bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_0837_orf2
Length=108
Score E
Sequences producing significant alignments: (Bits) Value
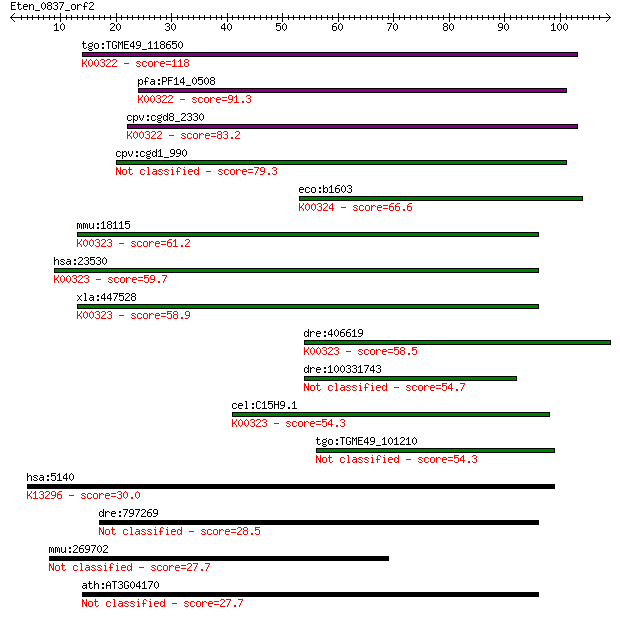
tgo:TGME49_118650 transhydrogenase, putative (EC:1.6.1.2); K00... 118 4e-27
pfa:PF14_0508 pyridine nucleotide transhydrogenase, putative (... 91.3 6e-19
cpv:cgd8_2330 pyridine nucleotide/ NAD(P) transhydrogenase alp... 83.2 2e-16
cpv:cgd1_990 pyridine nucleotide/ NAD(P) transhydrogenase alph... 79.3 3e-15
eco:b1603 pntA, ECK1598, JW1595; pyridine nucleotide transhydr... 66.6 2e-11
mmu:18115 Nnt, 4930423F13Rik, AI323702, BB168308; nicotinamide... 61.2 7e-10
hsa:23530 NNT, MGC126502, MGC126503; nicotinamide nucleotide t... 59.7 2e-09
xla:447528 nnt, MGC83563; nicotinamide nucleotide transhydroge... 58.9 4e-09
dre:406619 nnt, wu:fa20d10, wu:fc86a04, zgc:76979; nicotinamid... 58.5 5e-09
dre:100331743 Nicotinamide Nucleotide Transhydrogenase family ... 54.7 6e-08
cel:C15H9.1 nnt-1; Nicotinamide Nucleotide Transhydrogenase fa... 54.3 8e-08
tgo:TGME49_101210 NAD(P) transhydrogenase, alpha subunit, puta... 54.3 1e-07
hsa:5140 PDE3B, HcGIP1, cGIPDE1; phosphodiesterase 3B, cGMP-in... 30.0 1.8
dre:797269 abcb11a; ATP-binding cassette, sub-family B (MDR/TA... 28.5 6.0
mmu:269702 Mphosph9, 4930548D04Rik, 9630025B04Rik, AW413446, A... 27.7 8.2
ath:AT3G04170 germin-like protein, putative 27.7 9.4
> tgo:TGME49_118650 transhydrogenase, putative (EC:1.6.1.2); K00322
NAD(P) transhydrogenase [EC:1.6.1.1]
Length=1013
Score = 118 bits (296), Expect = 4e-27, Method: Compositional matrix adjust.
Identities = 59/89 (66%), Positives = 68/89 (76%), Gaps = 0/89 (0%)
Query 14 PEKPGAFAQALASDAFFAMCLVVAAAVVGLLGIVLDPVELKHLTLLGLSLIVGYYCVWAV 73
P++ Q + SDAFFAM LV AA GLLG+ L +EL+ TLL LSLIVGYY VW+V
Sbjct 893 PKEVSLLDQLVTSDAFFAMSLVCTAAFAGLLGVTLKALELQQFTLLALSLIVGYYSVWSV 952
Query 74 TPSLHTPLMSVTNALSGVIVIGCMLEYLP 102
TP+LHTPLMSVTNALSGVI+IG MLEY P
Sbjct 953 TPALHTPLMSVTNALSGVIIIGSMLEYGP 981
> pfa:PF14_0508 pyridine nucleotide transhydrogenase, putative
(EC:1.6.1.2); K00322 NAD(P) transhydrogenase [EC:1.6.1.1]
Length=1176
Score = 91.3 bits (225), Expect = 6e-19, Method: Composition-based stats.
Identities = 43/77 (55%), Positives = 54/77 (70%), Gaps = 0/77 (0%)
Query 24 LASDAFFAMCLVVAAAVVGLLGIVLDPVELKHLTLLGLSLIVGYYCVWAVTPSLHTPLMS 83
+ SD FF + L+ + L L +L+ L L LS IVGYYCVW+VTP+LHTPLMS
Sbjct 1061 IESDTFFYISLLFVIILTFLAATYLSQSDLQSLFLFTLSTIVGYYCVWSVTPALHTPLMS 1120
Query 84 VTNALSGVIVIGCMLEY 100
+TNALSGVI+IG M+EY
Sbjct 1121 MTNALSGVIIIGSMIEY 1137
> cpv:cgd8_2330 pyridine nucleotide/ NAD(P) transhydrogenase alpha
plus beta subunits, duplicated gene, possible signal peptide
plus 12 transmembrane regions ; K00322 NAD(P) transhydrogenase
[EC:1.6.1.1]
Length=1143
Score = 83.2 bits (204), Expect = 2e-16, Method: Composition-based stats.
Identities = 41/81 (50%), Positives = 56/81 (69%), Gaps = 0/81 (0%)
Query 22 QALASDAFFAMCLVVAAAVVGLLGIVLDPVELKHLTLLGLSLIVGYYCVWAVTPSLHTPL 81
+ + S+ F + ++ V LG ++D L ++ + LS+IVGYYC+W VTPSLHTPL
Sbjct 1002 RVIYSNVSFGFFVFISVLVSIGLGYIIDHDTLGNILVFSLSVIVGYYCIWNVTPSLHTPL 1061
Query 82 MSVTNALSGVIVIGCMLEYLP 102
MSVTNALSG+I+IG MLE P
Sbjct 1062 MSVTNALSGIIIIGAMLECGP 1082
> cpv:cgd1_990 pyridine nucleotide/ NAD(P) transhydrogenase alpha
plus beta subunits, duplicated gene, 12 transmembrane domain
(EC:1.6.1.2)
Length=1147
Score = 79.3 bits (194), Expect = 3e-15, Method: Composition-based stats.
Identities = 35/81 (43%), Positives = 54/81 (66%), Gaps = 0/81 (0%)
Query 20 FAQALASDAFFAMCLVVAAAVVGLLGIVLDPVELKHLTLLGLSLIVGYYCVWAVTPSLHT 79
F + L F +++A +LG+ + ++++++ +S ++GYYCVW V P LHT
Sbjct 1028 FDKQLNGGVNFYAGVILATLFFTVLGLTMTTIQIQNIFSFIISTMLGYYCVWDVDPKLHT 1087
Query 80 PLMSVTNALSGVIVIGCMLEY 100
PLMSVTNALSGVI+IG M++Y
Sbjct 1088 PLMSVTNALSGVIIIGSMMQY 1108
> eco:b1603 pntA, ECK1598, JW1595; pyridine nucleotide transhydrogenase,
alpha subunit (EC:1.6.1.2); K00324 NAD(P) transhydrogenase
subunit alpha [EC:1.6.1.2]
Length=510
Score = 66.6 bits (161), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 29/51 (56%), Positives = 39/51 (76%), Gaps = 0/51 (0%)
Query 53 LKHLTLLGLSLIVGYYCVWAVTPSLHTPLMSVTNALSGVIVIGCMLEYLPG 103
L H T+ L+ +VGYY VW V+ +LHTPLMSVTNA+SG+IV+G +L+ G
Sbjct 425 LGHFTVFALACVVGYYVVWNVSHALHTPLMSVTNAISGIIVVGALLQIGQG 475
> mmu:18115 Nnt, 4930423F13Rik, AI323702, BB168308; nicotinamide
nucleotide transhydrogenase (EC:1.6.1.2); K00323 NAD(P) transhydrogenase
[EC:1.6.1.2]
Length=1086
Score = 61.2 bits (147), Expect = 7e-10, Method: Compositional matrix adjust.
Identities = 40/94 (42%), Positives = 53/94 (56%), Gaps = 11/94 (11%)
Query 13 APEKPGAFAQALASDA----FFAMCLVVA----AAVVGLLG--IVLDPVELKHL-TLLGL 61
AP KP A+ A A + L A A + G+LG IV V + T GL
Sbjct 448 APVKPKTVAELEAEKAGTVSMYTKTLTTASVYSAGLTGMLGLGIVAPNVAFSQMVTTFGL 507
Query 62 SLIVGYYCVWAVTPSLHTPLMSVTNALSGVIVIG 95
+ I+GY+ VW VTP+LH+PLMSVTNA+SG+ +G
Sbjct 508 AGIIGYHTVWGVTPALHSPLMSVTNAISGLTAVG 541
> hsa:23530 NNT, MGC126502, MGC126503; nicotinamide nucleotide
transhydrogenase (EC:1.6.1.2); K00323 NAD(P) transhydrogenase
[EC:1.6.1.2]
Length=1086
Score = 59.7 bits (143), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 39/96 (40%), Positives = 57/96 (59%), Gaps = 9/96 (9%)
Query 9 SGAPAPEKPGAF--AQALASDAFFAMCLVVAAA----VVGLLGIVLDPVEL---KHLTLL 59
GAP +K A A+ A+ F + A+A + G+LG+ + L + +T
Sbjct 446 QGAPVKQKTVAELEAEKAATITPFRKTMSTASAYTAGLTGILGLGIAAPNLAFSQMVTTF 505
Query 60 GLSLIVGYYCVWAVTPSLHTPLMSVTNALSGVIVIG 95
GL+ IVGY+ VW VTP+LH+PLMSVTNA+SG+ +G
Sbjct 506 GLAGIVGYHTVWGVTPALHSPLMSVTNAISGLTAVG 541
> xla:447528 nnt, MGC83563; nicotinamide nucleotide transhydrogenase
(EC:1.6.1.2); K00323 NAD(P) transhydrogenase [EC:1.6.1.2]
Length=1086
Score = 58.9 bits (141), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 39/94 (41%), Positives = 51/94 (54%), Gaps = 11/94 (11%)
Query 13 APEKPGAFAQALASDAF----FAMCLVVAAAVVGLLGIVLD-------PVELKHLTLLGL 61
AP K + AQ A A F + AAA LG +L + +T GL
Sbjct 448 APVKQKSVAQIEAEKAASISPFRKTMNGAAAYTAGLGTLLSLGIASPHSAFTQMVTTFGL 507
Query 62 SLIVGYYCVWAVTPSLHTPLMSVTNALSGVIVIG 95
+ IVGY+ VW VTP+LH+PLMSVTNA+SG+ +G
Sbjct 508 AGIVGYHTVWGVTPALHSPLMSVTNAISGLTAVG 541
> dre:406619 nnt, wu:fa20d10, wu:fc86a04, zgc:76979; nicotinamide
nucleotide transhydrogenase (EC:1.6.1.2); K00323 NAD(P)
transhydrogenase [EC:1.6.1.2]
Length=1079
Score = 58.5 bits (140), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 29/59 (49%), Positives = 40/59 (67%), Gaps = 4/59 (6%)
Query 54 KHLTLLGLSLIVGYYCVWAVTPSLHTPLMSVTNALSGVIVIGCML----EYLPGRPLES 108
+ +T GL+ IVGY+ VW VTP+LH+PLMSVTNA+SG+ +G + YLP E+
Sbjct 496 QMVTTFGLAGIVGYHTVWGVTPALHSPLMSVTNAISGLTAVGGLSLMGGGYLPSSTAET 554
> dre:100331743 Nicotinamide Nucleotide Transhydrogenase family
member (nnt-1)-like
Length=518
Score = 54.7 bits (130), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 24/38 (63%), Positives = 32/38 (84%), Gaps = 0/38 (0%)
Query 54 KHLTLLGLSLIVGYYCVWAVTPSLHTPLMSVTNALSGV 91
+ +T GL+ IVGY+ VW VTP+LH+PLMSVTNA+SG+
Sbjct 200 QMVTTFGLAGIVGYHTVWGVTPALHSPLMSVTNAISGL 237
> cel:C15H9.1 nnt-1; Nicotinamide Nucleotide Transhydrogenase
family member (nnt-1); K00323 NAD(P) transhydrogenase [EC:1.6.1.2]
Length=1041
Score = 54.3 bits (129), Expect = 8e-08, Method: Compositional matrix adjust.
Identities = 29/58 (50%), Positives = 37/58 (63%), Gaps = 1/58 (1%)
Query 41 VGLLGIV-LDPVELKHLTLLGLSLIVGYYCVWAVTPSLHTPLMSVTNALSGVIVIGCM 97
V LLGI +P T L+ +VGY+ VW VTP+LH+PLMSVTNA+SG G +
Sbjct 440 VSLLGIAGTNPQISSMSTTFALAGLVGYHTVWGVTPALHSPLMSVTNAISGTTAAGAL 497
> tgo:TGME49_101210 NAD(P) transhydrogenase, alpha subunit, putative
(EC:1.6.1.2)
Length=1165
Score = 54.3 bits (129), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 25/43 (58%), Positives = 32/43 (74%), Gaps = 0/43 (0%)
Query 56 LTLLGLSLIVGYYCVWAVTPSLHTPLMSVTNALSGVIVIGCML 98
L + L+ VGY VW V P+LHTPLMSVTNA+SG +++G ML
Sbjct 1052 LFIFFLACFVGYLLVWNVAPALHTPLMSVTNAISGTVLVGGML 1094
> hsa:5140 PDE3B, HcGIP1, cGIPDE1; phosphodiesterase 3B, cGMP-inhibited
(EC:3.1.4.17); K13296 cGMP-inhibited 3',5'-cyclic
phosphodiesterase [EC:3.1.4.17]
Length=1112
Score = 30.0 bits (66), Expect = 1.8, Method: Composition-based stats.
Identities = 26/96 (27%), Positives = 44/96 (45%), Gaps = 13/96 (13%)
Query 4 WSRPRSGAPAPEKPGAFAQALASDAFFAMCLVVAAAVVGLLGIVLDPVELKHLTLLGLSL 63
W +G+ AP P A A L++ + VGLL + P+ L+H L+ L
Sbjct 176 WGDGDAGSAAPHTPPEAA---------AGRLLLVLSCVGLLLTLAHPLRLRHCVLV---L 223
Query 64 IVGYYCVWAVTPSLHTPLMSVTNALSGVI-VIGCML 98
++ + W SL + ++ LSG++ GC+L
Sbjct 224 LLASFVWWVSFTSLGSLPSALRPLLSGLVGGAGCLL 259
> dre:797269 abcb11a; ATP-binding cassette, sub-family B (MDR/TAP),
member 11a
Length=1325
Score = 28.5 bits (62), Expect = 6.0, Method: Composition-based stats.
Identities = 20/81 (24%), Positives = 38/81 (46%), Gaps = 12/81 (14%)
Query 17 PGAFAQALASDAFFAMCLVVAAAVVGLLGIVLDPVELKHLTLLGLSLIVGYYCVW--AVT 74
PGA LA++A +V A +G++++ LT +G S I+ YY W ++
Sbjct 858 PGALTTRLATNAS-----MVQGATGSQIGMIVNS-----LTNIGASFIIAYYFSWKLSLV 907
Query 75 PSLHTPLMSVTNALSGVIVIG 95
+ PL+ ++ ++ G
Sbjct 908 VTCFLPLIGLSGVFQSKMLTG 928
> mmu:269702 Mphosph9, 4930548D04Rik, 9630025B04Rik, AW413446,
AW547060, B930097C17Rik, C87456, MPP-9, MPP9; M-phase phosphoprotein
9
Length=1114
Score = 27.7 bits (60), Expect = 8.2, Method: Composition-based stats.
Identities = 19/61 (31%), Positives = 26/61 (42%), Gaps = 0/61 (0%)
Query 8 RSGAPAPEKPGAFAQALASDAFFAMCLVVAAAVVGLLGIVLDPVELKHLTLLGLSLIVGY 67
R+GA PE+P SD L+ A G LDP + K L +L+ GY
Sbjct 735 RNGAKVPERPSRSNSVATSDVSRRKWLIPGAEYSIFTGQPLDPRDRKLDKQLEEALVPGY 794
Query 68 Y 68
+
Sbjct 795 H 795
> ath:AT3G04170 germin-like protein, putative
Length=227
Score = 27.7 bits (60), Expect = 9.4, Method: Compositional matrix adjust.
Identities = 21/91 (23%), Positives = 37/91 (40%), Gaps = 9/91 (9%)
Query 14 PEKPGAFAQA--------LASDAFFAMCLVVAAAVVGLLGIVLDPVELKHLTLLGLSLI- 64
PEK G F + SD FFA L + LG ++P + L LG+ +
Sbjct 38 PEKNGVFVNGEFCKDPKLVTSDDFFASGLNIPGNTNKRLGSFVNPANIPGLNTLGVGIAR 97
Query 65 VGYYCVWAVTPSLHTPLMSVTNALSGVIVIG 95
+ + + P +H + + G +++G
Sbjct 98 IDFAPGGLIPPHIHPRASEIILVIKGKLLVG 128
Lambda K H
0.324 0.139 0.435
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2003209916
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40