bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_0754_orf1
Length=356
Score E
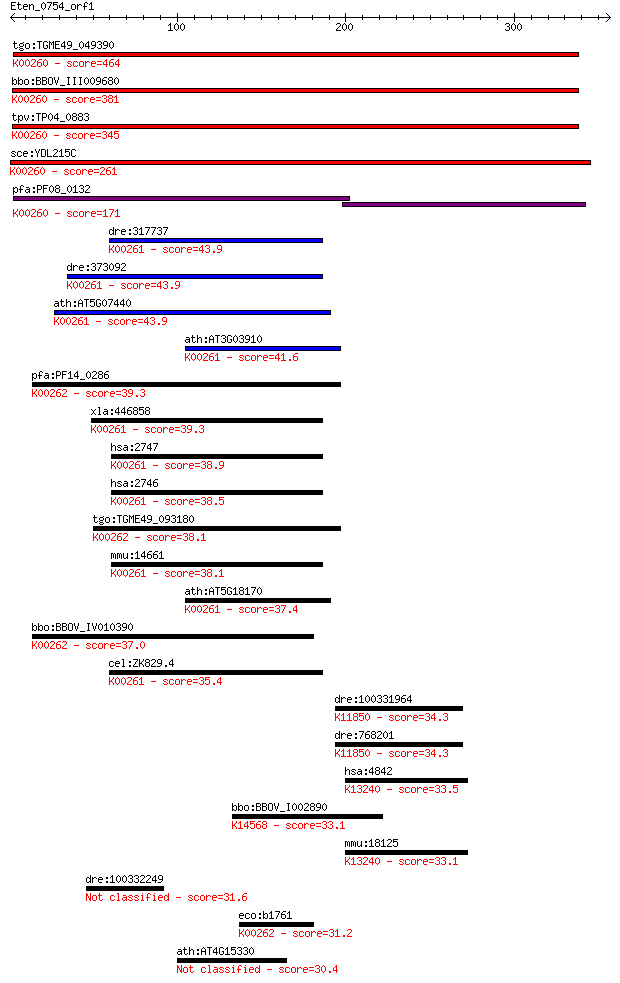
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_049390 NAD-specific glutamate dehydrogenase, putati... 464 2e-130
bbo:BBOV_III009680 17.m07840; Glutamate/Leucine/Phenylalanine/... 381 3e-105
tpv:TP04_0883 NAD-specific glutamate dehydrogenase (EC:1.4.1.2... 345 1e-94
sce:YDL215C GDH2; Gdh2p (EC:1.4.1.2); K00260 glutamate dehydro... 261 2e-69
pfa:PF08_0132 glutamate dehydrogenase, putative (EC:1.4.1.2); ... 171 4e-42
dre:317737 glud1a, cb622, glud1, wu:fc33g09, wu:fc66a10, zgc:7... 43.9 0.001
dre:373092 glud1b, MGC192851, cb719, glud1, wu:fb16e02, wu:fb5... 43.9 0.001
ath:AT5G07440 GDH2; GDH2 (GLUTAMATE DEHYDROGENASE 2); ATP bind... 43.9 0.001
ath:AT3G03910 GDH3; GDH3 (GLUTAMATE DEHYDROGENASE 3); binding ... 41.6 0.005
pfa:PF14_0286 glutamate dehydrogenase, putative (EC:1.4.1.3); ... 39.3 0.023
xla:446858 glud1, MGC80801; glutamate dehydrogenase 1 (EC:1.4.... 39.3 0.026
hsa:2747 GLUD2, GDH2, GLUDP1; glutamate dehydrogenase 2 (EC:1.... 38.9 0.028
hsa:2746 GLUD1, GDH, GDH1, GLUD, MGC132003; glutamate dehydrog... 38.5 0.041
tgo:TGME49_093180 NADP-specific glutamate dehydrogenase, putat... 38.1 0.050
mmu:14661 Glud1, AI118167, Gdh-X, Glud, Gludl; glutamate dehyd... 38.1 0.051
ath:AT5G18170 GDH1; GDH1 (GLUTAMATE DEHYDROGENASE 1); ATP bind... 37.4 0.100
bbo:BBOV_IV010390 23.m06170; glutamate dehydrogenase (EC:1.4.1... 37.0 0.11
cel:ZK829.4 hypothetical protein; K00261 glutamate dehydrogena... 35.4 0.32
dre:100331964 ubiquitin specific peptidase 29-like; K11850 ubi... 34.3 0.65
dre:768201 usp37, MGC153999, wu:fi15b04, wu:fi38d03, zgc:15288... 34.3 0.70
hsa:4842 NOS1, IHPS1, N-NOS, NC-NOS, NOS, bNOS, nNOS; nitric o... 33.5 1.4
bbo:BBOV_I002890 19.m02290; suppressor Mra1 family domain cont... 33.1 1.6
mmu:18125 Nos1, 2310005C01Rik, NO, NOS-I, Nos-1, bNOS, nNOS; n... 33.1 1.7
dre:100332249 Extracellular matrix protein 1-like 31.6 5.5
eco:b1761 gdhA, ECK1759, JW1750; glutamate dehydrogenase, NADP... 31.2 6.4
ath:AT4G15330 CYP705A1; electron carrier/ heme binding / iron ... 30.4 9.9
> tgo:TGME49_049390 NAD-specific glutamate dehydrogenase, putative
(EC:1.4.1.2); K00260 glutamate dehydrogenase [EC:1.4.1.2]
Length=1113
Score = 464 bits (1194), Expect = 2e-130, Method: Compositional matrix adjust.
Identities = 219/335 (65%), Positives = 270/335 (80%), Gaps = 1/335 (0%)
Query 3 GGPDGDLGSNALLQTKSKTLAVVDGAGVVYDPEGLNAEELRRLARLRFKGEKTSSMLFDS 62
GGPDGDLGSNALL++ +KT ++VDG+GV++DPEGL+ ELRRLA+ RF+G +TS+ML+D
Sbjct 767 GGPDGDLGSNALLKSNTKTTSIVDGSGVLHDPEGLDINELRRLAKRRFEGLQTSAMLYDE 826
Query 63 SLLSPKGFKVPQDARDITLPDGTYVASGIEFRNNFHLSKFAVCDLFNPCGGRPASVNPRN 122
LLSP GFKV QD RD+ LPDGT VASG EFR FHL DLFNPCGGRPASV P N
Sbjct 827 KLLSPMGFKVSQDDRDVVLPDGTAVASGFEFRGRFHLDPRGSADLFNPCGGRPASVTPFN 886
Query 123 VEQLFEQQTGLPKFKFIVEGANVFITDEARRELENRGVILFKDASTNKGGVTSSSMEVLA 182
V+++F+++ G P+FKFIVEGANVFITDEARR LE RGVILFKDASTNKGGVTSSS EVLA
Sbjct 887 VDKMFDEK-GKPRFKFIVEGANVFITDEARRMLEERGVILFKDASTNKGGVTSSSHEVLA 945
Query 183 ALSLTEEEFDRHMRVKDPNNPPKFYKDYVEAILQRIQENARLEFEAIWNEAERTGMHKCD 242
AL++T+EEF HM+V DP+NPP FY+ YV +++RI+ENARLEF A+W E+ RTG +CD
Sbjct 946 ALAMTDEEFAEHMQVTDPSNPPAFYQTYVRQVMERIRENARLEFNALWEESLRTGKPRCD 1005
Query 243 LTDILSQKINQLKTDMAGSSALWKDEALVNFVLTKSIPDVLVPGLISVETFRQRVPETYQ 302
LTD+LS KI +L D+ S +LW+D+ LV+ VL K++PDVLVPGL+S+ET RQRVPE+Y
Sbjct 1006 LTDVLSAKILRLNQDIHKSDSLWQDDELVSCVLLKALPDVLVPGLMSIETLRQRVPESYL 1065
Query 303 RAIFTTFLASRFLYKQPFTAEASAFAFIAYLDEIR 337
+AIF FLASRF Y Q FT SAFAF Y+ ++
Sbjct 1066 QAIFQCFLASRFYYSQQFTDNLSAFAFFDYVRHLK 1100
> bbo:BBOV_III009680 17.m07840; Glutamate/Leucine/Phenylalanine/Valine
dehydrogenase family protein (EC:1.4.1.2); K00260 glutamate
dehydrogenase [EC:1.4.1.2]
Length=1025
Score = 381 bits (978), Expect = 3e-105, Method: Compositional matrix adjust.
Identities = 193/336 (57%), Positives = 245/336 (72%), Gaps = 3/336 (0%)
Query 2 TGGPDGDLGSNALLQTKSKTLAVVDGAGVVYDPEGLNAEELRRLARLRFKGEKTSSMLFD 61
TGGPDGDLGSNALL +K+KTL V+D +GV++DPEGL+ EL+RLA R KG TS+M ++
Sbjct 693 TGGPDGDLGSNALLCSKTKTLTVIDKSGVLHDPEGLDINELQRLAANRLKGLPTSAMHYN 752
Query 62 SSLLSPKGFKVPQDARDITLPDGTYVASGIEFRNNFHLSKFAVCDLFNPCGGRPASVNPR 121
+LLS KGFKVP+DA D+ LPDGT V G +FR+ FHL DLFNPCGGRP+S+ P
Sbjct 753 EALLSDKGFKVPEDAVDMVLPDGTKVKRGHKFRDEFHLGA-CPSDLFNPCGGRPSSITPF 811
Query 122 NVEQLFEQQTGLPKFKFIVEGANVFITDEARRELENRGVILFKDASTNKGGVTSSSMEVL 181
NV +LF+++ G +KFIVEGANVFIT +ARR LEN+GVILFKDASTNKGGVTSSS EVL
Sbjct 812 NVNRLFDEK-GKCIYKFIVEGANVFITQDARRILENKGVILFKDASTNKGGVTSSSFEVL 870
Query 182 AALSLTEEEFDRHMRVKDPNNPPKFYKDYVEAILQRIQENARLEFEAIWNEAERTGMHKC 241
AAL L ++ FD M VK+ P+F KDY+ IL I++NAR EF A+WNE R GM +C
Sbjct 871 AALVLDDDTFDEMMTVKEGGEFPQFRKDYINEILDIIKKNARREFHALWNEGLRIGMPRC 930
Query 242 DLTDILSQKINQLKTDMAGSSALWKDEALVNFVLTKSIPDVLVPGLISVETFRQRVPETY 301
DLTD+LS KI +LK D+ S++LW+D LV VL+K+IP L L+ +E R+P+ Y
Sbjct 931 DLTDVLSTKIIRLKKDIIDSNSLWEDTVLVRAVLSKAIPQSL-QKLVPIEDIMVRLPDRY 989
Query 302 QRAIFTTFLASRFLYKQPFTAEASAFAFIAYLDEIR 337
R++F + LAS F Y Q FT + S FAF YL ++
Sbjct 990 MRSMFASHLASTFYYSQQFTDDTSVFAFYEYLKALK 1025
> tpv:TP04_0883 NAD-specific glutamate dehydrogenase (EC:1.4.1.2);
K00260 glutamate dehydrogenase [EC:1.4.1.2]
Length=1178
Score = 345 bits (885), Expect = 1e-94, Method: Compositional matrix adjust.
Identities = 179/372 (48%), Positives = 250/372 (67%), Gaps = 39/372 (10%)
Query 2 TGGPDGDLGSNALLQTKSKTLAVVDGAGVVYDPEGLNAEELRRLARLR------------ 49
TGGPDGDLGSNA+ + +KTL V+D +GV++DP GL+ ELRRLA LR
Sbjct 781 TGGPDGDLGSNAIKVSNTKTLTVLDKSGVLHDPNGLDLNELRRLAFLRDTTHLSATDNPN 840
Query 50 ----------------FKGE--------KTSSMLFDSSLLSPKGFKVPQDARDITLPDGT 85
+G+ KT SM +D LLS KGF VP++A ++ LPDG
Sbjct 841 NVTLNHSNSSLQRSSSLEGDEELLGARLKTCSMGYDKRLLSSKGFMVPEEAMNVVLPDGF 900
Query 86 YVASGIEFRNNFHLSKFAVCDLFNPCGGRPASVNPRNVEQLFEQQTGLPKFKFIVEGANV 145
V +G +FR+ FHLS +A DLF PCGGRP+S+ P NV +LF+++ G +FKFIVEG+NV
Sbjct 901 VVKNGYKFRDEFHLSSYAKADLFCPCGGRPSSITPFNVNRLFDEK-GKCRFKFIVEGSNV 959
Query 146 FITDEARRELENRGVILFKDASTNKGGVTSSSMEVLAALSLTEEEFDRHMRVKDPNNPPK 205
+IT ARR LE++GVILFKDASTNKGGVTSSS EVL +L L +E ++R + V+ N P+
Sbjct 960 YITQNARRFLESKGVILFKDASTNKGGVTSSSYEVLLSLVLDDETYER-VAVERDGNVPE 1018
Query 206 FYKDYVEAILQRIQENARLEFEAIWNEAERTGMHKCDLTDILSQKINQLKTDMAGSSALW 265
F K YV I++ I++NA LEFEA+W+E RTG+ +CDLTD+LS KIN LK+ + S+ L+
Sbjct 1019 FRKKYVNDIMEIIRKNATLEFEALWSEGLRTGIPRCDLTDVLSDKINALKSRIKSSNTLF 1078
Query 266 KDEALVNFVLTKSIPDVLVPGLISVETFRQRVPETYQRAIFTTFLASRFLYKQPFTAEAS 325
D+ LV+ VL++ +P L+ L++VE +R+P+ Y RA+F +++AS F YK+ F+ + S
Sbjct 1079 NDKELVHTVLSRCVPASLLE-LVTVEKIVERLPQIYLRALFASYIASNFYYKEKFSDDTS 1137
Query 326 AFAFIAYLDEIR 337
FAF Y+ ++
Sbjct 1138 VFAFYEYITSLK 1149
> sce:YDL215C GDH2; Gdh2p (EC:1.4.1.2); K00260 glutamate dehydrogenase
[EC:1.4.1.2]
Length=1092
Score = 261 bits (668), Expect = 2e-69, Method: Compositional matrix adjust.
Identities = 149/352 (42%), Positives = 214/352 (60%), Gaps = 17/352 (4%)
Query 1 QTGGPDGDLGSNALLQTKSK--TLAVVDGAGVVYDPEGLNAEELRRLARLRFKGEKTSSM 58
QTGGPDGDLGSN +L + LA++DG+GV+ DP+GL+ +EL RLA E+
Sbjct 750 QTGGPDGDLGSNEILLSSPNECYLAILDGSGVLCDPKGLDKDELCRLAH-----ERKMIS 804
Query 59 LFDSSLLSPKGFKVPQDARDITLPDGTYVASGIEFRNNFHLSKFAVCD---LFNPCGGRP 115
FD+S LS GF V DA DI LP+GT VA+G FRN FH F D +F PCGGRP
Sbjct 805 DFDTSKLSNNGFFVSVDAMDIMLPNGTIVANGTTFRNTFHTQIFKFVDHVDIFVPCGGRP 864
Query 116 ASVNPRNVEQLFEQQTGLPKFKFIVEGANVFITDEARRELENRGVILFKDASTNKGGVTS 175
S+ N+ +++TG K +IVEGAN+FIT A+ LE G ILFKDAS NKGGVTS
Sbjct 865 NSITLNNLHYFVDEKTGKCKIPYIVEGANLFITQPAKNALEEHGCILFKDASANKGGVTS 924
Query 176 SSMEVLAALSLTEEEFDRHMRVKDPNNP-PKFYKDYVEAILQRIQENARLEFEAIWNEAE 234
SSMEVLA+L+L + +F H + D + YK YV + RIQ+NA LEF +WN +
Sbjct 925 SSMEVLASLALNDNDF-VHKFIGDVSGERSALYKSYVVEVQSRIQKNAELEFGQLWNLNQ 983
Query 235 RTGMHKCDLTDILSQKINQLKTDMAGSSALW-KDEALVNFVLT-KSIPDVLVPGLISVET 292
G H ++++ LS IN+L D+ S LW D L N++L K IP +L+ + ++
Sbjct 984 LNGTHISEISNQLSFTINKLNDDLVASQELWLNDLKLRNYLLLDKIIPKILI-DVAGPQS 1042
Query 293 FRQRVPETYQRAIFTTFLASRFLYKQPFTAEASAFAFIAYLDEIRSQMAAAA 344
+ +PE+Y + + +++L+S F+Y+ + + F+ ++ ++ + A+A
Sbjct 1043 VLENIPESYLKVLLSSYLSSTFVYQNGI--DVNIGKFLEFIGGLKREAEASA 1092
> pfa:PF08_0132 glutamate dehydrogenase, putative (EC:1.4.1.2);
K00260 glutamate dehydrogenase [EC:1.4.1.2]
Length=1397
Score = 171 bits (433), Expect = 4e-42, Method: Composition-based stats.
Identities = 98/203 (48%), Positives = 142/203 (69%), Gaps = 7/203 (3%)
Query 3 GGPDGDLGSNALLQTKSKTLAVVDGAGVVYDPEGLNAEELRRLARLRFKGEKTSSM---- 58
GGPDGDLGSNA+LQ+K+K ++++DG+G++YD +GLN EEL RLA+ R +K+ ++
Sbjct 983 GGPDGDLGSNAILQSKTKIISIIDGSGILYDKQGLNKEELIRLAKRRNNKDKSKAITCCT 1042
Query 59 LFDSSLLSPKGFKVPQDARDITLPDGTYVASGIEFRNNFHLSKFAVCDLFNPCGGRPASV 118
L+D S GFK+ + ++ + G + +G++FRN F L+ C+LFNPCGGRP S+
Sbjct 1043 LYDEKYFSKDGFKISIEDHNVDI-FGNKIRNGLDFRNTFFLNPLNKCELFNPCGGRPHSI 1101
Query 119 NPRNVEQLFEQQTGLPKFKFIVEGANVFITDEARRELENRGVILFKDASTNKGGVTSSSM 178
N NV + G +K+IVEGANVFI+D+AR LE++ VILFKDA+TNKGGV SSS+
Sbjct 1102 NIFNVNN--IIKNGECIYKYIVEGANVFISDDARNILESKNVILFKDAATNKGGVISSSL 1159
Query 179 EVLAALSLTEEEFDRHMRVKDPN 201
EVLA L L ++++ +M D +
Sbjct 1160 EVLAGLVLDDKQYIDYMCSPDSD 1182
Score = 99.8 bits (247), Expect = 2e-20, Method: Composition-based stats.
Identities = 55/147 (37%), Positives = 82/147 (55%), Gaps = 7/147 (4%)
Query 198 KDPNNPPKFYKDYVEAILQRIQENARLEFEAIWNEAERTGMHKCDLTDILSQKINQLKTD 257
+D + FYK YV+ I ++I LEFE++W E RT +ILS KI++LK D
Sbjct 1255 EDQQDVSDFYKAYVKEIQKKITHYCELEFESLWKETRRTKTPISKAINILSNKISELKKD 1314
Query 258 MAGSSALWKDEALVNFVLTKSIPDVLVPGLISVETFRQ---RVPETYQRAIFTTFLASRF 314
+ S L +D L+ VL + IP P L+ + TF Q RVP Y +++F + LAS +
Sbjct 1315 ILSSDTLCRDYKLLKKVLERVIP----PTLLKIVTFEQILERVPYVYIKSLFASSLASNY 1370
Query 315 LYKQPFTAEASAFAFIAYLDEIRSQMA 341
Y Q F + SAF F Y+ +++S+ A
Sbjct 1371 YYSQQFLNDLSAFNFFEYITKLQSESA 1397
> dre:317737 glud1a, cb622, glud1, wu:fc33g09, wu:fc66a10, zgc:77186;
glutamate dehydrogenase 1a; K00261 glutamate dehydrogenase
(NAD(P)+) [EC:1.4.1.3]
Length=544
Score = 43.9 bits (102), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 38/128 (29%), Positives = 53/128 (41%), Gaps = 15/128 (11%)
Query 60 FDSSLLSPKGFKVPQDARDITLPDGTYVA--SGIEFRNNFHLSKFAVCDLFNPCGGRPAS 117
D S+ +P G P++ D L +GT V + N A CD+ P
Sbjct 319 LDGSIWNPSGID-PKELEDYKLANGTIVGYPGATAYEGNI---LEAECDILIPAASE--- 371
Query 118 VNPRNVEQLFEQQTGLPKFKFIVEGANVFITDEARRELENRGVILFKDASTNKGGVTSSS 177
+QL ++ K K I EGAN T EA + R +++ D N GGVT S
Sbjct 372 ------KQLTKKNANNIKAKIIAEGANGPTTPEADKIFLERNIMVIPDMYLNAGGVTVSY 425
Query 178 MEVLAALS 185
E L L+
Sbjct 426 FEWLKNLN 433
> dre:373092 glud1b, MGC192851, cb719, glud1, wu:fb16e02, wu:fb58f12,
wu:fe37f03, wu:fj43f02, zgc:192851, zgc:55630; glutamate
dehydrogenase 1b (EC:1.4.1.3); K00261 glutamate dehydrogenase
(NAD(P)+) [EC:1.4.1.3]
Length=542
Score = 43.9 bits (102), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 43/154 (27%), Positives = 62/154 (40%), Gaps = 16/154 (10%)
Query 35 EGLNAEELRRLARLRFKGEKTSSML-FDSSLLSPKGFKVPQDARDITLPDGTYVA--SGI 91
+G L + L G K + D S+ +P G P++ D L GT V +
Sbjct 291 QGFGNVGLHSMRYLHRYGAKCVGIAEIDGSIWNPNGMD-PKELEDYKLQHGTIVGFPNSQ 349
Query 92 EFRNNFHLSKFAVCDLFNPCGGRPASVNPRNVEQLFEQQTGLPKFKFIVEGANVFITDEA 151
+ N ++ CD+ P G +QL + K K I EGAN T +A
Sbjct 350 PYEGNILEAQ---CDILIPAAGE---------KQLTRKNAHNIKAKIIAEGANGPTTPDA 397
Query 152 RRELENRGVILFKDASTNKGGVTSSSMEVLAALS 185
+ R V++ D N GGVT S E L L+
Sbjct 398 DKIFIERNVMVIPDMYLNAGGVTVSYFEWLKNLN 431
> ath:AT5G07440 GDH2; GDH2 (GLUTAMATE DEHYDROGENASE 2); ATP binding
/ glutamate dehydrogenase [NAD(P)+]/ glutamate dehydrogenase/
oxidoreductase (EC:1.4.1.3); K00261 glutamate dehydrogenase
(NAD(P)+) [EC:1.4.1.3]
Length=411
Score = 43.9 bits (102), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 55/187 (29%), Positives = 80/187 (42%), Gaps = 35/187 (18%)
Query 27 GAGVVYDPEGLNAEELRRLARLRFKGEKTSSM-LFDSSLLSPKGFKVPQDARDITL---- 81
G GVV+ E L AE + + L F + ++ + + L+ KG KV DIT
Sbjct 186 GRGVVFATEALLAEYGKSIQGLTFVIQGFGNVGTWAAKLIHEKGGKVVA-VSDITGAIRN 244
Query 82 PDGTYVASGIEFR------NNFHLSK--------FAVCDLFNPC--GGRPASVNPRNVEQ 125
P+G + + I+ + N+F+ CD+ PC GG
Sbjct 245 PEGIDINALIKHKDATGSLNDFNGGDAMNSDELLIHECDVLIPCALGG-----------V 293
Query 126 LFEQQTGLPKFKFIVEGANVFITDEARRELENRGVILFKDASTNKGGVTSSSMEVLAALS 185
L ++ G K KFIVE AN +A L +GVI+ D N GGVT S E + +
Sbjct 294 LNKENAGDVKAKFIVEAANHPTDPDADEILSKKGVIILPDIYANAGGVTVSYFEWVQNIQ 353
Query 186 --LTEEE 190
+ EEE
Sbjct 354 GFMWEEE 360
> ath:AT3G03910 GDH3; GDH3 (GLUTAMATE DEHYDROGENASE 3); binding
/ catalytic/ oxidoreductase/ oxidoreductase, acting on the
CH-NH2 group of donors, NAD or NADP as acceptor (EC:1.4.1.3);
K00261 glutamate dehydrogenase (NAD(P)+) [EC:1.4.1.3]
Length=411
Score = 41.6 bits (96), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 31/97 (31%), Positives = 45/97 (46%), Gaps = 16/97 (16%)
Query 105 CDLFNPC--GGRPASVNPRNVEQLFEQQTGLPKFKFIVEGANVFITDEARRELENRGVIL 162
CD+ P GG +N N ++ K KFI+EGAN EA L+ +GV++
Sbjct 282 CDILVPAALGG---VINRENANEI--------KAKFIIEGANHPTDPEADEILKKKGVMI 330
Query 163 FKDASTNKGGVTSSSMEV---LAALSLTEEEFDRHMR 196
D N GGVT S E + EE+ +R ++
Sbjct 331 LPDIYANSGGVTVSYFEWVQNIQGFMWDEEKVNRELK 367
> pfa:PF14_0286 glutamate dehydrogenase, putative (EC:1.4.1.3);
K00262 glutamate dehydrogenase (NADP+) [EC:1.4.1.4]
Length=510
Score = 39.3 bits (90), Expect = 0.023, Method: Compositional matrix adjust.
Identities = 42/186 (22%), Positives = 73/186 (39%), Gaps = 35/186 (18%)
Query 14 LLQTKSKTLAVVDGAGVVYDPEGLNAEELRRLARLRFKGEKTSSMLFDSSLLSPKGFKVP 73
L++ + L + D G + +P G E+L + ++ + L K K
Sbjct 312 LIEKGAIVLTMSDSNGYILEPNGFTKEQLNYIMDIK----NNQRLRLKEYLKYSKTAKYF 367
Query 74 QDARDITLPDGTYVASGIEFRNNFHLSKFAVCDLFNPCGGRPASVNPRNVEQLFEQQTGL 133
++ + +P CD+ PC + +N + + LF Q
Sbjct 368 ENQKPWNIP----------------------CDIAFPCATQ-NEINENDAD-LFIQN--- 400
Query 134 PKFKFIVEGANVFITDEARRELENRGVILFKDASTNKGGVTSSSMEV---LAALSLTEEE 190
K K IVEGAN+ +A +L+ +IL + N GGV S +E+ L T +E
Sbjct 401 -KCKMIVEGANMPTHIKALHKLKQNNIILCPSKAANAGGVAVSGLEMSQNSMRLQWTHQE 459
Query 191 FDRHMR 196
D ++
Sbjct 460 TDMKLQ 465
> xla:446858 glud1, MGC80801; glutamate dehydrogenase 1 (EC:1.4.1.3);
K00261 glutamate dehydrogenase (NAD(P)+) [EC:1.4.1.3]
Length=540
Score = 39.3 bits (90), Expect = 0.026, Method: Compositional matrix adjust.
Identities = 39/139 (28%), Positives = 54/139 (38%), Gaps = 15/139 (10%)
Query 49 RFKGEKTSSMLFDSSLLSPKGFKVPQDARDITLPDGTYVA--SGIEFRNNFHLSKFAVCD 106
RF + D ++ +P G P++ D L GT V + N A CD
Sbjct 304 RFGAKCVGIGEIDGTIWNPNGID-PKELEDYKLQHGTIVGFPKAQPYDGNI---LEADCD 359
Query 107 LFNPCGGRPASVNPRNVEQLFEQQTGLPKFKFIVEGANVFITDEARRELENRGVILFKDA 166
+ P +QL + K K I EGAN T EA + R +++ D
Sbjct 360 ILIPAASE---------KQLTKSNAHKIKAKIIAEGANGPTTPEADKIFLERNIMVIPDL 410
Query 167 STNKGGVTSSSMEVLAALS 185
N GGVT S E L L+
Sbjct 411 YLNAGGVTVSYFEWLKNLN 429
> hsa:2747 GLUD2, GDH2, GLUDP1; glutamate dehydrogenase 2 (EC:1.4.1.3);
K00261 glutamate dehydrogenase (NAD(P)+) [EC:1.4.1.3]
Length=558
Score = 38.9 bits (89), Expect = 0.028, Method: Compositional matrix adjust.
Identities = 37/126 (29%), Positives = 52/126 (41%), Gaps = 13/126 (10%)
Query 61 DSSLLSPKGFKVPQDARDITLPDGTYVASGIEFRNNFHLSKFAV-CDLFNPCGGRPASVN 119
D S+ +P G P++ D L G+ + G + S V CD+ P
Sbjct 334 DGSIWNPDGID-PKELEDFKLQHGSIL--GFPKAKPYEGSILEVDCDILIPAATE----- 385
Query 120 PRNVEQLFEQQTGLPKFKFIVEGANVFITDEARRELENRGVILFKDASTNKGGVTSSSME 179
+QL + K K I EGAN T EA + R +++ D N GGVT S E
Sbjct 386 ----KQLTKSNAPRVKAKIIAEGANGPTTPEADKIFLERNILVIPDLYLNAGGVTVSYFE 441
Query 180 VLAALS 185
L L+
Sbjct 442 WLKNLN 447
> hsa:2746 GLUD1, GDH, GDH1, GLUD, MGC132003; glutamate dehydrogenase
1 (EC:1.4.1.3); K00261 glutamate dehydrogenase (NAD(P)+)
[EC:1.4.1.3]
Length=558
Score = 38.5 bits (88), Expect = 0.041, Method: Compositional matrix adjust.
Identities = 37/126 (29%), Positives = 52/126 (41%), Gaps = 13/126 (10%)
Query 61 DSSLLSPKGFKVPQDARDITLPDGTYVASGIEFRNNFHLSKF-AVCDLFNPCGGRPASVN 119
D S+ +P G P++ D L G+ + G + S A CD+ P
Sbjct 334 DGSIWNPDGID-PKELEDFKLQHGSIL--GFPKAKPYEGSILEADCDILIPAASE----- 385
Query 120 PRNVEQLFEQQTGLPKFKFIVEGANVFITDEARRELENRGVILFKDASTNKGGVTSSSME 179
+QL + K K I EGAN T EA + R +++ D N GGVT S E
Sbjct 386 ----KQLTKSNAPRVKAKIIAEGANGPTTPEADKIFLERNIMVIPDLYLNAGGVTVSYFE 441
Query 180 VLAALS 185
L L+
Sbjct 442 WLKNLN 447
> tgo:TGME49_093180 NADP-specific glutamate dehydrogenase, putative
(EC:1.4.1.4); K00262 glutamate dehydrogenase (NADP+) [EC:1.4.1.4]
Length=489
Score = 38.1 bits (87), Expect = 0.050, Method: Compositional matrix adjust.
Identities = 41/166 (24%), Positives = 68/166 (40%), Gaps = 25/166 (15%)
Query 50 FKGEKTSS-----MLFDSS---LLSPKGFKVPQDARDITLPDGTYVASGIEFRNNFHLSK 101
+ GEK ++ M F S +++ KGF + Q R + + EF + K
Sbjct 284 YAGEKLATLGAKVMTFSDSSGYIVNEKGFPLGQIQRLKEMKETRSSTRVSEFAAKYSTVK 343
Query 102 FAV--------CDLFNPCGGRPASVNPRNVEQLFEQQTGLPKFKFIVEGANVFITDEARR 153
F CD+ PC + ++ + + L + + + EGAN+ T +A R
Sbjct 344 FVPDKRAWEVPCDIAFPCATQ-NEISEEDAQLLIDNGC-----RIVAEGANMPTTRDAIR 397
Query 154 ELENRGVILFKDASTNKGGVTSSSMEV---LAALSLTEEEFDRHMR 196
+ GVIL + N GGV S +E+ + T E D +R
Sbjct 398 LFKQHGVILCPGKAANAGGVAVSGLEMSQNAMRIEWTREVVDEKLR 443
> mmu:14661 Glud1, AI118167, Gdh-X, Glud, Gludl; glutamate dehydrogenase
1 (EC:1.4.1.3); K00261 glutamate dehydrogenase (NAD(P)+)
[EC:1.4.1.3]
Length=558
Score = 38.1 bits (87), Expect = 0.051, Method: Compositional matrix adjust.
Identities = 37/126 (29%), Positives = 52/126 (41%), Gaps = 13/126 (10%)
Query 61 DSSLLSPKGFKVPQDARDITLPDGTYVASGIEFRNNFHLSKF-AVCDLFNPCGGRPASVN 119
D S+ +P G P++ D L G+ + G + S A CD+ P
Sbjct 334 DGSIWNPDGID-PKELEDFKLQHGSIL--GFPKAKVYEGSILEADCDILIPAASE----- 385
Query 120 PRNVEQLFEQQTGLPKFKFIVEGANVFITDEARRELENRGVILFKDASTNKGGVTSSSME 179
+QL + K K I EGAN T EA + R +++ D N GGVT S E
Sbjct 386 ----KQLTKSNAPRVKAKIIAEGANGPTTPEADKIFLERNIMVIPDLYLNAGGVTVSYFE 441
Query 180 VLAALS 185
L L+
Sbjct 442 WLKNLN 447
> ath:AT5G18170 GDH1; GDH1 (GLUTAMATE DEHYDROGENASE 1); ATP binding
/ glutamate dehydrogenase [NAD(P)+]/ oxidoreductase (EC:1.4.1.3);
K00261 glutamate dehydrogenase (NAD(P)+) [EC:1.4.1.3]
Length=411
Score = 37.4 bits (85), Expect = 0.100, Method: Compositional matrix adjust.
Identities = 29/90 (32%), Positives = 41/90 (45%), Gaps = 15/90 (16%)
Query 105 CDLFNPC--GGRPASVNPRNVEQLFEQQTGLPKFKFIVEGANVFITDEARRELENRGVIL 162
CD+ P GG +N N ++ K KFI+E AN +A L +GV++
Sbjct 282 CDILVPAALGG---VINRENANEI--------KAKFIIEAANHPTDPDADEILSKKGVVI 330
Query 163 FKDASTNKGGVTSSSMEVLAALS--LTEEE 190
D N GGVT S E + + + EEE
Sbjct 331 LPDIYANSGGVTVSYFEWVQNIQGFMWEEE 360
> bbo:BBOV_IV010390 23.m06170; glutamate dehydrogenase (EC:1.4.1.4);
K00262 glutamate dehydrogenase (NADP+) [EC:1.4.1.4]
Length=455
Score = 37.0 bits (84), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 41/169 (24%), Positives = 66/169 (39%), Gaps = 36/169 (21%)
Query 14 LLQTKSKTLAVVDGAGVVYDPEGLNAEELRRLARLRFKGEKTSSMLFDSSLLSP--KGFK 71
L++ + + + D +G + +PEG+ E LR + + + S + S + G K
Sbjct 257 LIELGAVPITMSDSSGYIIEPEGITLEGLRYIMAFKNPHSRRISEYLNYSKTATFHPGDK 316
Query 72 VPQDARDITLPDGTYVASGIEFRNNFHLSKFAVCDLFNPCGGRPASVNPRNVEQLFEQQT 131
++ DI P T +N L R+ E L +
Sbjct 317 PWGESADIAFPCAT--------QNEILL---------------------RDAETLVKGGV 347
Query 132 GLPKFKFIVEGANVFITDEARRELENRGVILFKDASTNKGGVTSSSMEV 180
K +VEGAN+ EA L+ GVIL + N GGV S +E+
Sbjct 348 -----KLVVEGANMPTHSEAVHYLKENGVILCPGKAANAGGVLVSGLEM 391
> cel:ZK829.4 hypothetical protein; K00261 glutamate dehydrogenase
(NAD(P)+) [EC:1.4.1.3]
Length=536
Score = 35.4 bits (80), Expect = 0.32, Method: Compositional matrix adjust.
Identities = 36/128 (28%), Positives = 55/128 (42%), Gaps = 12/128 (9%)
Query 60 FDSSLLSPKGFKVPQDARDITLPDGTYVA-SGIEFRNNFHLSKFAVCDLFNPCGGRPASV 118
+D ++ +P G P++ D +GT G + + F + CD+F P S+
Sbjct 311 YDCAVYNPDGIH-PKELEDWKDANGTIKNFPGAKNFDPFTELMYEKCDIFVPAACE-KSI 368
Query 119 NPRNVEQLFEQQTGLPKFKFIVEGANVFITDEARRELENRG-VILFKDASTNKGGVTSSS 177
+ N ++ + K I E AN T A R L RG ++ D N GGVT S
Sbjct 369 HKENASRI--------QAKIIAEAANGPTTPAADRILLARGDCLIIPDMYVNSGGVTVSY 420
Query 178 MEVLAALS 185
E L L+
Sbjct 421 FEWLKNLN 428
> dre:100331964 ubiquitin specific peptidase 29-like; K11850 ubiquitin
carboxyl-terminal hydrolase 26/29/37 [EC:3.1.2.15]
Length=935
Score = 34.3 bits (77), Expect = 0.65, Method: Compositional matrix adjust.
Identities = 25/76 (32%), Positives = 41/76 (53%), Gaps = 12/76 (15%)
Query 194 HMRVKDPNNPPKFYKDYVEAILQRIQENARLEFEAIWNEAER-TGMHKCDLTDILSQKIN 252
H+ K +PP+ KD +L+R++ AI + AER +G + D + LSQ ++
Sbjct 371 HLLAKKDISPPEVKKD----LLRRVKN-------AISSTAERFSGYMQNDAHEFLSQCLD 419
Query 253 QLKTDMAGSSALWKDE 268
QLK D+ + WK+E
Sbjct 420 QLKEDVEKINKSWKNE 435
> dre:768201 usp37, MGC153999, wu:fi15b04, wu:fi38d03, zgc:152882,
zgc:153999; ubiquitin specific peptidase 37 (EC:3.1.2.15);
K11850 ubiquitin carboxyl-terminal hydrolase 26/29/37 [EC:3.1.2.15]
Length=938
Score = 34.3 bits (77), Expect = 0.70, Method: Compositional matrix adjust.
Identities = 25/76 (32%), Positives = 41/76 (53%), Gaps = 12/76 (15%)
Query 194 HMRVKDPNNPPKFYKDYVEAILQRIQENARLEFEAIWNEAER-TGMHKCDLTDILSQKIN 252
H+ K +PP+ KD +L+R++ AI + AER +G + D + LSQ ++
Sbjct 371 HLLAKKDISPPEVKKD----LLRRVKN-------AISSTAERFSGYMQNDAHEFLSQCLD 419
Query 253 QLKTDMAGSSALWKDE 268
QLK D+ + WK+E
Sbjct 420 QLKEDVEKINKSWKNE 435
> hsa:4842 NOS1, IHPS1, N-NOS, NC-NOS, NOS, bNOS, nNOS; nitric
oxide synthase 1 (neuronal) (EC:1.14.13.39); K13240 nitric-oxide
synthase, brain [EC:1.14.13.39]
Length=1434
Score = 33.5 bits (75), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 28/87 (32%), Positives = 41/87 (47%), Gaps = 15/87 (17%)
Query 200 PNNPPKF--YKDY------VEAILQRIQENARLEFEAI----WNEAERTGMHK-CDLT-- 244
P PKF +KD + A+ + E LEF A W G+ CD +
Sbjct 550 PIRHPKFEWFKDLGLKWYGLPAVSNMLLEIGGLEFSACPFSGWYMGTEIGVRDYCDNSRY 609
Query 245 DILSQKINQLKTDMAGSSALWKDEALV 271
+IL + ++ DM +S+LWKD+ALV
Sbjct 610 NILEEVAKKMNLDMRKTSSLWKDQALV 636
> bbo:BBOV_I002890 19.m02290; suppressor Mra1 family domain containing
protein; K14568 essential for mitotic growth 1
Length=188
Score = 33.1 bits (74), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 31/107 (28%), Positives = 51/107 (47%), Gaps = 27/107 (25%)
Query 133 LPKFKFIVEGANVFITD-EARRELENRGVILFKDASTNKGGVTSSSMEVLAALSLTEE-- 189
LPK ++E A++ TD E+R ++ ++ +++ D+ NK G L L T++
Sbjct 3 LPKLVIVLEDASLLSTDKESRVDILHQSLLVLLDSPLNKNG-------FLKVLIRTDDGN 55
Query 190 --EFDRHMRVKDPNNPPKF--------YKDYV-----EAILQRIQEN 221
E H+RV P P +F YK +V +AIL R +N
Sbjct 56 LVEVSPHLRV--PRTPKQFESLLVYLLYKRHVKSQERDAILMRFIKN 100
> mmu:18125 Nos1, 2310005C01Rik, NO, NOS-I, Nos-1, bNOS, nNOS;
nitric oxide synthase 1, neuronal (EC:1.14.13.39); K13240 nitric-oxide
synthase, brain [EC:1.14.13.39]
Length=1429
Score = 33.1 bits (74), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 28/87 (32%), Positives = 41/87 (47%), Gaps = 15/87 (17%)
Query 200 PNNPPKF--YKDY------VEAILQRIQENARLEFEAI----WNEAERTGMHK-CDLT-- 244
P PKF +KD + A+ + E LEF A W G+ CD +
Sbjct 545 PIRHPKFDWFKDLGLKWYGLPAVSNMLLEIGGLEFSACPFSGWYMGTEIGVRDYCDNSRY 604
Query 245 DILSQKINQLKTDMAGSSALWKDEALV 271
+IL + ++ DM +S+LWKD+ALV
Sbjct 605 NILEEVAKKMDLDMRKTSSLWKDQALV 631
> dre:100332249 Extracellular matrix protein 1-like
Length=499
Score = 31.6 bits (70), Expect = 5.5, Method: Compositional matrix adjust.
Identities = 19/46 (41%), Positives = 23/46 (50%), Gaps = 0/46 (0%)
Query 46 ARLRFKGEKTSSMLFDSSLLSPKGFKVPQDARDITLPDGTYVASGI 91
A +FKG K +S SL SPK K A D+T P G +S I
Sbjct 238 ASQKFKGFKYNSSACKGSLASPKALKKQTAAPDLTFPPGRPESSNI 283
> eco:b1761 gdhA, ECK1759, JW1750; glutamate dehydrogenase, NADP-specific
(EC:1.4.1.4); K00262 glutamate dehydrogenase (NADP+)
[EC:1.4.1.4]
Length=447
Score = 31.2 bits (69), Expect = 6.4, Method: Compositional matrix adjust.
Identities = 16/44 (36%), Positives = 24/44 (54%), Gaps = 0/44 (0%)
Query 137 KFIVEGANVFITDEARRELENRGVILFKDASTNKGGVTSSSMEV 180
K + EGAN+ T EA + GV+ + N GGV +S +E+
Sbjct 341 KAVAEGANMPTTIEATELFQQAGVLFAPGKAANAGGVATSGLEM 384
> ath:AT4G15330 CYP705A1; electron carrier/ heme binding / iron
ion binding / monooxygenase/ oxygen binding
Length=513
Score = 30.4 bits (67), Expect = 9.9, Method: Compositional matrix adjust.
Identities = 20/65 (30%), Positives = 28/65 (43%), Gaps = 5/65 (7%)
Query 100 SKFAVCDLFNPCGGRPASVNPRNVEQLFEQQTGLPKFKFIVEGANVFITDEARRELENRG 159
S+F L+ C GR SV VE++ E L F++ R+ LE G
Sbjct 183 SRFVNNSLYKMCTGRSFSVENNEVERIMELTADLGAL-----SQKFFVSKMFRKLLEKLG 237
Query 160 VILFK 164
+ LFK
Sbjct 238 ISLFK 242
Lambda K H
0.317 0.133 0.377
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 15323160248
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40