+GGDFK+QGEI V+G ++++
Sbjct 258 AVSYGKAAGVEAIEKAMETIKNVITQYKGGDFKRQGEIEVVGGDDEK 304
> sce:YJR007W SUI2; Alpha subunit of the translation initiation
factor eIF2, involved in the identification of the start codon;
phosphorylation of Ser51 is required for regulation of
translation by inhibiting the exchange of GDP for GTP; K03237
translation initiation factor 2 subunit 1
Length=304
Score = 140 bits (354), Expect = 4e-33, Method: Compositional matrix adjust.
Identities = 81/210 (38%), Positives = 122/210 (58%), Gaps = 33/210 (15%)
Query 1 IDLSKSRVSPEDIVKCEERYSKGKKVHQTVRHVALKHNLGVDELNAKVIWPLYKKYGHAL 60
IDLSK RVS EDI+KCEE+Y K K VH +R+ A K + ++EL + WPL +K+GHA
Sbjct 83 IDLSKRRVSSEDIIKCEEKYQKSKTVHSILRYCAEKFQIPLEELYKTIAWPLSRKFGHAY 142
Query 61 DALKAAALDPEEVFGGLDV-SEEVKKSLIQDIQLRLAPQALKLRARIDVWCFGKDGIDAV 119
+A K + +D E V+ G++ S++V L I RL PQA+K+RA ++V CF +GIDA+
Sbjct 143 EAFKLSIID-ETVWEGIEPPSKDVLDELKNYISKRLTPQAVKIRADVEVSCFSYEGIDAI 201
Query 120 KKALLAAKEKAAETPEKAAETPEKAAETPEKAAETPERAVDTPQTQDIAVKLIAPPQYVV 179
K AL +A++ + E Q Q + VKL+A P YV+
Sbjct 202 KDALKSAEDMSTE------------------------------QMQ-VKVKLVAAPLYVL 230
Query 180 VTSSFDKESGMQKIQQAISRISQTIKSYRG 209
T + DK+ G+++++ AI +I++ I Y G
Sbjct 231 TTQALDKQKGIEQLESAIEKITEVITKYGG 260
> xla:380444 eif2s1, MGC53429, eif2a; eukaryotic translation initiation
factor 2, subunit 1 alpha, 35kDa; K03237 translation
initiation factor 2 subunit 1
Length=315
Score = 135 bits (341), Expect = 1e-31, Method: Compositional matrix adjust.
Identities = 95/261 (36%), Positives = 142/261 (54%), Gaps = 45/261 (17%)
Query 1 IDLSKSRVSPEDIVKCEERYSKGKKVHQTVRHVA----LKHNLGVDELNAKVIWPL---Y 53
IDLSK RVSPE+ +KCE++++K K V+ +RHVA + +D L + W Y
Sbjct 83 IDLSKRRVSPEEALKCEDKFTKSKTVYSILRHVAEVLDYTKDEQLDSLFQRTAWVFDEKY 142
Query 54 KKYGH-ALDALKAAALDPEEVFGGLDVSEEVKKSLIQDIQLRLAPQALKLRARIDVWCFG 112
KK G+ A DA K A DP+ + GLD+SE+ ++ LI +I RL PQA+K+RA I+V C+G
Sbjct 143 KKPGYGAYDAFKNAVSDPD-ILDGLDLSEDERRVLIDNINRRLTPQAVKIRADIEVACYG 201
Query 113 KDGIDAVKKALLAAKEKAAETPEKAAETPEKAAETPEKAAETPERAVDTPQTQDIAVKLI 172
+GIDAVK AL A + E + P I + LI
Sbjct 202 YEGIDAVKDALRAGLSCSTE---------------------------NMP----IKINLI 230
Query 173 APPQYVVVTSSFDKESGMQKIQQAISRISQTIKSYRGGDFKQQGEIMVLG----SEEDRR 228
APP+YV+ T++ ++ G+ + QA+S I + I+ RG F Q E V+ +E R+
Sbjct 231 APPRYVMTTTTLERTEGLSVLNQAMSVIKERIEEKRGV-FNVQMEPKVVTDTDETELARQ 289
Query 229 LEELLLEVSESEEEEEEEEEE 249
LE L E +E + +++ +E E
Sbjct 290 LERLEKENAEVDGDDDADEME 310
> ath:AT5G05470 EIF2 ALPHA; RNA binding / translation initiation
factor; K03237 translation initiation factor 2 subunit 1
Length=344
Score = 134 bits (336), Expect = 4e-31, Method: Compositional matrix adjust.
Identities = 85/248 (34%), Positives = 124/248 (50%), Gaps = 52/248 (20%)
Query 1 IDLSKSRVSPEDIVKCEERYSKGKKVHQTVRHVALKHNLGVDELNAKVIWPLYKKYGHAL 60
IDLSK RVS ED CEERY+K K VH +RHVA + ++EL + WPLYKK+GHA
Sbjct 87 IDLSKRRVSDEDKEACEERYNKSKLVHSIMRHVAETVGVDLEELYVNIGWPLYKKHGHAF 146
Query 61 DALKAAALDPEEVFGGLD-------------------VSEEVKKSLIQDIQLRLAPQALK 101
+A K DP+ VF L VSEE+K + ++DI+ R+ PQ +K
Sbjct 147 EAFKIVVTDPDSVFDALTREVKETGPDGVEVTKVVPAVSEELKDAFLKDIRRRMTPQPMK 206
Query 102 LRARIDVWCFGKDGIDAVKKALLAAKEKAAETPEKAAETPEKAAETPEKAAETPERAVDT 161
+RA I++ CF DG+ +K+A+ A+ + D
Sbjct 207 IRADIELKCFQFDGVLHIKEAMKKAEAVGTD---------------------------DC 239
Query 162 PQTQDIAVKLIAPPQYVVVTSSFDKESGMQKIQQAISRISQTIKSYRGGDFKQQGEIMVL 221
P + +KL+APP YV+ T + KE G+ + +AI I+ ++G ++G V
Sbjct 240 P----VKIKLVAPPLYVLTTHTHYKEKGIVTLNKAIEACITAIEEHKGKLVVKEGARAV- 294
Query 222 GSEEDRRL 229
SE D +L
Sbjct 295 -SERDDKL 301
> dre:321564 eif2s1, MGC56347, eif2, fb19d01, wu:fb19d01; eukaryotic
translation initiation factor 2, subunit 1 alpha; K03237
translation initiation factor 2 subunit 1
Length=315
Score = 132 bits (333), Expect = 1e-30, Method: Compositional matrix adjust.
Identities = 94/261 (36%), Positives = 137/261 (52%), Gaps = 45/261 (17%)
Query 1 IDLSKSRVSPEDIVKCEERYSKGKKVHQTVRHVALKHNLGVDE----LNAKVIWPL---Y 53
IDLSK RVSPE+ +KCE++++K K V+ +RHVA DE L + W Y
Sbjct 83 IDLSKRRVSPEEAIKCEDKFTKSKTVYSILRHVAEVLEYTKDEQLESLFQRTAWVFDEKY 142
Query 54 KKYGH-ALDALKAAALDPEEVFGGLDVSEEVKKSLIQDIQLRLAPQALKLRARIDVWCFG 112
KK G+ A D K A DP + GLD++EE + LI +I RL PQA+K+RA I+V C+G
Sbjct 143 KKPGYGAYDVFKQAVSDPA-ILDGLDLTEEERNVLIDNINRRLTPQAVKIRADIEVACYG 201
Query 113 KDGIDAVKKALLAAKEKAAETPEKAAETPEKAAETPEKAAETPERAVDTPQTQDIAVKLI 172
+GIDAVK+AL A + E I + LI
Sbjct 202 YEGIDAVKEALRAGLNCSTE-------------------------------AMPIKINLI 230
Query 173 APPQYVVVTSSFDKESGMQKIQQAISRISQTIKSYRGGDFKQQGEIMVLG----SEEDRR 228
APP+YV+ T++ ++ G+ + QA++ I + I+ RG F Q E V+ +E R+
Sbjct 231 APPRYVMTTTTLERTEGLSVLNQAMAAIKERIEEKRGV-FNVQMEPKVVTDTDETELQRQ 289
Query 229 LEELLLEVSESEEEEEEEEEE 249
LE L E +E + +++ EE E
Sbjct 290 LERLERENAEVDGDDDAEEME 310
> mmu:13665 Eif2s1, 0910001O23Rik, 2410026C18Rik, 35kDa, Eif2a,
eIF2alpha; eukaryotic translation initiation factor 2, subunit
1 alpha; K03237 translation initiation factor 2 subunit
1
Length=315
Score = 131 bits (330), Expect = 2e-30, Method: Compositional matrix adjust.
Identities = 93/261 (35%), Positives = 139/261 (53%), Gaps = 45/261 (17%)
Query 1 IDLSKSRVSPEDIVKCEERYSKGKKVHQTVRHVALKHNLGVDE----LNAKVIWPL---Y 53
IDLSK RVSPE+ +KCE++++K K V+ +RHVA DE L + W Y
Sbjct 83 IDLSKRRVSPEEAIKCEDKFTKSKTVYSILRHVAEVLEYTKDEQLESLFQRTAWVFDDKY 142
Query 54 KKYGH-ALDALKAAALDPEEVFGGLDVSEEVKKSLIQDIQLRLAPQALKLRARIDVWCFG 112
K+ G+ A DA K A DP + LD++E+ ++ LI +I RL PQA+K+RA I+V C+G
Sbjct 143 KRPGYGAYDAFKHAVSDPS-ILDSLDLNEDEREVLINNINRRLTPQAVKIRADIEVACYG 201
Query 113 KDGIDAVKKALLAAKEKAAETPEKAAETPEKAAETPEKAAETPERAVDTPQTQDIAVKLI 172
+GIDAVK+AL A + E T I + LI
Sbjct 202 YEGIDAVKEALRAGLNCSTE-------------------------------TMPIKINLI 230
Query 173 APPQYVVVTSSFDKESGMQKIQQAISRISQTIKSYRGGDFKQQGEIMVLG----SEEDRR 228
APP+YV+ T++ ++ G+ + QA++ I + I+ RG F Q E V+ +E R+
Sbjct 231 APPRYVMTTTTLERTEGLSVLNQAMAVIKEKIEEKRGV-FNVQMEPKVVTDTDETELARQ 289
Query 229 LEELLLEVSESEEEEEEEEEE 249
LE L E +E + +++ EE E
Sbjct 290 LERLERENAEVDGDDDAEEME 310
> hsa:1965 EIF2S1, EIF-2, EIF-2A, EIF-2alpha, EIF2, EIF2A; eukaryotic
translation initiation factor 2, subunit 1 alpha, 35kDa;
K03237 translation initiation factor 2 subunit 1
Length=315
Score = 129 bits (323), Expect = 2e-29, Method: Compositional matrix adjust.
Identities = 92/261 (35%), Positives = 140/261 (53%), Gaps = 45/261 (17%)
Query 1 IDLSKSRVSPEDIVKCEERYSKGKKVHQTVRHVALKHNLGVDE----LNAKVIWPL---Y 53
IDLSK RVSPE+ +KCE++++K K V+ +RHVA DE L + W Y
Sbjct 83 IDLSKRRVSPEEAIKCEDKFTKSKTVYSILRHVAEVLEYTKDEQLESLFQRTAWVFDDKY 142
Query 54 KKYGH-ALDALKAAALDPEEVFGGLDVSEEVKKSLIQDIQLRLAPQALKLRARIDVWCFG 112
K+ G+ A DA K A DP + LD++E+ ++ LI +I RL PQA+K+RA I+V C+G
Sbjct 143 KRPGYGAYDAFKHAVSDPS-ILDSLDLNEDEREVLINNINRRLTPQAVKIRADIEVACYG 201
Query 113 KDGIDAVKKALLAAKEKAAETPEKAAETPEKAAETPEKAAETPERAVDTPQTQDIAVKLI 172
+GIDAVK+AL A + E + P I + LI
Sbjct 202 YEGIDAVKEALRAGLNCSTE---------------------------NMP----IKINLI 230
Query 173 APPQYVVVTSSFDKESGMQKIQQAISRISQTIKSYRGGDFKQQGEIMVLG----SEEDRR 228
APP+YV+ T++ ++ G+ + QA++ I + I+ RG F Q E V+ +E R+
Sbjct 231 APPRYVMTTTTLERTEGLSVLSQAMAVIKEKIEEKRGV-FNVQMEPKVVTDTDETELARQ 289
Query 229 LEELLLEVSESEEEEEEEEEE 249
+E L E +E + +++ EE E
Sbjct 290 MERLERENAEVDGDDDAEEME 310
> dre:321807 eif2s1l, Eif2s1, fb36a08, wu:fb36a08, zgc:56510;
eukaryotic translation initiation factor 2, subunit 1 alpha,
like
Length=315
Score = 125 bits (314), Expect = 1e-28, Method: Compositional matrix adjust.
Identities = 91/261 (34%), Positives = 136/261 (52%), Gaps = 45/261 (17%)
Query 1 IDLSKSRVSPEDIVKCEERYSKGKKVHQTVRHVALKHNLGVDE----LNAKVIWPL---Y 53
IDLSK RVSPE+ +KCE++++K K V+ +RHVA DE L + W Y
Sbjct 83 IDLSKRRVSPEEAIKCEDKFTKSKTVYSILRHVAEVLEYTKDEQLESLYQRTAWVFDEKY 142
Query 54 KKYGH-ALDALKAAALDPEEVFGGLDVSEEVKKSLIQDIQLRLAPQALKLRARIDVWCFG 112
K+ G+ A D K A DP + L+++EE + LI +I RL PQA+K+RA I+V C+G
Sbjct 143 KRPGYGAYDVFKQAVSDPA-ILDCLELTEEERAVLIDNINRRLTPQAVKIRADIEVACYG 201
Query 113 KDGIDAVKKALLAAKEKAAETPEKAAETPEKAAETPEKAAETPERAVDTPQTQDIAVKLI 172
+GIDAVK+AL A + E I + LI
Sbjct 202 YEGIDAVKEALRAGLTCSTE-------------------------------CMPIKINLI 230
Query 173 APPQYVVVTSSFDKESGMQKIQQAISRISQTIKSYRGGDFKQQGEIMVLG----SEEDRR 228
APP+YV+ T++ ++ G+ + QA++ I + I+ RG F Q E V+ +E R+
Sbjct 231 APPRYVMTTTTLERTEGLSVLNQAMAVIKEKIEEKRGV-FNIQMEPKVVTDTDETELARQ 289
Query 229 LEELLLEVSESEEEEEEEEEE 249
LE L E +E + +++ EE E
Sbjct 290 LERLERENAEVDGDDDAEEME 310
> ath:AT2G40290 eukaryotic translation initiation factor 2 subunit
1, putative / eIF-2A, putative / eIF-2-alpha, putative
Length=241
Score = 116 bits (290), Expect = 1e-25, Method: Compositional matrix adjust.
Identities = 58/139 (41%), Positives = 84/139 (60%), Gaps = 19/139 (13%)
Query 1 IDLSKSRVSPEDIVKCEERYSKGKKVHQTVRHVALKHNLGVDELNAKVIWPLYKKYGHAL 60
IDLSK RVS EDI CEERY+K K VH +RHVA ++ +++L + WPLY+++GHA
Sbjct 87 IDLSKRRVSEEDIQTCEERYNKSKLVHSIMRHVAETLSIDLEDLYVNIGWPLYRRHGHAF 146
Query 61 DALKAAALDPEEVFGGLD-------------------VSEEVKKSLIQDIQLRLAPQALK 101
+A K DP+ V G L V+EEVK +L+++I+ R+ PQ +K
Sbjct 147 EAFKILVTDPDSVLGPLTREIKEVGPDGQEVTKVVPAVTEEVKDALVKNIRRRMTPQPMK 206
Query 102 LRARIDVWCFGKDGIDAVK 120
+RA I++ CF DG+ +K
Sbjct 207 IRADIELKCFQFDGVVHIK 225
> cel:Y37E3.10 hypothetical protein; K03237 translation initiation
factor 2 subunit 1
Length=342
Score = 114 bits (286), Expect = 3e-25, Method: Compositional matrix adjust.
Identities = 72/216 (33%), Positives = 109/216 (50%), Gaps = 39/216 (18%)
Query 1 IDLSKSRVSPEDIVKCEERYSKGKKVHQTVRHVALKHNLGVDE----LNAKVIWPL---Y 53
IDLSK RV +D+ +C+ER++ K V+ +RHVA + DE L K W
Sbjct 80 IDLSKRRVYQKDLKQCDERFANAKMVNSILRHVAEQVGYTTDEELEDLYQKTAWHFDRKE 139
Query 54 KKYGHALDALKAAALDPEEVFGGLDVSEEVKKSLIQDIQLRLAPQALKLRARIDVWCFGK 113
K+ + DA K A +P + D+S ++K+ L++DI+ +L PQA+K+RA I+V CF
Sbjct 140 KRKAASYDAFKKAITEPT-ILDECDISADIKEKLLEDIRKKLTPQAVKIRADIEVSCFDY 198
Query 114 DGIDAVKKALLAAKEKAAETPEKAAETPEKAAETPEKAAETPERAVDTPQTQDIAVKLIA 173
DGIDAVK AL+A K + T P I + LIA
Sbjct 199 DGIDAVKAALIAGKNCSNGT------FP-------------------------IKINLIA 227
Query 174 PPQYVVVTSSFDKESGMQKIQQAISRISQTIKSYRG 209
P +VV T + D+E G++ + + I I+ ++G
Sbjct 228 APHFVVTTQTLDREGGIEAVNSILDTIKNAIEGFKG 263
> xla:394335 np-a; nuclear protein; K13114 pinin
Length=718
Score = 44.7 bits (104), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 19/36 (52%), Positives = 30/36 (83%), Gaps = 0/36 (0%)
Query 128 EKAAETPEKAAETPEKAAETPEKAAETPERAVDTPQ 163
E+ AE+PE+ AE+PE+ AE+PE+ AE+PER ++P+
Sbjct 464 EREAESPEREAESPEREAESPEREAESPEREAESPE 499
Score = 44.7 bits (104), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 19/36 (52%), Positives = 30/36 (83%), Gaps = 0/36 (0%)
Query 128 EKAAETPEKAAETPEKAAETPEKAAETPERAVDTPQ 163
E+ AE+PE+ AE+PE+ AE+PE+ AE+PER ++P+
Sbjct 471 EREAESPEREAESPEREAESPEREAESPEREAESPE 506
Score = 42.0 bits (97), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 19/40 (47%), Positives = 31/40 (77%), Gaps = 0/40 (0%)
Query 124 LAAKEKAAETPEKAAETPEKAAETPEKAAETPERAVDTPQ 163
L A+ +A + PE+ AE+PE+ AE+PE+ AE+PER ++P+
Sbjct 453 LEAETEATKEPEREAESPEREAESPEREAESPEREAESPE 492
Score = 41.6 bits (96), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 19/35 (54%), Positives = 27/35 (77%), Gaps = 0/35 (0%)
Query 128 EKAAETPEKAAETPEKAAETPEKAAETPERAVDTP 162
E+ AE+PE+ AE+PE+ AE+PE+ AE+PER P
Sbjct 478 EREAESPEREAESPEREAESPEREAESPERPQKEP 512
Score = 33.1 bits (74), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 15/35 (42%), Positives = 24/35 (68%), Gaps = 0/35 (0%)
Query 128 EKAAETPEKAAETPEKAAETPEKAAETPERAVDTP 162
E+ AE+PE+ AE+PE+ AE+PE+ + P+ P
Sbjct 485 EREAESPEREAESPEREAESPERPQKEPDLRFQPP 519
Lambda K H
0.308 0.128 0.345
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 10082348456
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40