bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_0734_orf1
Length=199
Score E
Sequences producing significant alignments: (Bits) Value
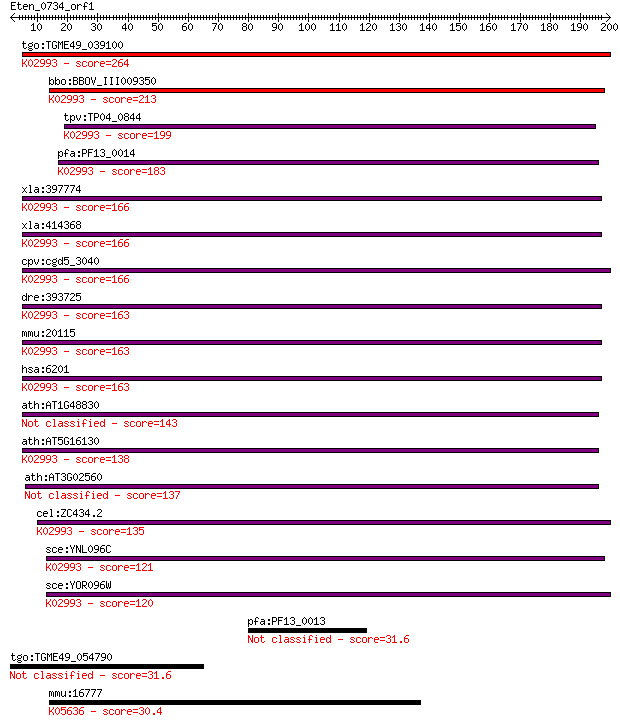
tgo:TGME49_039100 40S ribosomal protein S7, putative ; K02993 ... 264 1e-70
bbo:BBOV_III009350 17.m07812; ribosomal protein S7e family pro... 213 3e-55
tpv:TP04_0844 40S ribosomal protein S7; K02993 small subunit r... 199 7e-51
pfa:PF13_0014 40S ribosomal protein S7, putative; K02993 small... 183 4e-46
xla:397774 rps7, rpS8A; ribosomal protein S8 (EC:3.6.5.3); K02... 166 5e-41
xla:414368 rps7, MGC53555, dba8, rpS8B; ribosomal protein S7; ... 166 5e-41
cpv:cgd5_3040 40S ribosomal protein S7 ; K02993 small subunit ... 166 7e-41
dre:393725 rps7, MGC73216, zgc:73216; ribosomal protein S7 (EC... 163 3e-40
mmu:20115 Rps7, Rps7A, S7; ribosomal protein S7 (EC:3.6.5.3); ... 163 4e-40
hsa:6201 RPS7, DBA8; ribosomal protein S7 (EC:3.6.5.3); K02993... 163 4e-40
ath:AT1G48830 40S ribosomal protein S7 (RPS7A) 143 3e-34
ath:AT5G16130 40S ribosomal protein S7 (RPS7C); K02993 small s... 138 1e-32
ath:AT3G02560 40S ribosomal protein S7 (RPS7B) 137 3e-32
cel:ZC434.2 rps-7; Ribosomal Protein, Small subunit family mem... 135 1e-31
sce:YNL096C RPS7B; Rps7bp; K02993 small subunit ribosomal prot... 121 1e-27
sce:YOR096W RPS7A, RPS30; Rps7ap; K02993 small subunit ribosom... 120 2e-27
pfa:PF13_0013 PBS lyase HEAT-like protein, putative 31.6 2.0
tgo:TGME49_054790 hypothetical protein 31.6 2.1
mmu:16777 Lamb1, C77966, C80098, C81607, D130003D08Rik, Lamb-1... 30.4 4.3
> tgo:TGME49_039100 40S ribosomal protein S7, putative ; K02993
small subunit ribosomal protein S7e
Length=196
Score = 264 bits (675), Expect = 1e-70, Method: Compositional matrix adjust.
Identities = 124/195 (63%), Positives = 158/195 (81%), Gaps = 0/195 (0%)
Query 5 MTTRQRIVKVDGSQPTEVEKEVDRCLADIESSAQSELRAEVAELTICAVKPVEVPASGKT 64
M +++IVK G +PTE+E EV +CL DIE+S+QS+L+ +V L I +VK VEVP K
Sbjct 2 MNFKKKIVKPQGVEPTEIENEVAKCLFDIETSSQSDLKGDVRHLVISSVKEVEVPQGRKK 61
Query 65 ALVIFVPYRVYMNVVKRIHARLVQELEKKLKRHVVIIAQRSIVGLDYKRKNLKVRPRSRT 124
A V+FVP+ VY VK+I RLVQELEKKLK+HVV++AQR+++ LD+KRK LKVRPRSRT
Sbjct 62 AFVVFVPHAVYQKTVKKIQGRLVQELEKKLKKHVVLVAQRTMLPLDFKRKGLKVRPRSRT 121
Query 125 LTNVQDAMLEDIVAPSEVVGRRWRVRADGSKLMKVHLDPKDKAKDNLEEKLWTFAAVYKR 184
LT VQD MLEDIV+P+E+VG+R RVR D +K++KV LDP+DK KDN+E+KL TFAAVYK+
Sbjct 122 LTAVQDGMLEDIVSPTEIVGKRMRVRVDNTKMLKVFLDPRDKQKDNIEDKLETFAAVYKK 181
Query 185 LTNKDAAFLFPEHVY 199
LTNKDA F FP+HVY
Sbjct 182 LTNKDAVFSFPQHVY 196
> bbo:BBOV_III009350 17.m07812; ribosomal protein S7e family protein;
K02993 small subunit ribosomal protein S7e
Length=194
Score = 213 bits (543), Expect = 3e-55, Method: Compositional matrix adjust.
Identities = 100/184 (54%), Positives = 141/184 (76%), Gaps = 0/184 (0%)
Query 14 VDGSQPTEVEKEVDRCLADIESSAQSELRAEVAELTICAVKPVEVPASGKTALVIFVPYR 73
V +PT +EKE+ CL I++S ++L+ +V + I K VEV A K+A++IFVPYR
Sbjct 10 VGTHEPTALEKEIANCLVYIQTSGNADLKGDVQNVVISHCKEVEVGALKKSAVIIFVPYR 69
Query 74 VYMNVVKRIHARLVQELEKKLKRHVVIIAQRSIVGLDYKRKNLKVRPRSRTLTNVQDAML 133
VYM+ V+RIH RLV ELEKK K+HVVIIAQR+I+ +++ K ++RPR+R+LT+V +A+L
Sbjct 70 VYMSYVRRIHGRLVVELEKKTKKHVVIIAQRTILPKNFRSKQFRIRPRNRSLTSVHEAIL 129
Query 134 EDIVAPSEVVGRRWRVRADGSKLMKVHLDPKDKAKDNLEEKLWTFAAVYKRLTNKDAAFL 193
EDI AP+E+VG+ RV+ DGS++++V+LDPKDK DN+E+KL TFAAVYK+LT+KD F
Sbjct 130 EDIAAPTEIVGKVLRVKRDGSRILRVYLDPKDKQNDNVEDKLETFAAVYKKLTDKDVVFS 189
Query 194 FPEH 197
F H
Sbjct 190 FKLH 193
> tpv:TP04_0844 40S ribosomal protein S7; K02993 small subunit
ribosomal protein S7e
Length=202
Score = 199 bits (505), Expect = 7e-51, Method: Compositional matrix adjust.
Identities = 93/176 (52%), Positives = 133/176 (75%), Gaps = 0/176 (0%)
Query 19 PTEVEKEVDRCLADIESSAQSELRAEVAELTICAVKPVEVPASGKTALVIFVPYRVYMNV 78
PT +E+EV +CL I+SS ++L+ +V ++ + + + VEV A K A+V+FVPYRVY N
Sbjct 23 PTPLEREVAKCLLYIQSSGTADLKGDVKKIVVSSCREVEVGALKKNAVVVFVPYRVYQNH 82
Query 79 VKRIHARLVQELEKKLKRHVVIIAQRSIVGLDYKRKNLKVRPRSRTLTNVQDAMLEDIVA 138
V+RIH RL+ ELEKK KRHV I++QR I+ + K + RPR+RTLT+V +A+LEDI A
Sbjct 83 VRRIHGRLLIELEKKTKRHVFIVSQRKIMSKQFNNKAFRKRPRNRTLTSVHEAVLEDIAA 142
Query 139 PSEVVGRRWRVRADGSKLMKVHLDPKDKAKDNLEEKLWTFAAVYKRLTNKDAAFLF 194
P+E+VG+ R+R DG++++KV LDPKD+ DN+E+KL T +AVYK+LTNK+A F F
Sbjct 143 PTEIVGKVTRMRQDGTRVLKVFLDPKDRQNDNVEDKLETLSAVYKKLTNKEAVFTF 198
> pfa:PF13_0014 40S ribosomal protein S7, putative; K02993 small
subunit ribosomal protein S7e
Length=194
Score = 183 bits (464), Expect = 4e-46, Method: Compositional matrix adjust.
Identities = 80/179 (44%), Positives = 133/179 (74%), Gaps = 0/179 (0%)
Query 17 SQPTEVEKEVDRCLADIESSAQSELRAEVAELTICAVKPVEVPASGKTALVIFVPYRVYM 76
S P+++EKE+ +CL DIE S+ S+++ + E+ + + +EV K ++I++PY++Y
Sbjct 11 SNPSDLEKEIAQCLLDIELSSSSDIKTDAKEIKLLSCDLIEVEKLKKKTILIYIPYKIYT 70
Query 77 NVVKRIHARLVQELEKKLKRHVVIIAQRSIVGLDYKRKNLKVRPRSRTLTNVQDAMLEDI 136
V++I +L+ ELEKK K++VV++A+R+I+ K K+ K+ PRSRTLT+V D++LEDI
Sbjct 71 TYVRKIQRKLINELEKKTKKYVVLVAKRTILKGKQKNKSFKIIPRSRTLTSVYDSILEDI 130
Query 137 VAPSEVVGRRWRVRADGSKLMKVHLDPKDKAKDNLEEKLWTFAAVYKRLTNKDAAFLFP 195
V+PSE++G+R ++ADG ++ K+ LD K++ +DN+EEKL +FAAVYK++T +DA F P
Sbjct 131 VSPSEIIGKRISMKADGKRVFKIMLDSKERQRDNIEEKLISFAAVYKKITRRDAVFSLP 189
> xla:397774 rps7, rpS8A; ribosomal protein S8 (EC:3.6.5.3); K02993
small subunit ribosomal protein S7e
Length=194
Score = 166 bits (420), Expect = 5e-41, Method: Compositional matrix adjust.
Identities = 95/196 (48%), Positives = 133/196 (67%), Gaps = 10/196 (5%)
Query 5 MTTRQRIVKVDGSQPTEVEKEVDRCLADIESSAQSELRAEVAELTICAVKPVEVPASGKT 64
+T +IVK +G +P E E + + L ++E + S+L+A++ EL I A K +EV A G+
Sbjct 2 FSTSAKIVKPNGEKPDEFESGISQALLELEMN--SDLKAQLRELNITAAKEIEVGA-GRK 58
Query 65 ALVIFVPYRVYMNVVKRIHARLVQELEKKLK-RHVVIIAQRSIVGLDYKR---KNLKVRP 120
A++IFVP + ++I RLV+ELEKK +HVV IAQR I+ ++ KN + RP
Sbjct 59 AIIIFVPV-PQLKSFQKIQVRLVRELEKKFSGKHVVFIAQRRILPKPTRKSRTKNKQKRP 117
Query 121 RSRTLTNVQDAMLEDIVAPSEVVGRRWRVRADGSKLMKVHLDPKDKAKDNLEEKLWTFAA 180
RSRTLT V DA+LED+V PSE+VGRR RV+ DGS+L+KVHLD ++N+E K+ TF+
Sbjct 118 RSRTLTAVHDAILEDLVYPSEIVGRRIRVKLDGSRLIKVHLD--KAQQNNVEHKVETFSG 175
Query 181 VYKRLTNKDAAFLFPE 196
VYK+LT KD F FPE
Sbjct 176 VYKKLTGKDVVFEFPE 191
> xla:414368 rps7, MGC53555, dba8, rpS8B; ribosomal protein S7;
K02993 small subunit ribosomal protein S7e
Length=194
Score = 166 bits (420), Expect = 5e-41, Method: Compositional matrix adjust.
Identities = 95/196 (48%), Positives = 133/196 (67%), Gaps = 10/196 (5%)
Query 5 MTTRQRIVKVDGSQPTEVEKEVDRCLADIESSAQSELRAEVAELTICAVKPVEVPASGKT 64
+T +IVK +G +P E E + + L ++E + S+L+A++ EL I A K +EV A G+
Sbjct 2 FSTSAKIVKPNGEKPDEFESGISQALLELEMN--SDLKAQLRELNITAAKEIEVGA-GRK 58
Query 65 ALVIFVPYRVYMNVVKRIHARLVQELEKKLK-RHVVIIAQRSIVGLDYKR---KNLKVRP 120
A++IFVP + ++I RLV+ELEKK +HVV IAQR I+ ++ KN + RP
Sbjct 59 AIIIFVPV-PQLKSFQKIQVRLVRELEKKFSGKHVVFIAQRRILPKPTRKSRTKNKQKRP 117
Query 121 RSRTLTNVQDAMLEDIVAPSEVVGRRWRVRADGSKLMKVHLDPKDKAKDNLEEKLWTFAA 180
RSRTLT V DA+LED+V PSE+VGRR RV+ DGS+L+KVHLD ++N+E K+ TF+
Sbjct 118 RSRTLTAVHDAILEDLVYPSEIVGRRIRVKLDGSRLIKVHLD--KAQQNNVEHKVETFSG 175
Query 181 VYKRLTNKDAAFLFPE 196
VYK+LT KD F FPE
Sbjct 176 VYKKLTGKDVVFEFPE 191
> cpv:cgd5_3040 40S ribosomal protein S7 ; K02993 small subunit
ribosomal protein S7e
Length=204
Score = 166 bits (419), Expect = 7e-41, Method: Compositional matrix adjust.
Identities = 91/196 (46%), Positives = 128/196 (65%), Gaps = 4/196 (2%)
Query 5 MTTRQRIVKVDGSQPTEVEKEVDRCLADIESSAQSE-LRAEVAELTICAVKPVEVPASGK 63
+T + + +K + S T++EK+V+RC DIESS+ E L+A V + K E+ +
Sbjct 3 LTMKGKCIKKNDSPLTQLEKDVERCFQDIESSSNDEELKAIVQSVVFSTAK--EIDCGSR 60
Query 64 TALVIFVPYRVYMNVVKRIHARLVQELEKKLKRHVVIIAQRSIVGLDYKRKNLKVRPRSR 123
A+ +FVPY ++M V++I R++ ELEKKLK VV+I QRSI+ +Y++ K RPRSR
Sbjct 61 KAVAVFVPYAIFMAYVRKIQGRIIAELEKKLKCPVVMIGQRSILPRNYRKYGFKSRPRSR 120
Query 124 TLTNVQDAMLEDIVAPSEVVGRRWRVRADGSKLMKVHLDPKDKAKDNLEEKLWTFAAVYK 183
TLT V DA+LEDI PS++VG+R R R DGS L+KV LD ++ +K+ F AVYK
Sbjct 121 TLTAVHDALLEDICGPSDIVGKRLRYRVDGSTLLKVILD-PKDKDKDIADKVDVFGAVYK 179
Query 184 RLTNKDAAFLFPEHVY 199
RLTNK+A F FP Y
Sbjct 180 RLTNKEAEFSFPATNY 195
> dre:393725 rps7, MGC73216, zgc:73216; ribosomal protein S7 (EC:3.6.5.3);
K02993 small subunit ribosomal protein S7e
Length=194
Score = 163 bits (413), Expect = 3e-40, Method: Compositional matrix adjust.
Identities = 95/196 (48%), Positives = 132/196 (67%), Gaps = 10/196 (5%)
Query 5 MTTRQRIVKVDGSQPTEVEKEVDRCLADIESSAQSELRAEVAELTICAVKPVEVPASGKT 64
+T +IVK +G +P E E + + L ++E + S+L+A++ EL I A K +EV S K
Sbjct 2 FSTSAKIVKPNGEKPDEFESGISQALLELEMN--SDLKAQLRELHITAAKEIEVGGSRK- 58
Query 65 ALVIFVPYRVYMNVVKRIHARLVQELEKKLK-RHVVIIAQRSIVGLDYKR---KNLKVRP 120
A++IFVP + ++I RLV+ELEKK +HVV IAQR I+ ++ KN + RP
Sbjct 59 AIIIFVPV-PQLKSFQKIQVRLVRELEKKFSGKHVVFIAQRRILPKPTRKSRTKNKQKRP 117
Query 121 RSRTLTNVQDAMLEDIVAPSEVVGRRWRVRADGSKLMKVHLDPKDKAKDNLEEKLWTFAA 180
RSRTLTNV DA+LED+V PSE+VG+R RV+ D S+L+KVHLD ++N+E K+ TFA
Sbjct 118 RSRTLTNVHDAILEDLVFPSEIVGKRIRVKLDSSRLIKVHLD--KAQQNNVEHKVETFAG 175
Query 181 VYKRLTNKDAAFLFPE 196
VYK+LT KD F FPE
Sbjct 176 VYKKLTGKDVIFEFPE 191
> mmu:20115 Rps7, Rps7A, S7; ribosomal protein S7 (EC:3.6.5.3);
K02993 small subunit ribosomal protein S7e
Length=194
Score = 163 bits (412), Expect = 4e-40, Method: Compositional matrix adjust.
Identities = 92/196 (46%), Positives = 132/196 (67%), Gaps = 10/196 (5%)
Query 5 MTTRQRIVKVDGSQPTEVEKEVDRCLADIESSAQSELRAEVAELTICAVKPVEVPASGKT 64
++ +IVK +G +P E E + + L ++E + S+L+A++ EL I A K +EV G+
Sbjct 2 FSSSAKIVKPNGEKPDEFESGISQALLELEMN--SDLKAQLRELNITAAKEIEV-GGGRK 58
Query 65 ALVIFVPYRVYMNVVKRIHARLVQELEKKLK-RHVVIIAQRSIVGLDYKR---KNLKVRP 120
A++IFVP + ++I RLV+ELEKK +HVV IAQR I+ ++ KN + RP
Sbjct 59 AIIIFVPV-PQLKSFQKIQVRLVRELEKKFSGKHVVFIAQRRILPKPTRKSRTKNKQKRP 117
Query 121 RSRTLTNVQDAMLEDIVAPSEVVGRRWRVRADGSKLMKVHLDPKDKAKDNLEEKLWTFAA 180
RSRTLT V DA+LED+V PSE+VG+R RV+ DGS+L+KVHLD ++N+E K+ TF+
Sbjct 118 RSRTLTAVHDAILEDLVFPSEIVGKRIRVKLDGSRLIKVHLD--KAQQNNVEHKVETFSG 175
Query 181 VYKRLTNKDAAFLFPE 196
VYK+LT KD F FPE
Sbjct 176 VYKKLTGKDVNFEFPE 191
> hsa:6201 RPS7, DBA8; ribosomal protein S7 (EC:3.6.5.3); K02993
small subunit ribosomal protein S7e
Length=194
Score = 163 bits (412), Expect = 4e-40, Method: Compositional matrix adjust.
Identities = 92/196 (46%), Positives = 132/196 (67%), Gaps = 10/196 (5%)
Query 5 MTTRQRIVKVDGSQPTEVEKEVDRCLADIESSAQSELRAEVAELTICAVKPVEVPASGKT 64
++ +IVK +G +P E E + + L ++E + S+L+A++ EL I A K +EV G+
Sbjct 2 FSSSAKIVKPNGEKPDEFESGISQALLELEMN--SDLKAQLRELNITAAKEIEV-GGGRK 58
Query 65 ALVIFVPYRVYMNVVKRIHARLVQELEKKLK-RHVVIIAQRSIVGLDYKR---KNLKVRP 120
A++IFVP + ++I RLV+ELEKK +HVV IAQR I+ ++ KN + RP
Sbjct 59 AIIIFVPV-PQLKSFQKIQVRLVRELEKKFSGKHVVFIAQRRILPKPTRKSRTKNKQKRP 117
Query 121 RSRTLTNVQDAMLEDIVAPSEVVGRRWRVRADGSKLMKVHLDPKDKAKDNLEEKLWTFAA 180
RSRTLT V DA+LED+V PSE+VG+R RV+ DGS+L+KVHLD ++N+E K+ TF+
Sbjct 118 RSRTLTAVHDAILEDLVFPSEIVGKRIRVKLDGSRLIKVHLD--KAQQNNVEHKVETFSG 175
Query 181 VYKRLTNKDAAFLFPE 196
VYK+LT KD F FPE
Sbjct 176 VYKKLTGKDVNFEFPE 191
> ath:AT1G48830 40S ribosomal protein S7 (RPS7A)
Length=191
Score = 143 bits (361), Expect = 3e-34, Method: Compositional matrix adjust.
Identities = 82/192 (42%), Positives = 131/192 (68%), Gaps = 7/192 (3%)
Query 5 MTTRQRIVKVDGSQPTEVEKEVDRCLADIESSAQSELRAEVAELTICAVKPVEVPASGKT 64
+ + +I K G + +E++++V + D+E++ Q EL++E+ +L + + V++ + G+
Sbjct 2 FSAQHKIHKEKGVELSELDEQVAQAFFDLENTNQ-ELKSELKDLYVNSAVQVDI-SGGRK 59
Query 65 ALVIFVPYRVYMNVVKRIHARLVQELEKKLK-RHVVIIAQRSIVGLDYKRKNLKVRPRSR 123
A+V+ VPYR+ ++IH RLV+ELEKK + V++IA R IV K K RPR+R
Sbjct 60 AIVVNVPYRL-RKAYRKIHVRLVRELEKKFSGKDVILIATRRIVRPPKKGSAAK-RPRNR 117
Query 124 TLTNVQDAMLEDIVAPSEVVGRRWRVRADGSKLMKVHLDPKDKAKDNLEEKLWTFAAVYK 183
TLT+V +A+L+D+V P+E+VG+R R R DG+K+MKV LDPK+ ++N E K+ F+AVYK
Sbjct 118 TLTSVHEAILDDVVLPAEIVGKRTRYRLDGTKIMKVFLDPKE--RNNTEYKVEAFSAVYK 175
Query 184 RLTNKDAAFLFP 195
+LT KD F FP
Sbjct 176 KLTGKDVVFEFP 187
> ath:AT5G16130 40S ribosomal protein S7 (RPS7C); K02993 small
subunit ribosomal protein S7e
Length=190
Score = 138 bits (347), Expect = 1e-32, Method: Compositional matrix adjust.
Identities = 81/192 (42%), Positives = 126/192 (65%), Gaps = 7/192 (3%)
Query 5 MTTRQRIVKVDGSQPTEVEKEVDRCLADIESSAQSELRAEVAELTICAVKPVEVPASGKT 64
+ + +I K ++PTE E++V + L D+E++ Q EL++E+ +L I +++ + K
Sbjct 2 FSAQNKIKKDKNAEPTECEEQVAQALFDLENTNQ-ELKSELKDLYINQAVHMDISGNRK- 59
Query 65 ALVIFVPYRVYMNVVKRIHARLVQELEKKLK-RHVVIIAQRSIVGLDYKRKNLKVRPRSR 123
A+VI+VP+R+ ++IH RLV+ELEKK + V+ + R I+ K ++ RPR+R
Sbjct 60 AVVIYVPFRL-RKAFRKIHPRLVRELEKKFSGKDVIFVTTRRIMRPPKKGAAVQ-RPRNR 117
Query 124 TLTNVQDAMLEDIVAPSEVVGRRWRVRADGSKLMKVHLDPKDKAKDNLEEKLWTFAAVYK 183
TLT+V +AMLED+ P+E+VG+R R R DGSK+MKV LD K+ K+N E KL T VY+
Sbjct 118 TLTSVHEAMLEDVAFPAEIVGKRTRYRLDGSKIMKVFLDAKE--KNNTEYKLETMVGVYR 175
Query 184 RLTNKDAAFLFP 195
+LT KD F +P
Sbjct 176 KLTGKDVVFEYP 187
> ath:AT3G02560 40S ribosomal protein S7 (RPS7B)
Length=191
Score = 137 bits (345), Expect = 3e-32, Method: Compositional matrix adjust.
Identities = 83/192 (43%), Positives = 127/192 (66%), Gaps = 9/192 (4%)
Query 6 TTRQRIVKVDGSQPTEVEKEVDRCLADIESSAQSELRAEVAELTICAVKPVEVPASG-KT 64
+ + +I K G PTE E++V + L D+E++ Q EL++E+ +L I + V++ SG +
Sbjct 3 SGQNKIHKDKGVAPTEFEEQVTQALFDLENTNQ-ELKSELKDLYIN--QAVQMDISGNRK 59
Query 65 ALVIFVPYRVYMNVVKRIHARLVQELEKKLK-RHVVIIAQRSIVGLDYKRKNLKVRPRSR 123
A+VI+VP+R+ ++IH RLV+ELEKK + V+ +A R I+ K ++ RPR+R
Sbjct 60 AVVIYVPFRL-RKAFRKIHLRLVRELEKKFSGKDVIFVATRRIMRPPKKGSAVQ-RPRNR 117
Query 124 TLTNVQDAMLEDIVAPSEVVGRRWRVRADGSKLMKVHLDPKDKAKDNLEEKLWTFAAVYK 183
TLT+V +AMLED+ P+E+VG+R R R DG+K+MKV LD K K++ E KL T VY+
Sbjct 118 TLTSVHEAMLEDVAYPAEIVGKRTRYRLDGTKIMKVFLD--SKLKNDTEYKLETMVGVYR 175
Query 184 RLTNKDAAFLFP 195
+LT KD F +P
Sbjct 176 KLTGKDVVFEYP 187
> cel:ZC434.2 rps-7; Ribosomal Protein, Small subunit family member
(rps-7); K02993 small subunit ribosomal protein S7e
Length=194
Score = 135 bits (340), Expect = 1e-31, Method: Compositional matrix adjust.
Identities = 80/195 (41%), Positives = 126/195 (64%), Gaps = 12/195 (6%)
Query 10 RIVKVDGSQPTEVEKEVDRCLADIESSAQSELRAEVAELTICAVKPVEVPASGKTALVIF 69
+++K DG +E+EK+V + L D+E++ ++++++ EL I VK VE+ K+A++I+
Sbjct 7 KLLKSDGKVVSEIEKQVSQALIDLETN--DDVQSQLKELYIVGVKEVEL--GNKSAIIIY 62
Query 70 VPYRVYMNVVKRIHARLVQELEKKLK-RHVVIIAQRSIVGLDYK----RKNLKVRPRSRT 124
VP + +IH LV+ELEKK R ++I+A+R I+ + R + RPRSRT
Sbjct 63 VPVP-QLKAFHKIHPALVRELEKKFGGRDILILAKRRILPKPQRGSKARPQKQKRPRSRT 121
Query 125 LTNVQDAMLEDIVAPSEVVGRRWRVRADGSKLMKVHLDPKDKAKDNLEEKLWTFAAVYKR 184
LT V DA L+++V P+EVVGRR RV+ DG K+ KVHLD + N+ K+ FA+VY++
Sbjct 122 LTAVHDAWLDELVYPAEVVGRRIRVKLDGKKVYKVHLDKSHQT--NVGHKIGVFASVYRK 179
Query 185 LTNKDAAFLFPEHVY 199
LT KD F FP+ ++
Sbjct 180 LTGKDVTFEFPDPIF 194
> sce:YNL096C RPS7B; Rps7bp; K02993 small subunit ribosomal protein
S7e
Length=190
Score = 121 bits (304), Expect = 1e-27, Method: Compositional matrix adjust.
Identities = 74/187 (39%), Positives = 116/187 (62%), Gaps = 7/187 (3%)
Query 13 KVDGSQPTEVEKEVDRCLADIESSAQSELRAEVAELTICAVKPVEVPASGKTALVIFVPY 72
K+ P+E+E +V + D+ESS+ EL+A++ L I +++ ++V GK ALV+FVP
Sbjct 7 KILSQAPSELELQVAKTFIDLESSS-PELKADLRPLQIKSIREIDV-TGGKKALVLFVPV 64
Query 73 RVYMNVVKRIHARLVQELEKKLK-RHVVIIAQRSIVGLDYK-RKNLKVRPRSRTLTNVQD 130
++ ++ +L +ELEKK RHV+ +A+R I+ + + ++ RPRSRTLT V D
Sbjct 65 PA-LSAYHKVQTKLTRELEKKFPDRHVIFLAERRILPKPSRTSRQVQKRPRSRTLTAVHD 123
Query 131 AMLEDIVAPSEVVGRRWRVRADGSKLMKVHLDPKDKAKDNLEEKLWTFAAVYKRLTNKDA 190
+LED+V P+E+VG+R R G+K+ KV LD KD + ++ KL +F AVY +LT K
Sbjct 124 KVLEDMVFPTEIVGKRVRYLVGGNKIQKVLLDSKDVQQ--IDYKLESFQAVYNKLTGKQI 181
Query 191 AFLFPEH 197
F P
Sbjct 182 VFEIPSQ 188
> sce:YOR096W RPS7A, RPS30; Rps7ap; K02993 small subunit ribosomal
protein S7e
Length=190
Score = 120 bits (302), Expect = 2e-27, Method: Compositional matrix adjust.
Identities = 75/189 (39%), Positives = 114/189 (60%), Gaps = 7/189 (3%)
Query 13 KVDGSQPTEVEKEVDRCLADIESSAQSELRAEVAELTICAVKPVEVPASGKTALVIFVPY 72
K+ PTE+E +V + ++E+S+ EL+AE+ L +++ ++V A GK AL IFVP
Sbjct 7 KILSQAPTELELQVAQAFVELENSS-PELKAELRPLQFKSIREIDV-AGGKKALAIFVPV 64
Query 73 RVYMNVVKRIHARLVQELEKKLK-RHVVIIAQRSIVGLDYK-RKNLKVRPRSRTLTNVQD 130
K + +L +ELEKK + RHV+ +A+R I+ + + ++ RPRSRTLT V D
Sbjct 65 PSLAGFHK-VQTKLTRELEKKFQDRHVIFLAERRILPKPSRTSRQVQKRPRSRTLTAVHD 123
Query 131 AMLEDIVAPSEVVGRRWRVRADGSKLMKVHLDPKDKAKDNLEEKLWTFAAVYKRLTNKDA 190
+LED+V P+E+VG+R R G+K+ KV LD KD ++ KL +F AVY +LT K
Sbjct 124 KILEDLVFPTEIVGKRVRYLVGGNKIQKVLLDSKD--VQQIDYKLESFQAVYNKLTGKQI 181
Query 191 AFLFPEHVY 199
F P +
Sbjct 182 VFEIPSETH 190
> pfa:PF13_0013 PBS lyase HEAT-like protein, putative
Length=411
Score = 31.6 bits (70), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 14/39 (35%), Positives = 24/39 (61%), Gaps = 0/39 (0%)
Query 80 KRIHARLVQELEKKLKRHVVIIAQRSIVGLDYKRKNLKV 118
K I +++ L+K K V++A+ +VGLDY +NL +
Sbjct 368 KIIQEQIIDTLKKYSKDECVVVAESCLVGLDYISENLNI 406
> tgo:TGME49_054790 hypothetical protein
Length=1216
Score = 31.6 bits (70), Expect = 2.1, Method: Composition-based stats.
Identities = 23/67 (34%), Positives = 33/67 (49%), Gaps = 4/67 (5%)
Query 1 RVQKMTTRQRIVKVDGSQPTEVEKEVDRCLADIESSAQSELRAEVAELTICAVKPVE--- 57
RVQK T + K DG TEVE++V + D E + E E +C +P+
Sbjct 71 RVQKTCTDVSLKKADGECETEVEEKVCETVKDAEPDT-CYMVEEFDETYVCQSEPMPRQL 129
Query 58 VPASGKT 64
VP++ KT
Sbjct 130 VPSNCKT 136
> mmu:16777 Lamb1, C77966, C80098, C81607, D130003D08Rik, Lamb-1,
Lamb1-1; laminin B1; K05636 laminin, beta 1
Length=1834
Score = 30.4 bits (67), Expect = 4.3, Method: Composition-based stats.
Identities = 35/130 (26%), Positives = 61/130 (46%), Gaps = 13/130 (10%)
Query 14 VDGSQP--TEVEKEVDRCLADIESSAQ--SELRAEVAELTICAVKPVEVPASGKTALVIF 69
+ G+Q T +E E + +++Q S+L V EL A + SG+ +
Sbjct 1668 IQGTQNLLTSIESETAASEETLTNASQRISKLERNVEELKRKAAQ-----NSGEAEYIEK 1722
Query 70 VPYRVYMNVVKRIHARLVQELEKKLKRHVVIIAQRSIVGLDYKRKNLKVRPRSRTL---T 126
V Y V N + L EL++K K+ +IAQ++ D +RK ++ ++TL
Sbjct 1723 VVYSVKQNA-DDVKKTLDGELDEKYKKVESLIAQKTEESADARRKAELLQNEAKTLLAQA 1781
Query 127 NVQDAMLEDI 136
N + +LED+
Sbjct 1782 NSKLQLLEDL 1791
Lambda K H
0.320 0.133 0.373
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5931269072
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40