bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
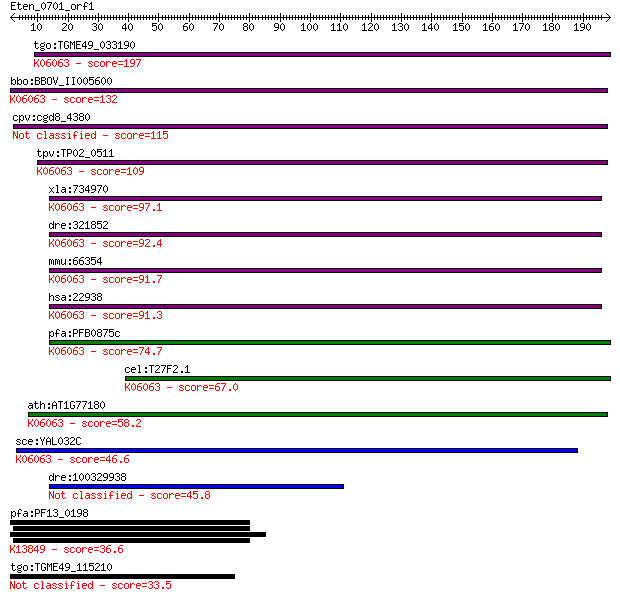
Query= Eten_0701_orf1
Length=198
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_033190 hypothetical protein ; K06063 SNW domain-con... 197 2e-50
bbo:BBOV_II005600 18.m06466; ski-interacting protein; K06063 S... 132 8e-31
cpv:cgd8_4380 SNW family nuclear protein 115 9e-26
tpv:TP02_0511 hypothetical protein; K06063 SNW domain-containi... 109 8e-24
xla:734970 snw1, MGC132028, bx42, ncoa-62, prp45, prpf45, skii... 97.1 3e-20
dre:321852 snw1, MGC123090, fb37b08, skiip, wu:fb37b08, zgc:12... 92.4 1e-18
mmu:66354 Snw1, 2310008B08Rik, AW048543, MGC55033, NCoA-62, SK... 91.7 2e-18
hsa:22938 SNW1, Bx42, MGC119379, NCOA-62, PRPF45, Prp45, SKIIP... 91.3 2e-18
pfa:PFB0875c chromatin-binding protein, putative; K06063 SNW d... 74.7 2e-13
cel:T27F2.1 skp-1; mammalian SKIP (Ski interacting protein) ho... 67.0 4e-11
ath:AT1G77180 chromatin protein family; K06063 SNW domain-cont... 58.2 2e-08
sce:YAL032C PRP45, FUN20; Protein required for pre-mRNA splici... 46.6 6e-05
dre:100329938 SKI-interacting protein-like 45.8 1e-04
pfa:PF13_0198 PfRh2a; reticulocyte binding protein 2 homolog A... 36.6 0.062
tgo:TGME49_115210 rhoptry protein, putative (EC:2.7.11.1) 33.5 0.50
> tgo:TGME49_033190 hypothetical protein ; K06063 SNW domain-containing
protein 1
Length=557
Score = 197 bits (501), Expect = 2e-50, Method: Compositional matrix adjust.
Identities = 113/191 (59%), Positives = 141/191 (73%), Gaps = 5/191 (2%)
Query 9 LREQQLRQLAAQARAERAQLLQQSRRAAAGDAEEEAERQQREQIAREREKDIERAVRLER 68
+REQQLR LAAQARAER+ LLQQ + G E+E R++RE +ARER+++IER +RLER
Sbjct 371 MREQQLRLLAAQARAERSNLLQQRKE---GQEEDEEARKKREAVARERQREIEREMRLER 427
Query 69 SRGKRRDDSQRDISELVALGLPVDAKKQRTDSMFDTRLFNQGGGTDSGYKGGEDDTDTLY 128
++ + R+D RD+SE VALGLP K + +FDTRLFNQ G DSG+ GG D+ LY
Sbjct 428 NK-RGRNDEDRDVSERVALGLPPTKKAAAGEGVFDTRLFNQSAGVDSGFDGGNDEAYNLY 486
Query 129 DRPLFAQR-GGAGIYQFSRDRFSTSVGEGSELAAFAGADKTRYMRTGPVEFEKDVSDPFG 187
D+PLFA R A IYQFSR+R SVG ++ +FAGAD++ + RT PVEFEKDVSDPFG
Sbjct 487 DQPLFANRSNNAAIYQFSRERLINSVGHTGDVPSFAGADRSTFTRTAPVEFEKDVSDPFG 546
Query 188 LDNLLSEAHKK 198
LDNLLSEA KK
Sbjct 547 LDNLLSEAKKK 557
> bbo:BBOV_II005600 18.m06466; ski-interacting protein; K06063
SNW domain-containing protein 1
Length=458
Score = 132 bits (332), Expect = 8e-31, Method: Compositional matrix adjust.
Identities = 86/199 (43%), Positives = 115/199 (57%), Gaps = 21/199 (10%)
Query 1 EQKKSAEELREQQLRQLAAQARAERAQLLQQSRRAAAGDAEEEAERQQREQIAREREKDI 60
E+ K A+E +E+QLR LAA+AR ER+++ S AA ER+++I
Sbjct 278 EKLKEAQE-KEEQLRALAARAREERSRMGLDSELQAA---------------ELERKREI 321
Query 61 ERAVRLERSRGKRRDDSQRD--ISELVALGLPVDAKKQRTDSMFDTRLFNQGGGTDSGYK 118
ER +RLER+ K + + RD I+E VALG PV +K DTRL N G DSG+
Sbjct 322 EREMRLERAGKKIKAAAARDRDITERVALGQPVPSK---LSDAHDTRLLNTAAGIDSGFD 378
Query 119 GGEDDTDTLYDRPLFAQRGGAGIYQFSRDRFSTSVGEGSELAAFAGADKTRYMRTGPVEF 178
GGED+ +YD+PLFA R GIYQ S +RF S+G + + A+ T RT PVEF
Sbjct 379 GGEDENYNIYDKPLFADRSIVGIYQHSSERFQQSLGVNEAMRVPSFANATMVQRTTPVEF 438
Query 179 EKDVSDPFGLDNLLSEAHK 197
K+ +DPFGL LL +A K
Sbjct 439 VKETADPFGLGTLLDKAKK 457
> cpv:cgd8_4380 SNW family nuclear protein
Length=431
Score = 115 bits (288), Expect = 9e-26, Method: Compositional matrix adjust.
Identities = 82/209 (39%), Positives = 116/209 (55%), Gaps = 16/209 (7%)
Query 2 QKKSAEEL-REQQLRQLAAQARAERA--QLLQQ----SRRAAAGDAEEEAERQQREQIAR 54
QKK EE+ RE++LR+LA +R ER+ LL + + G E+ E +I
Sbjct 222 QKKIREEMEREEKLRKLAESSRYERSSHHLLSKIPEFYQDKKEGSDEDNDEFDHIRRIEL 281
Query 55 EREKDIERAVRLERS--RGKRRDDSQRDISELVALGLPVDAKK-QRTDSMFDTRLFNQGG 111
E+ ++IER R ER+ + K DS RDISE +ALG A ++S FD RLFN+
Sbjct 282 EKRREIEREFRQERAGKKSKTLRDSDRDISEHIALGQAQSANIGSSSESQFDARLFNKVS 341
Query 112 GTDSGYKGGEDDTDTLYDRPLFAQRG--GAGIYQFSRDRFSTSVGEGSELAAFAGADKTR 169
G DSG+ +DT ++YD+PLF G+Y F R S+G + +F+G D ++
Sbjct 342 GLDSGF---SNDTISVYDKPLFNTNNLKHRGLYTFEESRVEESIGGRVHVPSFSGTDNSK 398
Query 170 Y-MRTGPVEFEKDVSDPFGLDNLLSEAHK 197
+RT PVEFE+D DPFGLD L+ K
Sbjct 399 TALRTKPVEFERDEDDPFGLDKLIDSVRK 427
> tpv:TP02_0511 hypothetical protein; K06063 SNW domain-containing
protein 1
Length=455
Score = 109 bits (272), Expect = 8e-24, Method: Compositional matrix adjust.
Identities = 77/197 (39%), Positives = 106/197 (53%), Gaps = 15/197 (7%)
Query 10 REQQLRQLAAQARAERAQLLQQSRRAAAGDAEEEAERQQREQIAREREKDIERAVRLERS 69
RE++LR+LAA+AR ER QL +S + R + ERE+ + R
Sbjct 264 REEELRELAAKAREERHQLYSKSHSKDDSKDRDRRRRHKDRDSDEEREELLRIEAERRRE 323
Query 70 RG---------KRRDDSQRDISELVALGLPVDAKKQRTDSMFDTRLFNQGGGTDSGYKGG 120
K+R +RDISE +ALG AK + ++DTRL G SG+ GG
Sbjct 324 LEREWRIEKNIKKRRTGERDISEKIALG---QAKPTKITDLYDTRLLQGDAGISSGFDGG 380
Query 121 EDDTDTLYDRPLFAQRGGAGIYQFSRDRFSTSVGEGSELAAFAGADKTRYMRTGPVEFEK 180
+D+ +YD+PLFA R A IYQ S++RF S G+ + +A+FA AD+ R PVEF K
Sbjct 381 DDEGYNIYDKPLFADRSTANIYQHSKERFQKSTGDVN-VASFANADRG-VQRNTPVEFVK 438
Query 181 DVSDPFGLDNLLSEAHK 197
D SDPFG + LL + K
Sbjct 439 D-SDPFGFEKLLEKVKK 454
> xla:734970 snw1, MGC132028, bx42, ncoa-62, prp45, prpf45, skiip;
SNW domain containing 1; K06063 SNW domain-containing protein
1
Length=535
Score = 97.1 bits (240), Expect = 3e-20, Method: Compositional matrix adjust.
Identities = 73/195 (37%), Positives = 108/195 (55%), Gaps = 24/195 (12%)
Query 14 LRQLAAQARAERAQLLQQSRRAAAGDAEEEAERQQREQIAREREKDIERAVRLERSRGKR 73
LR+LA AR RA + + + G+A+E + + + R+ E++I RA +RS+ +R
Sbjct 324 LRELAQIARERRAGIKSHTEKED-GEAKERDDIRHERRKERQHERNISRAAPDKRSKLQR 382
Query 74 RDDSQRDISELVALGLPVDAKKQRTDS--MFDTRLFNQGGGTDSGYKGGEDDTDTLYDRP 131
+ +RDISE +ALG+P QRT S +D RLFNQ G DSG+ GGED+ +YD+P
Sbjct 383 --NEERDISEQIALGIP----NQRTSSEIQYDQRLFNQSRGMDSGFAGGEDEVYNVYDQP 436
Query 132 LFAQRGGA-GIYQFSRDRFSTSVGEGSELAA----------FAGADKTRYMRTGPVEFEK 180
+ A IY+ S++ + G+ + F+GAD+ R GPV+FE+
Sbjct 437 WLGNKKLAQNIYRPSKNTDNDVYGDDLDTLVKTNRFVPDKDFSGADR-RQRHEGPVQFEE 495
Query 181 DVSDPFGLDNLLSEA 195
DPFGLD L EA
Sbjct 496 ---DPFGLDKFLEEA 507
> dre:321852 snw1, MGC123090, fb37b08, skiip, wu:fb37b08, zgc:123090;
SNW domain containing 1; K06063 SNW domain-containing
protein 1
Length=536
Score = 92.4 bits (228), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 75/202 (37%), Positives = 106/202 (52%), Gaps = 36/202 (17%)
Query 14 LRQLAAQARAERAQLLQQSRRAAAGDAEEEAERQQREQIAREREKD------IERAVRLE 67
LR+LA AR RA + + E++E ++R++I +R KD I RA +
Sbjct 324 LRELAKMARDRRAGIKPHGDKGG-----EDSEVRERDEIRHDRRKDRQHDRNISRAAPDK 378
Query 68 RSRGKRRDDSQRDISELVALGLPVDAKKQRTDSMFDTRLFNQGGGTDSGYKGGEDDTDTL 127
RS+ +R D RDISEL+ALG P + +++ +D RLFNQ G DSG+ GGED+ +
Sbjct 379 RSKLQR--DQDRDISELIALGQP--NPRTSSEAQYDQRLFNQSKGMDSGFAGGEDEMYNV 434
Query 128 YDRPLFAQRGG----AGIYQFSRDRFSTSVGEGSELAA----------FAGADKTRYMRT 173
YD+P RGG IY+ S++ G+ + F+GAD R
Sbjct 435 YDQPF---RGGRDMAQNIYRPSKNVDKDMYGDDLDTLMQNNRFVPDRDFSGADHGP-RRD 490
Query 174 GPVEFEKDVSDPFGLDNLLSEA 195
GPV+FE+ DPFGLD L EA
Sbjct 491 GPVQFEE---DPFGLDKFLEEA 509
> mmu:66354 Snw1, 2310008B08Rik, AW048543, MGC55033, NCoA-62,
SKIP, Skiip; SNW domain containing 1; K06063 SNW domain-containing
protein 1
Length=536
Score = 91.7 bits (226), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 69/196 (35%), Positives = 107/196 (54%), Gaps = 25/196 (12%)
Query 14 LRQLAAQARAERAQLLQQSRRAAAGDAEEEAERQQREQIAREREKDIERAVRLERSRGKR 73
LR++A +AR RA + + G+A E E + + R+ ++++ RA +RS+ +R
Sbjct 324 LREMAQKARERRAGIKTHVEKED-GEARERDEIRHDRRKERQHDRNLSRAAPDKRSKLQR 382
Query 74 RDDSQRDISELVALGLPVDAKKQRTDSMFDTRLFNQGGGTDSGYKGGEDDTDTLYDRPLF 133
++ RDISE++ALG+P + + +D RLFNQ G DSG+ GGED+ +YD+
Sbjct 383 NEN--RDISEVIALGVP--NPRTSNEVQYDQRLFNQSKGMDSGFAGGEDEIYNVYDQ--- 435
Query 134 AQRGGA----GIYQFSRDRFSTSVGEGSEL----------AAFAGADKTRYMRTGPVEFE 179
A RGG IY+ S++ G+ E F+G+D+ + R GPV+FE
Sbjct 436 AWRGGKDMAQSIYRPSKNLDKDMYGDDLEARIKTNRFVPDKEFSGSDRKQRGREGPVQFE 495
Query 180 KDVSDPFGLDNLLSEA 195
+ DPFGLD L EA
Sbjct 496 E---DPFGLDKFLEEA 508
> hsa:22938 SNW1, Bx42, MGC119379, NCOA-62, PRPF45, Prp45, SKIIP,
SKIP; SNW domain containing 1; K06063 SNW domain-containing
protein 1
Length=536
Score = 91.3 bits (225), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 69/196 (35%), Positives = 107/196 (54%), Gaps = 25/196 (12%)
Query 14 LRQLAAQARAERAQLLQQSRRAAAGDAEEEAERQQREQIAREREKDIERAVRLERSRGKR 73
LR++A +AR RA + + G+A E E + + R+ ++++ RA +RS+ +R
Sbjct 324 LREMAQKARERRAGIKTHVEKED-GEARERDEIRHDRRKERQHDRNLSRAAPDKRSKLQR 382
Query 74 RDDSQRDISELVALGLPVDAKKQRTDSMFDTRLFNQGGGTDSGYKGGEDDTDTLYDRPLF 133
++ RDISE++ALG+P + + +D RLFNQ G DSG+ GGED+ +YD+
Sbjct 383 NEN--RDISEVIALGVP--NPRTSNEVQYDQRLFNQSKGMDSGFAGGEDEIYNVYDQ--- 435
Query 134 AQRGGA----GIYQFSRDRFSTSVGEGSEL----------AAFAGADKTRYMRTGPVEFE 179
A RGG IY+ S++ G+ E F+G+D+ + R GPV+FE
Sbjct 436 AWRGGKDMAQSIYRPSKNLDKDMYGDDLEARIKTNRFVPDKEFSGSDRRQRGREGPVQFE 495
Query 180 KDVSDPFGLDNLLSEA 195
+ DPFGLD L EA
Sbjct 496 E---DPFGLDKFLEEA 508
> pfa:PFB0875c chromatin-binding protein, putative; K06063 SNW
domain-containing protein 1
Length=482
Score = 74.7 bits (182), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 57/188 (30%), Positives = 90/188 (47%), Gaps = 41/188 (21%)
Query 14 LRQLAAQARAERAQLLQQSRRAAAGDAEEEAERQQREQIAREREKDIERAVRLERSRGKR 73
L+ LA QAR E+ L S + +R+++IER ++E++ K
Sbjct 333 LKNLAIQARKEKG--LAHS------------------SLINDRKREIEREYKIEKNLKKM 372
Query 74 RDDSQRDISELVALGLPVDAKKQRTDSMFDTRLFN---QGGGTDSGYKGGEDDTDTLYDR 130
++ R + E +AL + +++ D LFN Q T + +DDT +YD
Sbjct 373 KNYENRYVEEQIALN---KVNVSKNNNIHDITLFNINEQNNVTTTQ----DDDTYQIYDT 425
Query 131 PLFAQRGGAGIYQFSRDRFSTSVGEGSELAAFAGADKTRYMRTGPVEFEKDVSDPFGLDN 190
LF + A IY+FS +R +V + D + PV++ KD+SDPFGLD+
Sbjct 426 ALFNNKNNANIYKFSSERLRKNVQK------IETRDTMQ-----PVKYIKDISDPFGLDS 474
Query 191 LLSEAHKK 198
LLS+A KK
Sbjct 475 LLSQAKKK 482
> cel:T27F2.1 skp-1; mammalian SKIP (Ski interacting protein)
homolog family member (skp-1); K06063 SNW domain-containing
protein 1
Length=535
Score = 67.0 bits (162), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 62/186 (33%), Positives = 89/186 (47%), Gaps = 40/186 (21%)
Query 39 DAEEEAERQQREQIAREREKDIERAVRLERSRG----KRRDDSQRDISELVALGLPVDAK 94
D E++ + + RE+I R+R DI + + RSR K R + +RDISE + LGLP +
Sbjct 345 DDEDDEQVKVREEIRRDRLDDIRKERNIARSRPDKADKLRKERERDISEKIVLGLPDTNQ 404
Query 95 KQRTDSMFDTRLFNQGGGTDSGYKGGEDDTDTLYDRPLFAQRGGAGIYQF---------- 144
K+ + FD RLF++ G DSG +DDT YD A RGG + Q
Sbjct 405 KRTGEPQFDQRLFDKTQGLDSG--AMDDDTYNPYD---AAWRGGDSVQQHVYRPSKNLDN 459
Query 145 ------------SRDRFSTSVGEGSELAAFAGADKTRYMRTGPVEFEKDVSDPFGLDNLL 192
++RF G F+GA+ + +GPV+FEKD D FGL +L
Sbjct 460 DVYGGDLDKIIEQKNRFVADKG-------FSGAEGSSRG-SGPVQFEKD-QDVFGLSSLF 510
Query 193 SEAHKK 198
+K
Sbjct 511 EHTKEK 516
> ath:AT1G77180 chromatin protein family; K06063 SNW domain-containing
protein 1
Length=613
Score = 58.2 bits (139), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 76/245 (31%), Positives = 105/245 (42%), Gaps = 58/245 (23%)
Query 7 EELREQQLRQLAAQARAERA----QLLQQSRRAAAGDAEEEAERQQREQ----------- 51
+E +EQ+LR LA +AR+ER + S R + + + +Q
Sbjct 328 KERKEQELRALAQKARSERTGAAMSMPVSSDRGRSESVDPRGDYDNYDQDRGREREREEP 387
Query 52 --IAREREKDIER---------------------AVRLERSRGKRRDDSQRDISELVALG 88
EREK I+R A ++S+ R D RDISE VALG
Sbjct 388 QETREEREKRIQREKIREERRRERERERRLDAKDAAMGKKSKITR--DRDRDISEKVALG 445
Query 89 LPVDAKKQRTDSMFDTRLFNQGGGTDSGYKGGEDDTDTLYDRPLF-AQRGGAGIYQFSRD 147
+ K + M+D RLFNQ G DSG+ DD LYD+ LF AQ + +Y+ +D
Sbjct 446 MASTGGKGGGEVMYDQRLFNQDKGMDSGF--AADDQYNLYDKGLFTAQPTLSTLYKPKKD 503
Query 148 RFSTSVGEGSEL-------------AAFAGA-DKTRYMRTGPVEFEK-DVSDPFGLDNLL 192
G E AF GA ++ R PVEFEK + DPFGL+ +
Sbjct 504 NDEEMYGNADEQLDKIKNTERFKPDKAFTGASERVGSKRDRPVEFEKEEEQDPFGLEKWV 563
Query 193 SEAHK 197
S+ K
Sbjct 564 SDLKK 568
> sce:YAL032C PRP45, FUN20; Protein required for pre-mRNA splicing;
associates with the spliceosome and interacts with splicing
factors Prp22p and Prp46p; orthologous to human transcriptional
coactivator SKIP and can activate transcription of
a reporter gene; K06063 SNW domain-containing protein 1
Length=379
Score = 46.6 bits (109), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 53/197 (26%), Positives = 90/197 (45%), Gaps = 43/197 (21%)
Query 3 KKSAEELREQ-QLRQLAAQ----ARAERAQLLQQSRRAAAGDAEEEA---ERQQREQIAR 54
KK+ +E+R + +L++LA + A+ + + L Q R G + A ++Q +AR
Sbjct 213 KKARQEIRSKMELKRLAMEQEMLAKESKLKELSQRARYHNGTPQTGAIVKPKKQTSTVAR 272
Query 55 EREKDIERAVRLERSRGKRRDDSQRDISELVALGLPVDAKKQRTDSMFDTRLFNQGGGTD 114
+E L S+G RD+SE + LG + ++ D +D+R F +G
Sbjct 273 LKE--------LAYSQG-------RDVSEKIILG--AAKRSEQPDLQYDSRFFTRGANA- 314
Query 115 SGYKGGEDDTDTLYDRPLFAQRGGAGIYQFSRDRFSTSVGEGSELAAFAGADKTRYMRTG 174
+ D +YD PLF Q+ IY+ + ++ +V SE GA + G
Sbjct 315 ----SAKRHEDQVYDNPLFVQQDIESIYKTNYEKLDEAVNVKSE-----GASGSH----G 361
Query 175 PVEFEK----DVSDPFG 187
P++F K D SD +G
Sbjct 362 PIQFTKAESDDKSDNYG 378
> dre:100329938 SKI-interacting protein-like
Length=464
Score = 45.8 bits (107), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 38/103 (36%), Positives = 57/103 (55%), Gaps = 15/103 (14%)
Query 14 LRQLAAQARAERAQLLQQSRRAAAGDAEEEAERQQREQIAREREKD------IERAVRLE 67
LR+LA AR RA + + E++E ++R++I +R KD I RA +
Sbjct 324 LRELAKMARDRRAGI-----KPHGDKGGEDSEVRERDEIRHDRRKDRQHDRNISRAAPDK 378
Query 68 RSRGKRRDDSQRDISELVALGLPVDAKKQRTDSMFDTRLFNQG 110
RS+ +R D RDISEL+ALG P + +++ +D RLFNQ
Sbjct 379 RSKLQR--DQDRDISELIALGQP--NPRTSSEAQYDQRLFNQS 417
> pfa:PF13_0198 PfRh2a; reticulocyte binding protein 2 homolog
A; K13849 reticulocyte-binding protein
Length=3130
Score = 36.6 bits (83), Expect = 0.062, Method: Composition-based stats.
Identities = 23/82 (28%), Positives = 42/82 (51%), Gaps = 5/82 (6%)
Query 1 EQKKSAEELREQQ---LRQLAAQARAERAQLLQQSRRAAAGDAEEEAERQQREQIARERE 57
EQ + EEL+ Q+ L++ A R E+ +L Q +E ER+++EQ+ +E E
Sbjct 2767 EQLQKEEELKRQEQERLQKEEALKRQEQERL--QKEEELKRQEQERLEREKQEQLQKEEE 2824
Query 58 KDIERAVRLERSRGKRRDDSQR 79
+ RL++ +R + +R
Sbjct 2825 LKRQEQERLQKEEALKRQEQER 2846
Score = 32.7 bits (73), Expect = 0.79, Method: Composition-based stats.
Identities = 18/78 (23%), Positives = 42/78 (53%), Gaps = 1/78 (1%)
Query 2 QKKSAEELREQQLRQLAAQARAERAQLLQQSRRAAAGDAEEEAERQQREQIAREREKDIE 61
QK+ A + +EQ+ Q + + + + L++ ++ EEE +RQ++E++ +E +
Sbjct 2784 QKEEALKRQEQERLQKEEELKRQEQERLEREKQEQL-QKEEELKRQEQERLQKEEALKRQ 2842
Query 62 RAVRLERSRGKRRDDSQR 79
RL++ +R + +R
Sbjct 2843 EQERLQKEEELKRQEQER 2860
Score = 31.6 bits (70), Expect = 1.7, Method: Composition-based stats.
Identities = 22/85 (25%), Positives = 45/85 (52%), Gaps = 1/85 (1%)
Query 1 EQKKSAEELREQQLRQLAAQARAERAQLLQQSRRAAAGDAEEEA-ERQQREQIAREREKD 59
E+ + EEL+ Q+ +L + + + + + R+ +EEA +RQ++E++ +E E
Sbjct 2795 ERLQKEEELKRQEQERLEREKQEQLQKEEELKRQEQERLQKEEALKRQEQERLQKEEELK 2854
Query 60 IERAVRLERSRGKRRDDSQRDISEL 84
+ RLER + + + Q S+L
Sbjct 2855 RQEQERLERKKIELAEREQHIKSKL 2879
Score = 31.6 bits (70), Expect = 2.0, Method: Composition-based stats.
Identities = 18/78 (23%), Positives = 40/78 (51%), Gaps = 7/78 (8%)
Query 2 QKKSAEELREQQLRQLAAQARAERAQLLQQSRRAAAGDAEEEAERQQREQIAREREKDIE 61
+++ E L++++ + Q R ER + Q + EEE +RQ++E++ +E +
Sbjct 2740 KRQEQERLQKEEELKRQEQERLEREKQEQLQK-------EEELKRQEQERLQKEEALKRQ 2792
Query 62 RAVRLERSRGKRRDDSQR 79
RL++ +R + +R
Sbjct 2793 EQERLQKEEELKRQEQER 2810
> tgo:TGME49_115210 rhoptry protein, putative (EC:2.7.11.1)
Length=958
Score = 33.5 bits (75), Expect = 0.50, Method: Composition-based stats.
Identities = 23/79 (29%), Positives = 44/79 (55%), Gaps = 5/79 (6%)
Query 1 EQKKSAEELREQQLR--QLAAQARAERAQLLQQSRRAAAGD--AEEEAERQQREQIARE- 55
+++K+ EEL ++L+ Q A+ RAE +++ + R + +EE E ++R Q +E
Sbjct 739 QEQKAQEELLRKELKEEQRKARERAEAEKMVAEEERKKLLEQRTQEELETKRRIQEEKEL 798
Query 56 REKDIERAVRLERSRGKRR 74
REK E + E + K++
Sbjct 799 REKQAEEMRKQEEEQRKKQ 817
Lambda K H
0.312 0.130 0.349
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5866798756
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40