bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_0717_orf1
Length=118
Score E
Sequences producing significant alignments: (Bits) Value
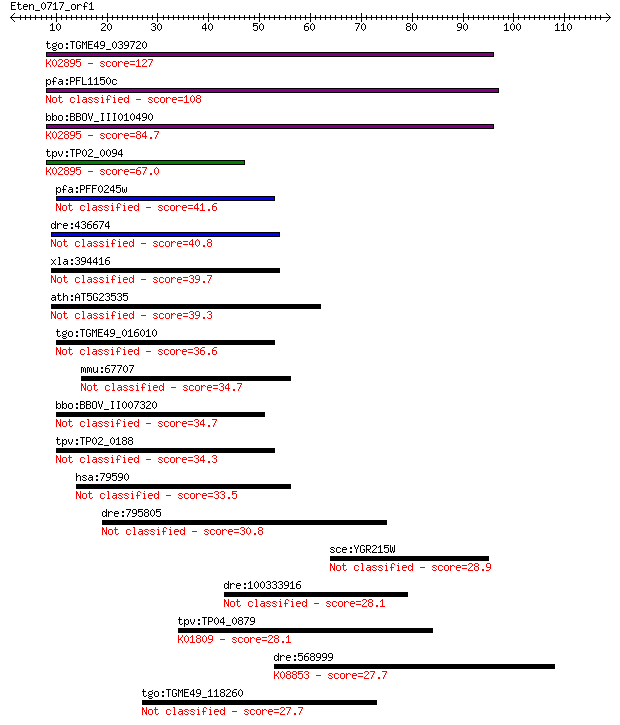
tgo:TGME49_039720 ribosomal protein L24, putative ; K02895 lar... 127 6e-30
pfa:PFL1150c mitochondrial ribosomal protein L24-2 precursor, ... 108 5e-24
bbo:BBOV_III010490 17.m07907; ribosomal protein L24; K02895 la... 84.7 6e-17
tpv:TP02_0094 60S ribosomal protein L24; K02895 large subunit ... 67.0 1e-11
pfa:PFF0245w mitochondrial ribosomal protein L24 precursor, pu... 41.6 7e-04
dre:436674 mrpl24, zgc:92702; mitochondrial ribosomal protein L24 40.8 0.001
xla:394416 mrpl24, L24, rpl24, rpl24-A; mitochondrial ribosoma... 39.7 0.002
ath:AT5G23535 KOW domain-containing protein 39.3 0.003
tgo:TGME49_016010 50S ribosomal protein L24, putative 36.6 0.018
mmu:67707 Mrpl24, 2010005E08Rik, 2810470K06Rik, 6720473G22Rik,... 34.7 0.073
bbo:BBOV_II007320 18.m06608; 50S ribosomal subunit L24 34.7 0.074
tpv:TP02_0188 60S ribosomal protein L24 34.3 0.11
hsa:79590 MRPL24, FLJ20917, L24mt, MGC22737, MGC9831, MRP-L18,... 33.5 0.17
dre:795805 NLR family, CARD domain containing 30.8 1.1
sce:YGR215W RSM27; Mitochondrial ribosomal protein of the smal... 28.9 4.3
dre:100333916 myomegalin-like 28.1 6.4
tpv:TP04_0879 mannose-6-phosphate isomerase (EC:5.3.1.8); K018... 28.1 7.4
dre:568999 si:dkey-71c4.3; K08853 AP2-associated kinase [EC:2.... 27.7 8.2
tgo:TGME49_118260 hypothetical protein 27.7 9.3
> tgo:TGME49_039720 ribosomal protein L24, putative ; K02895 large
subunit ribosomal protein L24
Length=229
Score = 127 bits (320), Expect = 6e-30, Method: Compositional matrix adjust.
Identities = 61/89 (68%), Positives = 77/89 (86%), Gaps = 1/89 (1%)
Query 8 LPIHLTNLALLDPIVKRPTRVKRRYMMNGEVVRIFKLSGCAMPEPL-TPLPRPPSLYQQF 66
+PIH+TN+ALLDP+VKRPTRVKRR+MMNGE VRI KLSG AMPEP+ T R P+LYQ++
Sbjct 112 MPIHITNVALLDPVVKRPTRVKRRFMMNGECVRISKLSGSAMPEPVSTSALRQPNLYQEY 171
Query 67 LREKAKGPPIKEKYANPDPLHMQLLKQLA 95
LR+KA GPP+K YA PDPLH+++L++LA
Sbjct 172 LRQKALGPPLKASYARPDPLHLKILQRLA 200
> pfa:PFL1150c mitochondrial ribosomal protein L24-2 precursor,
putative
Length=227
Score = 108 bits (269), Expect = 5e-24, Method: Compositional matrix adjust.
Identities = 50/90 (55%), Positives = 69/90 (76%), Gaps = 1/90 (1%)
Query 8 LPIHLTNLALLDPIVKRPTRVKRRYMMNGEVVRIFKLSGCAMPEPL-TPLPRPPSLYQQF 66
+PIH+TN++LLDPI K+PT VKRRYMMNGE VRI K+SGCAMPEP+ + + + Y++F
Sbjct 113 MPIHITNVSLLDPISKKPTVVKRRYMMNGECVRISKISGCAMPEPVHKNILKEQNNYERF 172
Query 67 LREKAKGPPIKEKYANPDPLHMQLLKQLAF 96
+ +K GPPIK+ YA D + LLK++A+
Sbjct 173 MHKKKIGPPIKDIYAEKDYKNFNLLKKIAY 202
> bbo:BBOV_III010490 17.m07907; ribosomal protein L24; K02895
large subunit ribosomal protein L24
Length=218
Score = 84.7 bits (208), Expect = 6e-17, Method: Compositional matrix adjust.
Identities = 44/91 (48%), Positives = 61/91 (67%), Gaps = 12/91 (13%)
Query 8 LPIHLTNLALLDPIVKRPTRVKRRYMMNGEVVRIFKLSGCAMPEPLT---PLPRPPSLYQ 64
+PIH++N+AL+DP+VK+PT VKRRYMMNGE VRI KLSGCA+P+P+ L +PP
Sbjct 115 MPIHISNVALVDPVVKKPTIVKRRYMMNGECVRICKLSGCALPKPVEVVKGLYKPPP--- 171
Query 65 QFLREKAKGPPIKEKYANPDPLHMQLLKQLA 95
+ PIK+ YA+ D + +L +A
Sbjct 172 ------KRTVPIKDDYAHKDYDNFNMLVNIA 196
> tpv:TP02_0094 60S ribosomal protein L24; K02895 large subunit
ribosomal protein L24
Length=154
Score = 67.0 bits (162), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 28/39 (71%), Positives = 36/39 (92%), Gaps = 0/39 (0%)
Query 8 LPIHLTNLALLDPIVKRPTRVKRRYMMNGEVVRIFKLSG 46
+PIH+TN++L+DP++K+PT VKRRYMMNGE VRI KLSG
Sbjct 114 MPIHVTNISLVDPVMKKPTIVKRRYMMNGECVRICKLSG 152
> pfa:PFF0245w mitochondrial ribosomal protein L24 precursor,
putative
Length=194
Score = 41.6 bits (96), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 18/43 (41%), Positives = 27/43 (62%), Gaps = 0/43 (0%)
Query 10 IHLTNLALLDPIVKRPTRVKRRYMMNGEVVRIFKLSGCAMPEP 52
IH +N+ L+D +K T+V RY + +V+RI K SG +P P
Sbjct 99 IHYSNVQLIDNFLKTNTKVALRYTDDNQVIRISKKSGTVIPWP 141
> dre:436674 mrpl24, zgc:92702; mitochondrial ribosomal protein
L24
Length=216
Score = 40.8 bits (94), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 17/45 (37%), Positives = 30/45 (66%), Gaps = 0/45 (0%)
Query 9 PIHLTNLALLDPIVKRPTRVKRRYMMNGEVVRIFKLSGCAMPEPL 53
P+ L ++AL+DP ++PT ++ RY GE VR+ +G +P+P+
Sbjct 114 PLLLKDIALIDPTDRKPTEIQWRYTEEGERVRVSVRTGRIIPKPV 158
> xla:394416 mrpl24, L24, rpl24, rpl24-A; mitochondrial ribosomal
protein L24
Length=276
Score = 39.7 bits (91), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 17/45 (37%), Positives = 29/45 (64%), Gaps = 0/45 (0%)
Query 9 PIHLTNLALLDPIVKRPTRVKRRYMMNGEVVRIFKLSGCAMPEPL 53
P+ L ++L+DP ++PT ++ RY GE VR+ SG +P+P+
Sbjct 114 PLLLNQVSLIDPTDRKPTEIEWRYTEEGERVRVSARSGRIIPKPV 158
> ath:AT5G23535 KOW domain-containing protein
Length=159
Score = 39.3 bits (90), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 19/60 (31%), Positives = 34/60 (56%), Gaps = 7/60 (11%)
Query 9 PIHLTNLALLDPIVKRPTRVKRRYMMNGEVVRIFK---LSGCAMPEP----LTPLPRPPS 61
P+H +N+ ++DP+ RP +V +Y+ +G VR+ + SG +P P + PRP +
Sbjct 73 PLHASNVQVVDPVTGRPCKVGVKYLEDGTKVRVARGTGTSGSIIPRPEILKIRATPRPTT 132
> tgo:TGME49_016010 50S ribosomal protein L24, putative
Length=438
Score = 36.6 bits (83), Expect = 0.018, Method: Composition-based stats.
Identities = 20/43 (46%), Positives = 26/43 (60%), Gaps = 0/43 (0%)
Query 10 IHLTNLALLDPIVKRPTRVKRRYMMNGEVVRIFKLSGCAMPEP 52
IH++N+ L+D + PTRV R G VVRI K SG +P P
Sbjct 319 IHVSNVMLIDQAIDLPTRVALRVDDRGNVVRISKKSGLVIPWP 361
> mmu:67707 Mrpl24, 2010005E08Rik, 2810470K06Rik, 6720473G22Rik,
AA407670, MGC25749, MGC38056, MGC7749; mitochondrial ribosomal
protein L24
Length=216
Score = 34.7 bits (78), Expect = 0.073, Method: Compositional matrix adjust.
Identities = 15/41 (36%), Positives = 26/41 (63%), Gaps = 0/41 (0%)
Query 15 LALLDPIVKRPTRVKRRYMMNGEVVRIFKLSGCAMPEPLTP 55
+ L+DP+ ++PT ++ R+ GE VR+ SG +P+P P
Sbjct 120 VKLVDPVDRKPTEIQWRFTEAGERVRVSTRSGRIIPKPEFP 160
> bbo:BBOV_II007320 18.m06608; 50S ribosomal subunit L24
Length=200
Score = 34.7 bits (78), Expect = 0.074, Method: Compositional matrix adjust.
Identities = 16/41 (39%), Positives = 26/41 (63%), Gaps = 0/41 (0%)
Query 10 IHLTNLALLDPIVKRPTRVKRRYMMNGEVVRIFKLSGCAMP 50
IH +N+ L+DP++ TRV R+ + E +R+ K SG +P
Sbjct 144 IHYSNVQLIDPLLNCATRVAIRFNKDNEPLRVSKKSGYVIP 184
> tpv:TP02_0188 60S ribosomal protein L24
Length=233
Score = 34.3 bits (77), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 16/43 (37%), Positives = 26/43 (60%), Gaps = 0/43 (0%)
Query 10 IHLTNLALLDPIVKRPTRVKRRYMMNGEVVRIFKLSGCAMPEP 52
IH +N+ L+D ++ TRV RY + + +R+ K SG +P P
Sbjct 150 IHYSNVQLVDQLLNVGTRVSIRYSHDNKPLRVSKKSGYVIPWP 192
> hsa:79590 MRPL24, FLJ20917, L24mt, MGC22737, MGC9831, MRP-L18,
MRP-L24; mitochondrial ribosomal protein L24
Length=216
Score = 33.5 bits (75), Expect = 0.17, Method: Compositional matrix adjust.
Identities = 15/42 (35%), Positives = 26/42 (61%), Gaps = 0/42 (0%)
Query 14 NLALLDPIVKRPTRVKRRYMMNGEVVRIFKLSGCAMPEPLTP 55
+ L+DP+ ++PT ++ R+ GE VR+ SG +P+P P
Sbjct 119 QVKLVDPMDRKPTEIEWRFTEAGERVRVSTRSGRIIPKPEFP 160
> dre:795805 NLR family, CARD domain containing
Length=1033
Score = 30.8 bits (68), Expect = 1.1, Method: Composition-based stats.
Identities = 16/56 (28%), Positives = 29/56 (51%), Gaps = 1/56 (1%)
Query 19 DPIVKRPTRVKRRYMMNGEVVRIFKLSGCAMPEPLTPLPRPPSLYQQFLREKAKGP 74
DP+ P R+KR+ + + E+ + SG +M +P+ P + Q RE+ + P
Sbjct 29 DPVTSDP-RIKRQRLESSEISDVSVKSGRSMEQPMRFSDAPVTSNPQMRRERLESP 83
> sce:YGR215W RSM27; Mitochondrial ribosomal protein of the small
subunit
Length=110
Score = 28.9 bits (63), Expect = 4.3, Method: Compositional matrix adjust.
Identities = 13/31 (41%), Positives = 17/31 (54%), Gaps = 0/31 (0%)
Query 64 QQFLREKAKGPPIKEKYANPDPLHMQLLKQL 94
+ L E+ KGP + Y NPD L + LK L
Sbjct 32 SKILNERLKGPSVASYYGNPDILKFRHLKTL 62
> dre:100333916 myomegalin-like
Length=636
Score = 28.1 bits (61), Expect = 6.4, Method: Composition-based stats.
Identities = 10/36 (27%), Positives = 19/36 (52%), Gaps = 0/36 (0%)
Query 43 KLSGCAMPEPLTPLPRPPSLYQQFLREKAKGPPIKE 78
+L C+ P P +P P +Q L + + PP+++
Sbjct 340 QLEQCSEPRPSPTIPASPLYRRQLLHDPSPSPPVRD 375
> tpv:TP04_0879 mannose-6-phosphate isomerase (EC:5.3.1.8); K01809
mannose-6-phosphate isomerase [EC:5.3.1.8]
Length=454
Score = 28.1 bits (61), Expect = 7.4, Method: Composition-based stats.
Identities = 17/50 (34%), Positives = 22/50 (44%), Gaps = 0/50 (0%)
Query 34 MNGEVVRIFKLSGCAMPEPLTPLPRPPSLYQQFLREKAKGPPIKEKYANP 83
++G+ V I K S + LT PR L L E K P K+ Y P
Sbjct 289 VSGDCVEIMKCSDNVIRCGLTSKPRDAKLCISLLEENLKNPSAKDFYVTP 338
> dre:568999 si:dkey-71c4.3; K08853 AP2-associated kinase [EC:2.7.11.1]
Length=802
Score = 27.7 bits (60), Expect = 8.2, Method: Composition-based stats.
Identities = 23/66 (34%), Positives = 28/66 (42%), Gaps = 11/66 (16%)
Query 53 LTPLPRP---PSLYQQFLREKAKGPPIKEKYANPD-----PLHMQLLKQ---LAFFFLNK 101
LTP RP P Q A PP+ ++ A P P H QL Q AFF +
Sbjct 385 LTPRKRPSAPPGPSQPISVNMASQPPVAQQQATPSPPQTTPQHAQLFVQQQNTAFFTAQQ 444
Query 102 QPAAAA 107
Q AA +
Sbjct 445 QSAAGS 450
> tgo:TGME49_118260 hypothetical protein
Length=959
Score = 27.7 bits (60), Expect = 9.3, Method: Compositional matrix adjust.
Identities = 17/50 (34%), Positives = 28/50 (56%), Gaps = 7/50 (14%)
Query 27 RVKRRYMMNGEVVRIFKLSGCAMPEPLTPLPRPPS----LYQQFLREKAK 72
R KR ++ GE +R +L A P L PLP PP LY++ ++++ +
Sbjct 319 RTKRVRLLGGENLRETEL---AEPNSLLPLPEPPKDSFLLYRRLIKQQTE 365
Lambda K H
0.322 0.138 0.414
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2027872200
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40