bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_0893_orf4
Length=310
Score E
Sequences producing significant alignments: (Bits) Value
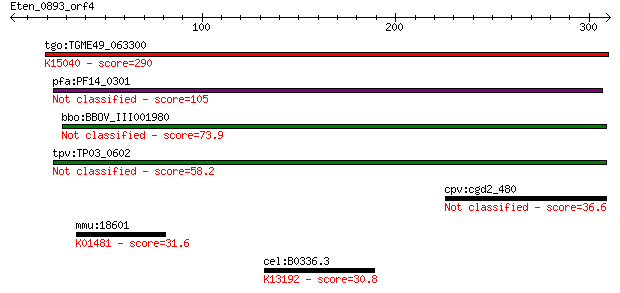
tgo:TGME49_063300 porin, putative ; K15040 voltage-dependent a... 290 4e-78
pfa:PF14_0301 conserved protein, unknown function 105 1e-22
bbo:BBOV_III001980 17.m07193; hypothetical protein 73.9 8e-13
tpv:TP03_0602 hypothetical protein 58.2 4e-08
cpv:cgd2_480 hypothetical protein 36.6 0.13
mmu:18601 Padi3, AI507004, KIAA4171, Pad3, Pdi3, mKIAA4171; pe... 31.6 3.6
cel:B0336.3 hypothetical protein; K13192 RNA-binding protein 26 30.8
> tgo:TGME49_063300 porin, putative ; K15040 voltage-dependent
anion channel protein 2
Length=290
Score = 290 bits (743), Expect = 4e-78, Method: Compositional matrix adjust.
Identities = 135/291 (46%), Positives = 191/291 (65%), Gaps = 2/291 (0%)
Query 19 MVLFKDIGKPASDLLGKGFPHEKPWELEYKYKSKNPQITNVAGVGSHGGFDASSSLVYSA 78
MVLFKD+ K +DLL KG+PHEK W+LEYKYKSKNP+I N A V G FDASS L Y
Sbjct 1 MVLFKDLNKSCADLLTKGYPHEKAWDLEYKYKSKNPEIVNTASVSPAGAFDASSKLKYCV 60
Query 79 GEAKAEVKVFSAGRSYVDLSYKIPQLPNLIISTKYERKGEKNTSDLFDFSTEYVTPRFHS 138
+ EVK+ + G+S +D+ Y P+L L + K++R+G+K + D D EY P HS
Sbjct 61 SDVTTEVKMMAMGKSTIDVKYAAPKLKGLTLGAKFDRRGDKTSKDSLDIVAEYKLPTIHS 120
Query 139 ILNVNPLLRTFSSSGTFTYDRFRFGGEVSGKLDASGMKYALAASYSAPTPGMKGGSWMAA 198
+VNPL +F+ Y FR G EVSGK DAS MKYA+ ASY+ K G + +
Sbjct 121 FFSVNPLASSFNFGNVVEYKAFRIGSEVSGKFDASAMKYAVGASYTGVAS--KAGEFTLS 178
Query 199 VKTAPAGSLMFGKVIGSLNGRSVEGRGAELAPEVEYNIAGNKSNISFGGLWHLSEEKDPI 258
+KTAP+G MFG++IGS++G++ + + AELA EV+ N+ ++NI FGGLW+L+++KD
Sbjct 179 LKTAPSGDAMFGRMIGSVHGKTTDNKSAELAAEVDCNLLDGRTNIQFGGLWYLNDKKDTF 238
Query 259 VKSKISQNGLFGGALTPRFCSNLQAPIGTQLEVPKAQNADNCKYGIKIEVV 309
+K+K++QN ALT + C + A IG+Q++V K N D K+G+K+E+
Sbjct 239 LKAKLTQNARVSVALTHKVCDYVSATIGSQIDVSKPSNPDAVKHGLKLEIC 289
> pfa:PF14_0301 conserved protein, unknown function
Length=289
Score = 105 bits (263), Expect = 1e-22, Method: Compositional matrix adjust.
Identities = 76/289 (26%), Positives = 146/289 (50%), Gaps = 13/289 (4%)
Query 23 KDIGKPASDLLGKGFPHEKPWELEYKYKSKNPQITNVAGVGSHGGFDASSSLVYSAGEAK 82
K + KP+ DLL FP +ELE+ SK+P + + S+ + ++ + +K
Sbjct 5 KLLNKPSLDLLKNDFPLSSKFELEHSSISKHPFLKS-GFTFSNNTYSIYTNFKNNIYNSK 63
Query 83 AEVKVFSAGRSYVDLSYKIPQLPNLIISTKYERKGEKNTSDLFDFSTEYVTPRFHSILNV 142
E+K ++G S +D+ Y+ + NL + KY + K D F+ +EY T + +V
Sbjct 64 NEIKFDNSGISLLDIKYEPGFIKNLNLCGKYTKTNGK-EDDTFEVYSEYTTDDMNIFSSV 122
Query 143 NPLLRTFS-----SSGTFTYDRFRFGGEVSGKLDASGMKYALAASYSAPTPGMKGGSWMA 197
N +R F+ S Y F+FGG + G +D + ++Y+ +Y T K ++
Sbjct 123 N--VRNFAFKYIHVSSHPKYRNFKFGGLLEGNMDINNLQYSFGGAY---TKHHKDNLYIF 177
Query 198 AVKTAPAGSLMFGKVIGSLNGRSVEGRGAELAPEVEYNIAGNKSNISFGGLWHLSEEKDP 257
++++ P+ +G + +L ++ ++ EV NI K+NI+ +W+L + K+
Sbjct 178 SLRSIPSNKHFYGSLALNLFFQNKSINDNAISVEVIQNIMEKKANINVASIWYL-DNKNT 236
Query 258 IVKSKISQNGLFGGALTPRFCSNLQAPIGTQLEVPKAQNADNCKYGIKI 306
VK+KIS + +LT ++ + +G+Q+++ K DN K+G+K+
Sbjct 237 FVKTKISNDTKVALSLTHKYNEFVTITLGSQVDISKMSLPDNTKFGMKL 285
> bbo:BBOV_III001980 17.m07193; hypothetical protein
Length=298
Score = 73.9 bits (180), Expect = 8e-13, Method: Compositional matrix adjust.
Identities = 76/299 (25%), Positives = 119/299 (39%), Gaps = 32/299 (10%)
Query 28 PASDLLGKGFPHEKPWELEYKYKSKNPQITNVAGVGSHGGFDASSSLVYSAGEAKAEVKV 87
PA D+ K F +E WE+E+ K TN + S+S+ G A E+K+
Sbjct 13 PAVDMFRKDFVNENIWEVEHADARKGASFTNTLTLNKDQKLVGSTSVNVDVGAANFEMKL 72
Query 88 FSAGRSYVDLSYKIPQLPNLIISTKYERKGEKNTSDLFDFSTEYVTPRFHSILNVNPLLR 147
+ G Y++LS P + + +KY N ++ + E V P + L + P R
Sbjct 73 GNKGDRYLELSSMNAHWPTMKVLSKY-MFNPPNKNNACEVVAENVHPINTTQLKIIPFAR 131
Query 148 TFSSSGTFTYDRF---RFGG-EVSGKLDASGMKYALAASY-----------SAPTPGMKG 192
+S F Y FGG EV+GK Y +A Y SA G KG
Sbjct 132 DYSMFSLFNYKLNCCQLFGGMEVTGKNCTLFSNYTMALGYKRIRDEKTYQLSARVFGDKG 191
Query 193 GSWMAAVKTAPAGSLMFGKVIGSLNGRSVEGRGAELAPEVEYNIAGNKSNISFGGLWHLS 252
A A S + +G+ +G + +A +E+ + + + F GLWHL+
Sbjct 192 ---------ALAKSFVGNVYVGTAHGDAQNA----MAVALEHQLKDGHTKLMFSGLWHLT 238
Query 253 E---EKDPIVKSKISQNGLFGGALTPRFCSNLQAPIGTQLEVPKAQNADNCKYGIKIEV 308
E VK K +G F + + RF N+ + + + YG K+ V
Sbjct 239 EPGHATPAFVKGKCDTDGQFALSYSQRFNKNIAGILTVGGNMNTTCDPATMNYGYKVTV 297
> tpv:TP03_0602 hypothetical protein
Length=297
Score = 58.2 bits (139), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 66/307 (21%), Positives = 121/307 (39%), Gaps = 40/307 (13%)
Query 23 KDIGKPASDLLGKGFPHEKPWELEYKYKSKNPQITNVAGVGSHGGFDASSSLVYSAGEAK 82
K + P+ DL K FPH W +E SK P + D SS + GE
Sbjct 8 KLLAGPSVDLFEKDFPHNNVWHVE--TLSKEPTL------------DMSSDFRLTRGEEL 53
Query 83 A--------------EVKVFSAGRSYVDLSYKIPQLPNLIISTKYERKGEKNTSDLFDFS 128
A E+ + S G ++D++ ++P P+ + +KY +KN F S
Sbjct 54 AGLVTLKNVFHNFATELSLNSTGVHHLDVTTRLPFFPDATVGSKYLFDTDKNHHH-FLLS 112
Query 129 TEYVTPRFHSILNVNPL---LRTFSSSGTFTYDRFRFGGEVSGKLDASGMKYALAASYSA 185
++ S L + PL FS + R G EVSG+ + + ++Y
Sbjct 113 SQVENKLLWSRLELKPLDFEYNFFSLLRFGSMHRCLLGAEVSGQ-KLGKLNVTVGSAYKR 171
Query 186 PTPGMKGGSWMAAVKTAPAGSLMFGKVIGSLNGRSVEG-RGAELAPEVEYNIAGNKSNIS 244
+++ A++ +++G+++ E + A L E+ + K+ ++
Sbjct 172 E---FGNNNFLLALRLFGNKDEYLNRLVGNVHVSKNESVKPAVLGVSFEHLLKEKKTKLA 228
Query 245 FGGLWHLSEEKDP---IVKSKISQNGLFGGALTPRFCSNLQAPIGTQLEVPKAQNADNCK 301
FG W+L+ DP VK+K+ + +F S + G + +
Sbjct 229 FGAHWNLTTLDDPHASFVKAKVDNEARVALSFCQKFNSLVTFLFGFDFNGKSPLHEPDVN 288
Query 302 YGIKIEV 308
YG+K+ +
Sbjct 289 YGLKLTL 295
> cpv:cgd2_480 hypothetical protein
Length=471
Score = 36.6 bits (83), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 23/83 (27%), Positives = 42/83 (50%), Gaps = 5/83 (6%)
Query 226 AELAPEVEYNIAGNKSNISFGGLWHLSEEKDPIVKSKISQNGLFGGALTPRFCSNLQAPI 285
+ + +V+Y I G ISFG + + + D ++ K+ N +LT F N+ A
Sbjct 392 TKFSDKVQYTIGGK---ISFGSMTNNMKSTD--LRFKVMNNLKMAYSLTHNFTKNISATF 446
Query 286 GTQLEVPKAQNADNCKYGIKIEV 308
G Q++ K + D+ KYG +++
Sbjct 447 GAQIDPRKLNDPDSVKYGFILDM 469
> mmu:18601 Padi3, AI507004, KIAA4171, Pad3, Pdi3, mKIAA4171;
peptidyl arginine deiminase, type III (EC:3.5.3.15); K01481
protein-arginine deiminase [EC:3.5.3.15]
Length=664
Score = 31.6 bits (70), Expect = 3.6, Method: Compositional matrix adjust.
Identities = 13/46 (28%), Positives = 29/46 (63%), Gaps = 0/46 (0%)
Query 35 KGFPHEKPWELEYKYKSKNPQITNVAGVGSHGGFDASSSLVYSAGE 80
+GFP+++ L++ Y ++ P+ ++V+G+ S G + S +V + E
Sbjct 377 QGFPYKRILGLDFGYVTREPKDSSVSGLDSFGNLEVSPPVVANGKE 422
> cel:B0336.3 hypothetical protein; K13192 RNA-binding protein
26
Length=719
Score = 30.8 bits (68), Expect = 7.1, Method: Compositional matrix adjust.
Identities = 16/57 (28%), Positives = 26/57 (45%), Gaps = 3/57 (5%)
Query 132 VTPRFHSILNVNPLLRTFSSSGTFTYDRFRFGGEVSGKLDASGMKYALAASYSAPTP 188
+ P ++I +N TF GT + R+ GE+ L K+ +Y +PTP
Sbjct 373 IPPEMNTIAKLNEHFATF---GTVDNIQVRYNGEIDSALVTYASKFDAGKAYKSPTP 426
Lambda K H
0.315 0.134 0.389
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 12602271528
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40