bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_0867_orf1
Length=108
Score E
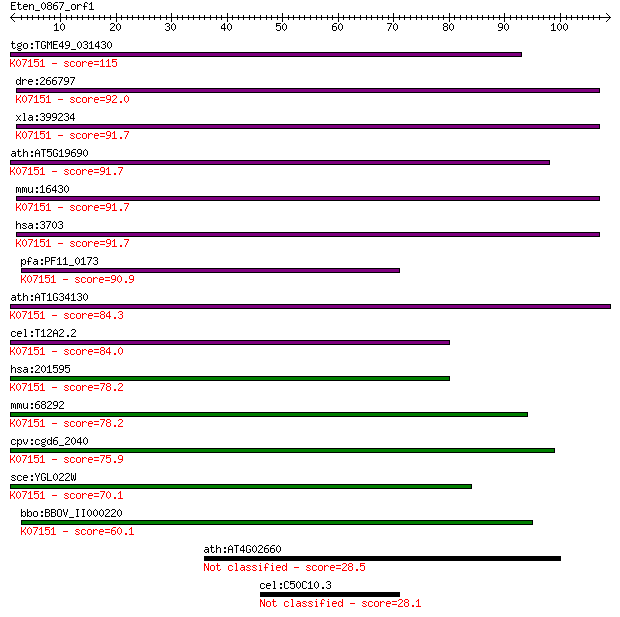
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_031430 oligosaccharyl transferase STT3, putative (E... 115 3e-26
dre:266797 itm1, cb437, wu:fb38c11; integral membrane protein ... 92.0 4e-19
xla:399234 stt3a, MGC98320, itm1; STT3, subunit of the oligosa... 91.7 5e-19
ath:AT5G19690 STT3A; STT3A (STAUROSPORIN AND TEMPERATURE SENSI... 91.7 5e-19
mmu:16430 Stt3a, AA408947, BB081708, Itm1; STT3, subunit of th... 91.7 6e-19
hsa:3703 STT3A, FLJ27038, ITM1, MGC9042, STT3-A, TMC; STT3, su... 91.7 6e-19
pfa:PF11_0173 oligosaccharyl transferase STT3 subunit, putativ... 90.9 1e-18
ath:AT1G34130 STT3B; STT3B (staurosporin and temperature sensi... 84.3 9e-17
cel:T12A2.2 hypothetical protein; K07151 dolichyl-diphosphooli... 84.0 1e-16
hsa:201595 STT3B, FLJ90106, SIMP, STT3-B; STT3, subunit of the... 78.2 5e-15
mmu:68292 Stt3b, 1300006C19Rik, Simp; STT3, subunit of the oli... 78.2 7e-15
cpv:cgd6_2040 oligosaccharyl transferase STT3 protein ; K07151... 75.9 3e-14
sce:YGL022W STT3; Stt3p (EC:2.4.1.119); K07151 dolichyl-diphos... 70.1 1e-12
bbo:BBOV_II000220 18.m05999; oligosaccharyl transferase STT3 s... 60.1 2e-09
ath:AT4G02660 WD-40 repeat family protein / beige-related 28.5 5.4
cel:C50C10.3 sru-30; Serpentine Receptor, class U family membe... 28.1 6.1
> tgo:TGME49_031430 oligosaccharyl transferase STT3, putative
(EC:2.4.1.119); K07151 dolichyl-diphosphooligosaccharide--protein
glycosyltransferase [EC:2.4.1.119]
Length=753
Score = 115 bits (289), Expect = 3e-26, Method: Compositional matrix adjust.
Identities = 57/93 (61%), Positives = 73/93 (78%), Gaps = 1/93 (1%)
Query 1 YIPIVASVSEHQPPTWSTYFLDLHMIMFLAPLGLILSLQR-GGGLFFVGLYGVLACYFSA 59
Y+PI+ASVSEHQP TW+TY DLH+ LAPLGLI+ L++ G F G+YGVLA YFS
Sbjct 387 YLPIIASVSEHQPATWATYVFDLHVATLLAPLGLIVCLRKPTDGSLFAGIYGVLAAYFSG 446
Query 60 VMVRLVLVLSPAASILAGIGAATLVAGVLSQCR 92
+MVRL+LVLSPAA++LAGIGA+ V+ ++S R
Sbjct 447 IMVRLMLVLSPAAAVLAGIGASRFVSSLMSYLR 479
> dre:266797 itm1, cb437, wu:fb38c11; integral membrane protein
1 (EC:2.4.1.-); K07151 dolichyl-diphosphooligosaccharide--protein
glycosyltransferase [EC:2.4.1.119]
Length=705
Score = 92.0 bits (227), Expect = 4e-19, Method: Compositional matrix adjust.
Identities = 44/106 (41%), Positives = 64/106 (60%), Gaps = 1/106 (0%)
Query 2 IPIVASVSEHQPPTWSTYFLDLHMIMFLAPLGLILSLQR-GGGLFFVGLYGVLACYFSAV 60
IPI+ASVSEHQP TWS+Y+ DL +++F+ P+GL F+ +YGV + YFSAV
Sbjct 343 IPIIASVSEHQPTTWSSYYFDLQLLVFMFPVGLYYCFNNLSDATIFIIMYGVTSMYFSAV 402
Query 61 MVRLVLVLSPAASILAGIGAATLVAGVLSQCRKVYPGDSGPARAAA 106
MVRL+LVL+P IL+GIG + ++ + P + +
Sbjct 403 MVRLMLVLAPVMCILSGIGVSQVLTTFMKNLDVSRPDKKTKKQQDS 448
> xla:399234 stt3a, MGC98320, itm1; STT3, subunit of the oligosaccharyltransferase
complex, homolog A; K07151 dolichyl-diphosphooligosaccharide--protein
glycosyltransferase [EC:2.4.1.119]
Length=705
Score = 91.7 bits (226), Expect = 5e-19, Method: Compositional matrix adjust.
Identities = 45/106 (42%), Positives = 64/106 (60%), Gaps = 1/106 (0%)
Query 2 IPIVASVSEHQPPTWSTYFLDLHMIMFLAPLGLILSLQR-GGGLFFVGLYGVLACYFSAV 60
IPI+ASVSEHQP TWS+Y+ DL ++F+ P+GL F+ +YGV + YFSAV
Sbjct 343 IPIIASVSEHQPTTWSSYYFDLQSLVFMFPVGLYYCFSNLSDARIFIIMYGVTSMYFSAV 402
Query 61 MVRLVLVLSPAASILAGIGAATLVAGVLSQCRKVYPGDSGPARAAA 106
MVRL+LVL+P IL+GIG + ++A + P + +
Sbjct 403 MVRLMLVLAPVMCILSGIGVSQVLATYMKNLDITRPDKKTKKQQDS 448
> ath:AT5G19690 STT3A; STT3A (STAUROSPORIN AND TEMPERATURE SENSITIVE
3-LIKE A); oligosaccharyl transferase; K07151 dolichyl-diphosphooligosaccharide--protein
glycosyltransferase [EC:2.4.1.119]
Length=779
Score = 91.7 bits (226), Expect = 5e-19, Method: Compositional matrix adjust.
Identities = 50/98 (51%), Positives = 65/98 (66%), Gaps = 9/98 (9%)
Query 1 YIPIVASVSEHQPPTWSTYFLDLHMIMFLAPLGLILSLQ-RGGGLFFVGLYGVLACYFSA 59
YIPI+ASVSEHQPPTW +YF+D++++ FL P G+I FV LY V++ YFS
Sbjct 350 YIPIIASVSEHQPPTWPSYFMDINVLAFLVPAGIIACFSPLSDASSFVVLYIVMSVYFSG 409
Query 60 VMVRLVLVLSPAASILAGIGAATLVAGVLSQCRKVYPG 97
VMVRL+LVL+PAA I++GI LSQ V+ G
Sbjct 410 VMVRLMLVLAPAACIMSGIA--------LSQAFDVFTG 439
> mmu:16430 Stt3a, AA408947, BB081708, Itm1; STT3, subunit of
the oligosaccharyltransferase complex, homolog A (S. cerevisiae)
(EC:2.4.1.119); K07151 dolichyl-diphosphooligosaccharide--protein
glycosyltransferase [EC:2.4.1.119]
Length=705
Score = 91.7 bits (226), Expect = 6e-19, Method: Compositional matrix adjust.
Identities = 44/106 (41%), Positives = 65/106 (61%), Gaps = 1/106 (0%)
Query 2 IPIVASVSEHQPPTWSTYFLDLHMIMFLAPLGLILSLQR-GGGLFFVGLYGVLACYFSAV 60
IPI+ASVSEHQP TWS+Y+ DL +++F+ P+GL F+ +YGV + YFSAV
Sbjct 343 IPIIASVSEHQPTTWSSYYFDLQLLVFMFPVGLYYCFSNLSDARIFIIMYGVTSMYFSAV 402
Query 61 MVRLVLVLSPAASILAGIGAATLVAGVLSQCRKVYPGDSGPARAAA 106
MVRL+LVL+P IL+GIG + +++ + P + +
Sbjct 403 MVRLMLVLAPVMCILSGIGVSQVLSTYMKNLDISRPDKKSKKQQDS 448
> hsa:3703 STT3A, FLJ27038, ITM1, MGC9042, STT3-A, TMC; STT3,
subunit of the oligosaccharyltransferase complex, homolog A
(S. cerevisiae) (EC:2.4.1.119); K07151 dolichyl-diphosphooligosaccharide--protein
glycosyltransferase [EC:2.4.1.119]
Length=705
Score = 91.7 bits (226), Expect = 6e-19, Method: Compositional matrix adjust.
Identities = 44/106 (41%), Positives = 65/106 (61%), Gaps = 1/106 (0%)
Query 2 IPIVASVSEHQPPTWSTYFLDLHMIMFLAPLGLILSLQR-GGGLFFVGLYGVLACYFSAV 60
IPI+ASVSEHQP TWS+Y+ DL +++F+ P+GL F+ +YGV + YFSAV
Sbjct 343 IPIIASVSEHQPTTWSSYYFDLQLLVFMFPVGLYYCFSNLSDARIFIIMYGVTSMYFSAV 402
Query 61 MVRLVLVLSPAASILAGIGAATLVAGVLSQCRKVYPGDSGPARAAA 106
MVRL+LVL+P IL+GIG + +++ + P + +
Sbjct 403 MVRLMLVLAPVMCILSGIGVSQVLSTYMKNLDISRPDKKSKKQQDS 448
> pfa:PF11_0173 oligosaccharyl transferase STT3 subunit, putative;
K07151 dolichyl-diphosphooligosaccharide--protein glycosyltransferase
[EC:2.4.1.119]
Length=856
Score = 90.9 bits (224), Expect = 1e-18, Method: Composition-based stats.
Identities = 38/70 (54%), Positives = 53/70 (75%), Gaps = 2/70 (2%)
Query 3 PIVASVSEHQPPTWSTYFLDLHMIMFLAPLGLILSLQRGGGL--FFVGLYGVLACYFSAV 60
PIVAS+SEHQP TWS+YF D+H+I+ PLGL ++ G + FF+G++ VL YFS++
Sbjct 376 PIVASISEHQPTTWSSYFFDIHLILLFFPLGLYECFKKNGSIESFFLGIFTVLCLYFSSL 435
Query 61 MVRLVLVLSP 70
MVRL+L+ SP
Sbjct 436 MVRLLLIFSP 445
> ath:AT1G34130 STT3B; STT3B (staurosporin and temperature sensitive
3-like b); oligosaccharyl transferase; K07151 dolichyl-diphosphooligosaccharide--protein
glycosyltransferase [EC:2.4.1.119]
Length=735
Score = 84.3 bits (207), Expect = 9e-17, Method: Compositional matrix adjust.
Identities = 42/110 (38%), Positives = 65/110 (59%), Gaps = 2/110 (1%)
Query 1 YIPIVASVSEHQPPTWSTYFLDLHMIMFLAPLGLILSLQR-GGGLFFVGLYGVLACYFSA 59
+IPI+ASVSEHQP WS++ D H+++FL P GL +R F+ +YG+ + YF+
Sbjct 366 HIPIIASVSEHQPTAWSSFMFDYHILLFLFPAGLYFCFKRLTDATIFIVMYGLTSLYFAG 425
Query 60 VMVRLVLVLSPAASILAGIGAATLVAGVLSQCR-KVYPGDSGPARAAARS 108
VMVRL+LV +PA +++ I + + + S R K +G + A S
Sbjct 426 VMVRLILVATPAVCLISAIAVSATIKNLTSLLRTKQKVSQTGSTKGAGSS 475
> cel:T12A2.2 hypothetical protein; K07151 dolichyl-diphosphooligosaccharide--protein
glycosyltransferase [EC:2.4.1.119]
Length=757
Score = 84.0 bits (206), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 41/80 (51%), Positives = 56/80 (70%), Gaps = 1/80 (1%)
Query 1 YIPIVASVSEHQPPTWSTYFLDLHMIMFLAPLGLILSLQR-GGGLFFVGLYGVLACYFSA 59
+IPI+ASVSEHQP TW ++F DLH+ + P+GL +++ F+ LY V A YF+
Sbjct 344 HIPIIASVSEHQPTTWVSFFFDLHITAAVFPVGLWYCIKKVNDERVFIILYAVSAVYFAG 403
Query 60 VMVRLVLVLSPAASILAGIG 79
VMVRL+L L+PA +LAGIG
Sbjct 404 VMVRLMLTLTPAVCVLAGIG 423
> hsa:201595 STT3B, FLJ90106, SIMP, STT3-B; STT3, subunit of the
oligosaccharyltransferase complex, homolog B (S. cerevisiae)
(EC:2.4.1.119); K07151 dolichyl-diphosphooligosaccharide--protein
glycosyltransferase [EC:2.4.1.119]
Length=826
Score = 78.2 bits (191), Expect = 5e-15, Method: Composition-based stats.
Identities = 37/80 (46%), Positives = 52/80 (65%), Gaps = 1/80 (1%)
Query 1 YIPIVASVSEHQPPTWSTYFLDLHMIMFLAPLGLILSLQR-GGGLFFVGLYGVLACYFSA 59
+IPI+ASVSEHQP TW ++F DLH+++ P GL ++ FV LY + A YF+
Sbjct 396 HIPIIASVSEHQPTTWVSFFFDLHILVCTFPAGLWFCIKNINDERVFVALYAISAVYFAG 455
Query 60 VMVRLVLVLSPAASILAGIG 79
VMVRL+L L+P +L+ I
Sbjct 456 VMVRLMLTLTPVVCMLSAIA 475
> mmu:68292 Stt3b, 1300006C19Rik, Simp; STT3, subunit of the oligosaccharyltransferase
complex, homolog B (S. cerevisiae)
(EC:2.4.1.119); K07151 dolichyl-diphosphooligosaccharide--protein
glycosyltransferase [EC:2.4.1.119]
Length=823
Score = 78.2 bits (191), Expect = 7e-15, Method: Compositional matrix adjust.
Identities = 39/94 (41%), Positives = 56/94 (59%), Gaps = 1/94 (1%)
Query 1 YIPIVASVSEHQPPTWSTYFLDLHMIMFLAPLGLILSLQR-GGGLFFVGLYGVLACYFSA 59
+IPI+ASVSEHQP TW ++F DLH+++ P GL ++ FV LY + A YF+
Sbjct 393 HIPIIASVSEHQPTTWVSFFFDLHILVCTFPAGLWFCIKNINDERVFVALYAISAVYFAG 452
Query 60 VMVRLVLVLSPAASILAGIGAATLVAGVLSQCRK 93
VMVRL+L L+P +L+ I + + L K
Sbjct 453 VMVRLMLTLTPVVCMLSAIAFSNVFEHYLGDDMK 486
> cpv:cgd6_2040 oligosaccharyl transferase STT3 protein ; K07151
dolichyl-diphosphooligosaccharide--protein glycosyltransferase
[EC:2.4.1.119]
Length=722
Score = 75.9 bits (185), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 44/122 (36%), Positives = 66/122 (54%), Gaps = 24/122 (19%)
Query 1 YIPIVASVSEHQPPTWSTYFLDLHMIMFLAPLGLI-------LSLQRG------------ 41
++PI+ASVSEHQ TWS Y DLH+ PLG+ +S ++
Sbjct 353 HVPIIASVSEHQATTWSQYIFDLHITAIFFPLGIFVCAYSIYISQKKSPKNSPFQTSHST 412
Query 42 ---GGLFFVGLYGVLACYFSAVMVRLVLVLSPAASILAGIGAATLVAGVL--SQCRKVYP 96
F+ +YGVLA YFS+VM+RL+LV PAA L+ +G + +++ ++ Q +K
Sbjct 413 CIPDTAIFLIVYGVLAVYFSSVMIRLMLVFGPAACCLSAVGFSFILSSLIGRRQTKKTTY 472
Query 97 GD 98
GD
Sbjct 473 GD 474
> sce:YGL022W STT3; Stt3p (EC:2.4.1.119); K07151 dolichyl-diphosphooligosaccharide--protein
glycosyltransferase [EC:2.4.1.119]
Length=718
Score = 70.1 bits (170), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 36/84 (42%), Positives = 51/84 (60%), Gaps = 1/84 (1%)
Query 1 YIPIVASVSEHQPPTWSTYFLDLHMIMFLAPLG-LILSLQRGGGLFFVGLYGVLACYFSA 59
+IPI+ASVSEHQP +W +F D H +++L P G +L L FV Y VL YF+
Sbjct 341 HIPIIASVSEHQPVSWPAFFFDTHFLIWLFPAGVFLLFLDLKDEHVFVIAYSVLCSYFAG 400
Query 60 VMVRLVLVLSPAASILAGIGAATL 83
VMVRL+L L+P + A + + +
Sbjct 401 VMVRLMLTLTPVICVSAAVALSKI 424
> bbo:BBOV_II000220 18.m05999; oligosaccharyl transferase STT3
subunit; K07151 dolichyl-diphosphooligosaccharide--protein
glycosyltransferase [EC:2.4.1.119]
Length=717
Score = 60.1 bits (144), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 28/94 (29%), Positives = 56/94 (59%), Gaps = 2/94 (2%)
Query 3 PIVASVSEHQPPTWSTYFLDLHMIMFLAPLGLILSL--QRGGGLFFVGLYGVLACYFSAV 60
P+VAS+SEHQP W + D + + P+G+ + + + G + +YG++ YF++
Sbjct 401 PLVASISEHQPTKWFDFVSDFWVALLFVPIGIYVCVMYKLDIGSMALLIYGIITAYFTSA 460
Query 61 MVRLVLVLSPAASILAGIGAATLVAGVLSQCRKV 94
M+RL ++ +P +IL+G+G + ++ + RK+
Sbjct 461 MIRLEVLFAPGVAILSGLGMSFVIERLSRDRRKM 494
> ath:AT4G02660 WD-40 repeat family protein / beige-related
Length=3471
Score = 28.5 bits (62), Expect = 5.4, Method: Composition-based stats.
Identities = 22/68 (32%), Positives = 32/68 (47%), Gaps = 4/68 (5%)
Query 36 LSLQRGGGLFFVGLYGVLACYFSAV----MVRLVLVLSPAASILAGIGAATLVAGVLSQC 91
LS+Q G G + + A +V LV LS AAS++ GI + G +S C
Sbjct 1293 LSIQLPGKKLIFAFDGTCSEFMRATGSFSLVNLVDPLSAAASLIGGIPRFGRLVGNVSLC 1352
Query 92 RKVYPGDS 99
R+ G+S
Sbjct 1353 RQNVIGNS 1360
> cel:C50C10.3 sru-30; Serpentine Receptor, class U family member
(sru-30)
Length=340
Score = 28.1 bits (61), Expect = 6.1, Method: Composition-based stats.
Identities = 12/25 (48%), Positives = 18/25 (72%), Gaps = 0/25 (0%)
Query 46 FVGLYGVLACYFSAVMVRLVLVLSP 70
+ G YG L+ F A ++RLV++LSP
Sbjct 119 YFGDYGTLSSPFLASLIRLVMILSP 143
Lambda K H
0.328 0.141 0.431
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2003209916
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40