bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_0796_orf2
Length=164
Score E
Sequences producing significant alignments: (Bits) Value
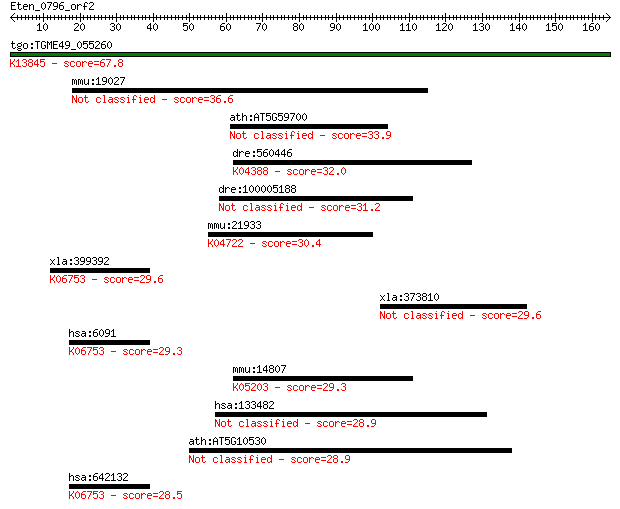
tgo:TGME49_055260 apical membrane antigen 1, putative ; K13845... 67.8 2e-11
mmu:19027 Sypl, AI314763, AI604763, D12Ertd446e, PanI, Pphn, S... 36.6 0.037
ath:AT5G59700 protein kinase, putative 33.9 0.25
dre:560446 similar to TGF-beta type II receptor; K04388 TGF-be... 32.0 1.00
dre:100005188 hypothetical LOC100005188 31.2 1.7
mmu:21933 Tnfrsf10b, DR5, KILLER, Ly98, MK, TRAILR2, TRICK2A, ... 30.4 2.9
xla:399392 robo1, dutt1, roundabout; roundabout, axon guidance... 29.6 4.5
xla:373810 syn2-a, synII, synIIa, synIIb; synapsin II 29.6 4.8
hsa:6091 ROBO1, DUTT1, FLJ21882, MGC131599, MGC133277, SAX3; r... 29.3 5.3
mmu:14807 Grik3, 9630027E11, GluR7-3, Glur-7, Glur7; glutamate... 29.3 5.7
hsa:133482 SLCO6A1, CT48, GST, MGC26949, OATP6A1, OATPY; solut... 28.9 7.5
ath:AT5G10530 lectin protein kinase, putative 28.9 8.2
hsa:642132 roundabout homolog 1-like; K06753 roundabout, axon ... 28.5 10.0
> tgo:TGME49_055260 apical membrane antigen 1, putative ; K13845
apical merozoite antigen 1
Length=569
Score = 67.8 bits (164), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 56/183 (30%), Positives = 83/183 (45%), Gaps = 25/183 (13%)
Query 1 CLLPRQGAAAFTSVGSLEEEELPHCDPT-------------------FPASLGSCDPSSC 41
CL+ A ++T+ GSL EE P+ FP S G+CD +C
Sbjct 393 CLVSDSAAVSYTAAGSLSEETPNFIIPSNPSVTPPTPETALQCTADKFPDSFGACDVQAC 452
Query 42 KAILTECRGGRLVEQQTDCVPEDGSKCESKGGGVFIGLAVAGGLLLLLLTGGAFFIYKQR 101
K T C GG++ DC ++ ++C S + LL LL G F R
Sbjct 453 KRQKTSCVGGQIQSTSVDCTADEQNECGSNTALIAGLAVGGVLLLALLGGGCYFAKRLDR 512
Query 102 QKALPKESSPQRTDFVQDEAATGRGKKRQSDLVQQAEPSLWEEAEADEPHADENTQVLLD 161
K + +++ +F D A KKR SDL+Q+AEPS W+EAE + D T V+++
Sbjct 513 NKGV--QAAHHEHEFQSDRGAR---KKRPSDLMQEAEPSFWDEAE-ENIEQDGETHVMVE 566
Query 162 QEY 164
+Y
Sbjct 567 GDY 569
> mmu:19027 Sypl, AI314763, AI604763, D12Ertd446e, PanI, Pphn,
Sypl1; synaptophysin-like protein
Length=261
Score = 36.6 bits (83), Expect = 0.037, Method: Compositional matrix adjust.
Identities = 33/109 (30%), Positives = 50/109 (45%), Gaps = 15/109 (13%)
Query 18 EEEELPHCD--PTFPASLGSCDPSSCKA-ILTECR---GGRLVEQQTDCVPEDGSKC--- 68
+ +LP D T A+ SS A LT+ + G R+VE+ C PE G C
Sbjct 146 DSRKLPMIDFIVTLVATFLWLVSSSAWAKALTDIKVATGHRIVEELEICNPESGVSCYFV 205
Query 69 --ESKGGGVFIGLAVAGGLLLLLLTGG-AFFIYKQRQKALPKESSPQRT 114
S G + ++V G L ++L GG A+F+YK+ P +S +
Sbjct 206 SVTSMGS---LNVSVIFGFLNMILWGGNAWFVYKETSLHSPSNTSASHS 251
> ath:AT5G59700 protein kinase, putative
Length=829
Score = 33.9 bits (76), Expect = 0.25, Method: Composition-based stats.
Identities = 17/43 (39%), Positives = 25/43 (58%), Gaps = 1/43 (2%)
Query 61 VPEDGSKCESKGGGVFIGLAVAGGLLLLLLTGGAFFIYKQRQK 103
+P S K G+ IGL + G LL L++ GG F +YK+R +
Sbjct 392 LPSGSSSTTKKNVGMIIGLTI-GSLLALVVLGGFFVLYKKRGR 433
> dre:560446 similar to TGF-beta type II receptor; K04388 TGF-beta
receptor type-2 [EC:2.7.11.30]
Length=576
Score = 32.0 bits (71), Expect = 1.00, Method: Composition-based stats.
Identities = 20/66 (30%), Positives = 35/66 (53%), Gaps = 1/66 (1%)
Query 62 PEDGSKCESKGGGVFIGLAVAGGLLLLLLTGGAFFIYKQRQKA-LPKESSPQRTDFVQDE 120
P SK +SK + +++ LL+ ++ AF++Y+ RQ PKE P+RT + +
Sbjct 118 PNGFSKLKSKDVIPVVVISLVPPLLVAVIATMAFYLYRTRQPGKKPKEWGPRRTHYQSLD 177
Query 121 AATGRG 126
A G+
Sbjct 178 PAEGQA 183
> dre:100005188 hypothetical LOC100005188
Length=829
Score = 31.2 bits (69), Expect = 1.7, Method: Composition-based stats.
Identities = 21/53 (39%), Positives = 27/53 (50%), Gaps = 7/53 (13%)
Query 58 TDCVPEDGSKCESKGGGVFIGLAVAGGLLLLLLTGGAFFIYKQRQKALPKESS 110
T VP+ G S G I + V LLL+ TG A F++ R K LPK+ S
Sbjct 113 TVSVPDSGL---SPGAAAGISVIV----LLLVFTGAAAFVFYHRHKFLPKKIS 158
> mmu:21933 Tnfrsf10b, DR5, KILLER, Ly98, MK, TRAILR2, TRICK2A,
TRICK2B, TRICKB; tumor necrosis factor receptor superfamily,
member 10b; K04722 tumor necrosis factor receptor superfamily
member 10
Length=381
Score = 30.4 bits (67), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 18/52 (34%), Positives = 25/52 (48%), Gaps = 12/52 (23%)
Query 55 EQQTDCVPEDGSKCESKGG-------GVFIGLAVAGGLLLLLLTGGAFFIYK 99
E+ T C P + KC SK G++IGL V LL+ GA ++K
Sbjct 156 EELTSCTPRENRKCVSKTAWASWHKLGLWIGLLVPVVLLI-----GALLVWK 202
> xla:399392 robo1, dutt1, roundabout; roundabout, axon guidance
receptor, homolog 1; K06753 roundabout, axon guidance receptor
1
Length=1614
Score = 29.6 bits (65), Expect = 4.5, Method: Composition-based stats.
Identities = 12/27 (44%), Positives = 16/27 (59%), Gaps = 0/27 (0%)
Query 12 TSVGSLEEEELPHCDPTFPASLGSCDP 38
T ++E+ LP+C PTFP S DP
Sbjct 1539 TITNQMQEDILPYCKPTFPTSNNPRDP 1565
> xla:373810 syn2-a, synII, synIIa, synIIb; synapsin II
Length=560
Score = 29.6 bits (65), Expect = 4.8, Method: Composition-based stats.
Identities = 16/40 (40%), Positives = 23/40 (57%), Gaps = 2/40 (5%)
Query 102 QKALPKESSPQRTDFVQDEAATGRGKKRQSDLVQQAEPSL 141
Q +P ++ Q+ DF Q+ A G G+ QSD Q+A P L
Sbjct 478 QTTIPHGAAQQQPDFGQE--ALGSGRVSQSDPPQKAHPQL 515
> hsa:6091 ROBO1, DUTT1, FLJ21882, MGC131599, MGC133277, SAX3;
roundabout, axon guidance receptor, homolog 1 (Drosophila);
K06753 roundabout, axon guidance receptor 1
Length=1551
Score = 29.3 bits (64), Expect = 5.3, Method: Compositional matrix adjust.
Identities = 11/22 (50%), Positives = 15/22 (68%), Gaps = 0/22 (0%)
Query 17 LEEEELPHCDPTFPASLGSCDP 38
++E+ LP+C PTFP S DP
Sbjct 1480 IQEDILPYCRPTFPTSNNPRDP 1501
> mmu:14807 Grik3, 9630027E11, GluR7-3, Glur-7, Glur7; glutamate
receptor, ionotropic, kainate 3; K05203 glutamate receptor,
ionotropic, kainate 3
Length=919
Score = 29.3 bits (64), Expect = 5.7, Method: Composition-based stats.
Identities = 22/54 (40%), Positives = 30/54 (55%), Gaps = 7/54 (12%)
Query 62 PEDGSKCES-----KGGGVFIGLAVAGGLLLLLLTGGAFFIYKQRQKALPKESS 110
PE+ +K S K GG+FI LA GL+L +L FIYK R+ A ++ S
Sbjct 806 PEEENKEASALGIQKIGGIFIVLA--AGLVLSVLVAVGEFIYKLRKTAEREQRS 857
> hsa:133482 SLCO6A1, CT48, GST, MGC26949, OATP6A1, OATPY; solute
carrier organic anion transporter family, member 6A1; K14357
solute carrier organic anion transporter family, member
6A
Length=719
Score = 28.9 bits (63), Expect = 7.5, Method: Composition-based stats.
Identities = 21/84 (25%), Positives = 36/84 (42%), Gaps = 18/84 (21%)
Query 57 QTDCVPEDGSKC----------ESKGGGVFIGLAVAGGLLLLLLTGGAFFIYKQRQKALP 106
+T C+ D +KC ++K + +G+ L ++ T AFFIYK+R
Sbjct 640 ETSCILRDVNKCGHTGRCWIYNKTKMAFLLVGICFLCKLCTIIFTTIAFFIYKRRLN--- 696
Query 107 KESSPQRTDFVQDEAATGRGKKRQ 130
+ TDF + KK++
Sbjct 697 -----ENTDFPDVTVKNPKVKKKE 715
> ath:AT5G10530 lectin protein kinase, putative
Length=651
Score = 28.9 bits (63), Expect = 8.2, Method: Composition-based stats.
Identities = 24/91 (26%), Positives = 38/91 (41%), Gaps = 3/91 (3%)
Query 50 GGRLVEQQTDCVPE--DGSKCESKGGGVFIGLAVAGGLLLLLLTGGAFFIYKQRQKALPK 107
G RL+ + E D K ++ G+ IG++V+G +LL K++Q+
Sbjct 242 GNRLLSWEFSSSLELIDIKKSQNDKKGMIIGISVSGFVLLTFFITSLIVFLKRKQQKKKA 301
Query 108 ESSPQRTDFVQD-EAATGRGKKRQSDLVQQA 137
E + T +D E G K DL A
Sbjct 302 EETENLTSINEDLERGAGPRKFTYKDLASAA 332
> hsa:642132 roundabout homolog 1-like; K06753 roundabout, axon
guidance receptor 1
Length=1615
Score = 28.5 bits (62), Expect = 10.0, Method: Composition-based stats.
Identities = 11/22 (50%), Positives = 15/22 (68%), Gaps = 0/22 (0%)
Query 17 LEEEELPHCDPTFPASLGSCDP 38
++E+ LP+C PTFP S DP
Sbjct 1544 IQEDILPYCRPTFPTSNNPRDP 1565
Lambda K H
0.314 0.133 0.396
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3897828240
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40