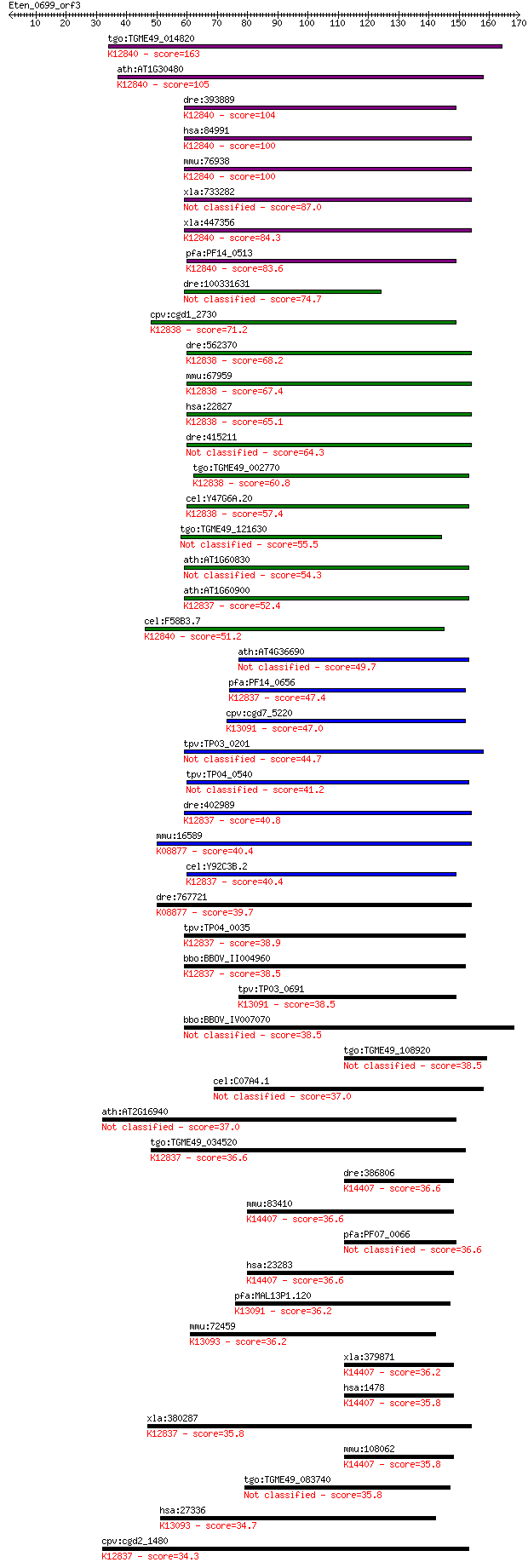
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_0699_orf3
Length=169
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_014820 DNA repair enzyme, putative ; K12840 splicin... 163 3e-40
ath:AT1G30480 DRT111; DRT111; nucleic acid binding / nucleotid... 105 7e-23
dre:393889 rbm17, MGC55840, zgc:55840; RNA binding motif prote... 104 2e-22
hsa:84991 RBM17, DKFZp686F13131, MGC14439, SPF45; RNA binding ... 100 2e-21
mmu:76938 Rbm17, 2700027J02Rik; RNA binding motif protein 17; ... 100 2e-21
xla:733282 rbm17; RNA binding motif protein 17 87.0
xla:447356 MGC130690; MGC84102 protein; K12840 splicing factor 45 84.3
pfa:PF14_0513 RNA binding protein, putative; K12840 splicing f... 83.6 3e-16
dre:100331631 RNA binding motif protein 17-like 74.7 1e-13
cpv:cgd1_2730 Ro ribonucleoprotein-binding protein 1, RNA bind... 71.2 2e-12
dre:562370 puf60a, fc21a05, wu:fc21a05, wu:fi43a08; poly-U bin... 68.2 1e-11
mmu:67959 Puf60, 2410104I19Rik, 2810454F19Rik, SIAHBP1; poly-U... 67.4 2e-11
hsa:22827 PUF60, FIR, FLJ31379, RoBPI, SIAHBP1; poly-U binding... 65.1 1e-10
dre:415211 puf60b, SIAHBP1, fe37c05, repressor, si:ch211-12p8.... 64.3 2e-10
tgo:TGME49_002770 RNA-binding protein, putative ; K12838 poly(... 60.8 2e-09
cel:Y47G6A.20 rnp-6; RNP (RRM RNA binding domain) containing f... 57.4 2e-08
tgo:TGME49_121630 hypothetical protein 55.5 9e-08
ath:AT1G60830 U2 snRNP auxiliary factor large subunit, putative 54.3 2e-07
ath:AT1G60900 U2 snRNP auxiliary factor large subunit, putativ... 52.4 7e-07
cel:F58B3.7 hypothetical protein; K12840 splicing factor 45 51.2
ath:AT4G36690 ATU2AF65A; RNA binding / nucleic acid binding / ... 49.7 4e-06
pfa:PF14_0656 U2 snRNP auxiliary factor, putative; K12837 spli... 47.4 2e-05
cpv:cgd7_5220 splicing factor with 3 RRM domains ; K13091 RNA-... 47.0 3e-05
tpv:TP03_0201 hypothetical protein 44.7 2e-04
tpv:TP04_0540 hypothetical protein 41.2 0.002
dre:402989 u2af2b, MGC77804, wu:fb73g02, zgc:77804; U2 small n... 40.8 0.002
mmu:16589 Uhmk1, 4732477C12Rik, 4930500M09Rik, AA673513, AI449... 40.4 0.003
cel:Y92C3B.2 uaf-1; U2AF splicing factor family member (uaf-1)... 40.4 0.003
dre:767721 uhmk1, MGC153241, zgc:153241; U2AF homology motif (... 39.7 0.005
tpv:TP04_0035 U2 small nuclear ribonucleoprotein, auxiliary fa... 38.9 0.009
bbo:BBOV_II004960 18.m06413; RNA recognition motif (RRM)-conta... 38.5 0.010
tpv:TP03_0691 splicing factor; K13091 RNA-binding protein 39 38.5
bbo:BBOV_IV007070 23.m05938; hypothetical protein 38.5 0.011
tgo:TGME49_108920 U2 small nuclear ribonucleoprotein auxiliary... 38.5 0.011
cel:C07A4.1 hypothetical protein 37.0 0.030
ath:AT2G16940 RNA recognition motif (RRM)-containing protein 37.0 0.034
tgo:TGME49_034520 U2 snRNP auxiliary factor or splicing factor... 36.6 0.039
dre:386806 cstf2, CstF-64, fb11e07, wu:fb11e07, zgc:56346, zgc... 36.6 0.040
mmu:83410 Cstf2t, 64kDa, C77975, tCstF-64, tauCstF-64; cleavag... 36.6 0.042
pfa:PF07_0066 conserved Plasmodium protein, unknown function 36.6 0.043
hsa:23283 CSTF2T, CstF-64T, DKFZp434C1013, KIAA0689; cleavage ... 36.6 0.044
pfa:MAL13P1.120 splicing factor, putative; K13091 RNA-binding ... 36.2 0.050
mmu:72459 Htatsf1, 1600023H17Rik, 2600017A12Rik, 2700077B20Rik... 36.2 0.051
xla:379871 cstf2, MGC53575, MGC83019, cstf-64; cleavage stimul... 36.2 0.056
hsa:1478 CSTF2, CstF-64; cleavage stimulation factor, 3' pre-R... 35.8 0.063
xla:380287 u2af2, MGC53441, u2af65; U2 small nuclear RNA auxil... 35.8 0.067
mmu:108062 Cstf2, 64kDa, C630034J23Rik, Cstf64; cleavage stimu... 35.8 0.071
tgo:TGME49_083740 mRNA processing protein, putative (EC:3.4.21... 35.8 0.076
hsa:27336 HTATSF1, TAT-SF1, dJ196E23.2; HIV-1 Tat specific fac... 34.7 0.16
cpv:cgd2_1480 splicing factor U2AF U2 SnRNP auxiliary factor l... 34.3 0.21
> tgo:TGME49_014820 DNA repair enzyme, putative ; K12840 splicing
factor 45
Length=466
Score = 163 bits (413), Expect = 3e-40, Method: Compositional matrix adjust.
Identities = 89/130 (68%), Positives = 100/130 (76%), Gaps = 1/130 (0%)
Query 34 CGVARQGQDVTTLGQVPRAVTFNMAPTRILLLLNMVGRGQVDSDLKEETEEEAAKLGNLL 93
GV QG +V RA FN PTR+LLLLNMVGRG+VD DLKEETEEEAAK GNLL
Sbjct 335 SGVIVQGAEVVQAAAQVRA-QFNRPPTRVLLLLNMVGRGEVDEDLKEETEEEAAKFGNLL 393
Query 94 QVRIFEAAGVPDDCAVRIFCEYERKEEATRALLTFNGRAFGGRTVKAKFYSEERFARGDL 153
QV+I EAA PDD AVRIFCEYERKEEATRA +T NGR FGGRTVK +FY EER+ + DL
Sbjct 394 QVKIVEAAEAPDDEAVRIFCEYERKEEATRAFMTMNGRVFGGRTVKGRFYCEERWGKDDL 453
Query 154 RPDPEQEIDQ 163
P+PE+E D+
Sbjct 454 MPNPEEEADK 463
> ath:AT1G30480 DRT111; DRT111; nucleic acid binding / nucleotide
binding; K12840 splicing factor 45
Length=387
Score = 105 bits (262), Expect = 7e-23, Method: Compositional matrix adjust.
Identities = 56/123 (45%), Positives = 78/123 (63%), Gaps = 2/123 (1%)
Query 37 ARQGQDVTTLGQVPRAVTFNMAPTRILLLLNMVGRGQVDSDLKEETEEEAAKLGNLLQVR 96
A + + + +V ++V N PTR+LLL NMVG GQVD +L++E E K G + +V
Sbjct 259 ASENKSSSAEKKVVKSVNINGEPTRVLLLRNMVGPGQVDDELEDEVGGECGKYGTVTRVL 318
Query 97 IFE--AAGVPDDCAVRIFCEYERKEEATRALLTFNGRAFGGRTVKAKFYSEERFARGDLR 154
IFE P AVRIF ++ R EE T+AL+ +GR FGGRTV+A FY EE+F++ +L
Sbjct 319 IFEITEPNFPVHEAVRIFVQFSRPEETTKALVDLDGRYFGGRTVRATFYDEEKFSKNELA 378
Query 155 PDP 157
P P
Sbjct 379 PVP 381
> dre:393889 rbm17, MGC55840, zgc:55840; RNA binding motif protein
17; K12840 splicing factor 45
Length=418
Score = 104 bits (259), Expect = 2e-22, Method: Compositional matrix adjust.
Identities = 52/90 (57%), Positives = 68/90 (75%), Gaps = 0/90 (0%)
Query 59 PTRILLLLNMVGRGQVDSDLKEETEEEAAKLGNLLQVRIFEAAGVPDDCAVRIFCEYERK 118
PT+++LL NMVGRG+VD DL+ ET+EE K G +++ IFE +GV DD AVRIF E+ER
Sbjct 320 PTKVVLLRNMVGRGEVDEDLEAETKEECEKYGKVVRCVIFEISGVTDDEAVRIFLEFERV 379
Query 119 EEATRALLTFNGRAFGGRTVKAKFYSEERF 148
E A +A++ NGR FGGR VKA FY+ +RF
Sbjct 380 ESAIKAVVDLNGRYFGGRVVKACFYNLDRF 409
> hsa:84991 RBM17, DKFZp686F13131, MGC14439, SPF45; RNA binding
motif protein 17; K12840 splicing factor 45
Length=401
Score = 100 bits (249), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 52/95 (54%), Positives = 67/95 (70%), Gaps = 0/95 (0%)
Query 59 PTRILLLLNMVGRGQVDSDLKEETEEEAAKLGNLLQVRIFEAAGVPDDCAVRIFCEYERK 118
PT+++LL NMVG G+VD DL+ ET+EE K G + + IFE G PDD AVRIF E+ER
Sbjct 303 PTKVVLLRNMVGAGEVDEDLEVETKEECEKYGKVGKCVIFEIPGAPDDEAVRIFLEFERV 362
Query 119 EEATRALLTFNGRAFGGRTVKAKFYSEERFARGDL 153
E A +A++ NGR FGGR VKA FY+ ++F DL
Sbjct 363 ESAIKAVVDLNGRYFGGRVVKACFYNLDKFRVLDL 397
> mmu:76938 Rbm17, 2700027J02Rik; RNA binding motif protein 17;
K12840 splicing factor 45
Length=405
Score = 100 bits (249), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 52/95 (54%), Positives = 67/95 (70%), Gaps = 0/95 (0%)
Query 59 PTRILLLLNMVGRGQVDSDLKEETEEEAAKLGNLLQVRIFEAAGVPDDCAVRIFCEYERK 118
PT+++LL NMVG G+VD DL+ ET+EE K G + + IFE G PDD AVRIF E+ER
Sbjct 307 PTKVVLLRNMVGAGEVDEDLEVETKEECEKYGKVGKCVIFEIPGAPDDEAVRIFLEFERV 366
Query 119 EEATRALLTFNGRAFGGRTVKAKFYSEERFARGDL 153
E A +A++ NGR FGGR VKA FY+ ++F DL
Sbjct 367 ESAIKAVVDLNGRYFGGRVVKACFYNLDKFRVLDL 401
> xla:733282 rbm17; RNA binding motif protein 17
Length=400
Score = 87.0 bits (214), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 50/95 (52%), Positives = 67/95 (70%), Gaps = 0/95 (0%)
Query 59 PTRILLLLNMVGRGQVDSDLKEETEEEAAKLGNLLQVRIFEAAGVPDDCAVRIFCEYERK 118
PT+++LL NMVG G+VD +L+ ET+EE K G + + IFE G PD+ AVRIF E+ER
Sbjct 304 PTKVVLLQNMVGAGEVDEELEGETKEECEKYGKVAKCVIFEIPGAPDEEAVRIFLEFERV 363
Query 119 EEATRALLTFNGRAFGGRTVKAKFYSEERFARGDL 153
E A +A++ NGR FGGR VKA FY+ ++F DL
Sbjct 364 ESAIKAVVDLNGRYFGGRIVKAGFYNLDKFRTLDL 398
> xla:447356 MGC130690; MGC84102 protein; K12840 splicing factor
45
Length=400
Score = 84.3 bits (207), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 49/95 (51%), Positives = 67/95 (70%), Gaps = 0/95 (0%)
Query 59 PTRILLLLNMVGRGQVDSDLKEETEEEAAKLGNLLQVRIFEAAGVPDDCAVRIFCEYERK 118
PT+++LL NMVG G+VD +L+ ET+EE K G + + IFE G P++ AVRIF E+ER
Sbjct 304 PTKVVLLRNMVGAGEVDEELEGETKEECEKYGKVGRCVIFEIPGAPEEEAVRIFLEFERV 363
Query 119 EEATRALLTFNGRAFGGRTVKAKFYSEERFARGDL 153
E A +A++ NGR FGGR VKA FY+ ++F DL
Sbjct 364 ESAIKAVVDLNGRYFGGRIVKASFYNLDKFRTLDL 398
> pfa:PF14_0513 RNA binding protein, putative; K12840 splicing
factor 45
Length=511
Score = 83.6 bits (205), Expect = 3e-16, Method: Composition-based stats.
Identities = 46/89 (51%), Positives = 59/89 (66%), Gaps = 0/89 (0%)
Query 60 TRILLLLNMVGRGQVDSDLKEETEEEAAKLGNLLQVRIFEAAGVPDDCAVRIFCEYERKE 119
TRI+ L N+V +VD LKEE EEEA+K GNLL + I + D AV+I+CEYE K+
Sbjct 416 TRIVHLTNLVTPEEVDETLKEEIEEEASKFGNLLNINIVVDKNLLDALAVKIYCEYESKD 475
Query 120 EATRALLTFNGRAFGGRTVKAKFYSEERF 148
+A AL TF GR F GR V+A F +EE +
Sbjct 476 QAQNALNTFKGRTFAGRKVQASFATEEEY 504
> dre:100331631 RNA binding motif protein 17-like
Length=418
Score = 74.7 bits (182), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 37/65 (56%), Positives = 49/65 (75%), Gaps = 0/65 (0%)
Query 59 PTRILLLLNMVGRGQVDSDLKEETEEEAAKLGNLLQVRIFEAAGVPDDCAVRIFCEYERK 118
PT+++LL NMVGRG+VD DL+ ET+EE K G +++ IFE +GV DD AVRIF E+ER
Sbjct 320 PTKVVLLRNMVGRGEVDEDLEAETKEECEKYGKVVRCVIFEISGVTDDEAVRIFLEFERV 379
Query 119 EEATR 123
E A +
Sbjct 380 ESAIK 384
> cpv:cgd1_2730 Ro ribonucleoprotein-binding protein 1, RNA binding
protein with 3x RRM domains ; K12838 poly(U)-binding-splicing
factor PUF60
Length=693
Score = 71.2 bits (173), Expect = 2e-12, Method: Composition-based stats.
Identities = 40/103 (38%), Positives = 58/103 (56%), Gaps = 3/103 (2%)
Query 48 QVPRAVTFNMAPTRILLLLNMVGRGQVDSDLKEETEEEAAKLGNLLQVRIFEAAGV--PD 105
Q+P A++ + + T I+LL NMVG ++D +LKEE + E +K G + VRI + + P
Sbjct 585 QIPGAISGSASSTNIILLTNMVGPDEIDDELKEEVKIECSKYGKVYDVRIHVSNNISKPS 644
Query 106 DCAVRIFCEYERKEEATRALLTFNGRAFGGRTVKAKFYSEERF 148
D VRIF +E A A+ N R FGG V Y+ ER+
Sbjct 645 D-RVRIFVVFESPSMAQIAVPALNNRWFGGNQVFCSLYNTERY 686
> dre:562370 puf60a, fc21a05, wu:fc21a05, wu:fi43a08; poly-U binding
splicing factor a; K12838 poly(U)-binding-splicing factor
PUF60
Length=518
Score = 68.2 bits (165), Expect = 1e-11, Method: Composition-based stats.
Identities = 38/97 (39%), Positives = 54/97 (55%), Gaps = 3/97 (3%)
Query 60 TRILLLLNMVGRGQVDSDLKEETEEEAAKLGNLLQVRIF-EAAGVPDDCAV--RIFCEYE 116
+ +++L NMVG +D DL+ E EE K G + +V I+ E G +D V +IF E+
Sbjct 420 STVMVLRNMVGPEDIDDDLEGEVTEECGKFGAVNRVIIYQEKQGEEEDAEVIVKIFVEFS 479
Query 117 RKEEATRALLTFNGRAFGGRTVKAKFYSEERFARGDL 153
E +A+ N R FGGR V A+ Y +ERF DL
Sbjct 480 AASEMNKAIQALNNRWFGGRKVIAEVYDQERFDNSDL 516
> mmu:67959 Puf60, 2410104I19Rik, 2810454F19Rik, SIAHBP1; poly-U
binding splicing factor 60; K12838 poly(U)-binding-splicing
factor PUF60
Length=499
Score = 67.4 bits (163), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 38/97 (39%), Positives = 54/97 (55%), Gaps = 3/97 (3%)
Query 60 TRILLLLNMVGRGQVDSDLKEETEEEAAKLGNLLQVRIF-EAAGVPDDCA--VRIFCEYE 116
+ +++L NMV +D DL+ E EE K G + +V I+ E G +D V+IF E+
Sbjct 401 STVMVLRNMVDPKDIDDDLEGEVTEECGKFGAVNRVIIYQEKQGEEEDAEIIVKIFVEFS 460
Query 117 RKEEATRALLTFNGRAFGGRTVKAKFYSEERFARGDL 153
E +A+ NGR FGGR V A+ Y +ERF DL
Sbjct 461 MASETHKAIQALNGRWFGGRKVVAEVYDQERFDNSDL 497
> hsa:22827 PUF60, FIR, FLJ31379, RoBPI, SIAHBP1; poly-U binding
splicing factor 60KDa; K12838 poly(U)-binding-splicing factor
PUF60
Length=516
Score = 65.1 bits (157), Expect = 1e-10, Method: Composition-based stats.
Identities = 37/97 (38%), Positives = 53/97 (54%), Gaps = 3/97 (3%)
Query 60 TRILLLLNMVGRGQVDSDLKEETEEEAAKLGNLLQVRIF-EAAGVPDDCA--VRIFCEYE 116
+ +++L NMV +D DL+ E EE K G + +V I+ E G +D V+IF E+
Sbjct 418 STVMVLRNMVDPKDIDDDLEGEVTEECGKFGAVNRVIIYQEKQGEEEDAEIIVKIFVEFS 477
Query 117 RKEEATRALLTFNGRAFGGRTVKAKFYSEERFARGDL 153
E +A+ NGR F GR V A+ Y +ERF DL
Sbjct 478 IASETHKAIQALNGRWFAGRKVVAEVYDQERFDNSDL 514
> dre:415211 puf60b, SIAHBP1, fe37c05, repressor, si:ch211-12p8.2,
si:zc12p8.2, wu:fb33e11, wu:fe37c05, zgc:86806; poly-U
binding splicing factor b
Length=502
Score = 64.3 bits (155), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 37/97 (38%), Positives = 53/97 (54%), Gaps = 3/97 (3%)
Query 60 TRILLLLNMVGRGQVDSDLKEETEEEAAKLGNLLQVRIF-EAAGVPDDCA--VRIFCEYE 116
+ +++L NMVG +D DL+ E EE K G + +V I+ E G DD V+IF E+
Sbjct 404 STVMVLRNMVGPEDIDDDLEGEVMEECGKYGAVNRVIIYQERQGEEDDAEIIVKIFVEFS 463
Query 117 RKEEATRALLTFNGRAFGGRTVKAKFYSEERFARGDL 153
E +A+ N R F GR V A+ Y ++RF DL
Sbjct 464 DAGEMNKAIQALNNRWFAGRKVVAELYDQDRFNSSDL 500
> tgo:TGME49_002770 RNA-binding protein, putative ; K12838 poly(U)-binding-splicing
factor PUF60
Length=532
Score = 60.8 bits (146), Expect = 2e-09, Method: Composition-based stats.
Identities = 33/91 (36%), Positives = 51/91 (56%), Gaps = 5/91 (5%)
Query 62 ILLLLNMVGRGQVDSDLKEETEEEAAKLGNLLQVRIFEAAGVPDDCAVRIFCEYERKEEA 121
++LL NMV +VD +LK+E EE +K G++ +V + VRIF E+ A
Sbjct 444 VVLLSNMVTPSEVDGELKDEVREECSKFGSIKRVEVHTLKET-----VRIFVEFSDLSGA 498
Query 122 TRALLTFNGRAFGGRTVKAKFYSEERFARGD 152
A+ + +GR FGGR + A Y +E F +G+
Sbjct 499 REAIPSLHGRWFGGRQIIANTYDQELFHQGE 529
> cel:Y47G6A.20 rnp-6; RNP (RRM RNA binding domain) containing
family member (rnp-6); K12838 poly(U)-binding-splicing factor
PUF60
Length=749
Score = 57.4 bits (137), Expect = 2e-08, Method: Composition-based stats.
Identities = 34/95 (35%), Positives = 51/95 (53%), Gaps = 7/95 (7%)
Query 60 TRILLLLNMVGRGQVDSDLKEETEEEAAKLGNLLQVRI--FEAAGVPDDCAVRIFCEYER 117
+ +++L NMV +D L+ E EE K GN++ V I F ++G+ V+IF +Y
Sbjct 657 SNVIVLRNMVTPQDIDEFLEGEIREECGKYGNVIDVVIANFASSGL-----VKIFVKYSD 711
Query 118 KEEATRALLTFNGRAFGGRTVKAKFYSEERFARGD 152
+ RA +GR FGG TVKA+ Y + F D
Sbjct 712 SMQVDRAKAALDGRFFGGNTVKAEAYDQILFDHAD 746
> tgo:TGME49_121630 hypothetical protein
Length=400
Score = 55.5 bits (132), Expect = 9e-08, Method: Compositional matrix adjust.
Identities = 30/86 (34%), Positives = 50/86 (58%), Gaps = 3/86 (3%)
Query 58 APTRILLLLNMVGRGQVDSDLKEETEEEAAKLGNLLQVRIFEAAGVPDDCAVRIFCEYER 117
AP+ +++L N++ ++D D+K+E +E K G +++VRI A + VR F Y+
Sbjct 306 APSTLIVLKNLMEVAELDDDVKDEIYQECLKHGKVVEVRIHVVASTQE---VRAFALYQL 362
Query 118 KEEATRALLTFNGRAFGGRTVKAKFY 143
E+A RA+ N R+F R VK + Y
Sbjct 363 PEQANRAVRVLNERSFAKRKVKCELY 388
> ath:AT1G60830 U2 snRNP auxiliary factor large subunit, putative
Length=111
Score = 54.3 bits (129), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 32/101 (31%), Positives = 52/101 (51%), Gaps = 7/101 (6%)
Query 59 PTRILLLLNMVGRGQVD-----SDLKEETEEEAAKLGNLLQVRIF--EAAGVPDDCAVRI 111
PT+I+ L +V + +D+ E+ +E K GNL+ V I P ++
Sbjct 8 PTKIVCLTQVVTADDLRDDAEYADIMEDMSQEGGKFGNLVNVVIPRPNPDHDPTPGVGKV 67
Query 112 FCEYERKEEATRALLTFNGRAFGGRTVKAKFYSEERFARGD 152
F EY + +++A NGR FGG V A +Y E+++A+GD
Sbjct 68 FLEYADVDGSSKARSGMNGRKFGGNQVVAVYYPEDKYAQGD 108
> ath:AT1G60900 U2 snRNP auxiliary factor large subunit, putative;
K12837 splicing factor U2AF 65 kDa subunit
Length=589
Score = 52.4 bits (124), Expect = 7e-07, Method: Composition-based stats.
Identities = 33/103 (32%), Positives = 52/103 (50%), Gaps = 11/103 (10%)
Query 59 PTRILLLLNMVGRGQVDSD-----LKEETEEEAAKLGNLLQVRIFEAAGVPDDCAV---- 109
PT+I+ L +V + D + E+ +E K GNL+ V I PD
Sbjct 486 PTKIVCLTQVVTADDLRDDEEYAEIMEDMRQEGGKFGNLVNVVIPRPN--PDHDPTPGVG 543
Query 110 RIFCEYERKEEATRALLTFNGRAFGGRTVKAKFYSEERFARGD 152
++F EY + +++A NGR FGG V A +Y E+++A+GD
Sbjct 544 KVFLEYADVDGSSKARSGMNGRKFGGNQVVAVYYPEDKYAQGD 586
> cel:F58B3.7 hypothetical protein; K12840 splicing factor 45
Length=371
Score = 51.2 bits (121), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 33/111 (29%), Positives = 53/111 (47%), Gaps = 12/111 (10%)
Query 46 LGQVPRAVTFN-------MAPTRILLLLNMVGRGQVD-----SDLKEETEEEAAKLGNLL 93
+ + P+A TF T+IL L N+ +V + +E +EE K G ++
Sbjct 252 VAEAPKAPTFATNSMEAVQNATKILQLWNLTDLSEVSGEEGKKEFADEIKEEMEKCGQVV 311
Query 94 QVRIFEAAGVPDDCAVRIFCEYERKEEATRALLTFNGRAFGGRTVKAKFYS 144
V + +D VR+F E+ +A +A + NGR FGGR+V A F +
Sbjct 312 NVIVHVDESQEEDRQVRVFVEFTNNAQAIKAFVMMNGRFFGGRSVSAGFQN 362
> ath:AT4G36690 ATU2AF65A; RNA binding / nucleic acid binding
/ nucleotide binding
Length=573
Score = 49.7 bits (117), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 28/78 (35%), Positives = 41/78 (52%), Gaps = 2/78 (2%)
Query 77 DLKEETEEEAAKLGNLLQVRIFEAA--GVPDDCAVRIFCEYERKEEATRALLTFNGRAFG 134
D+ E+ +E K G L V I + G P ++F +Y + +TRA NGR FG
Sbjct 493 DIMEDMRQEGGKFGALTNVVIPRPSPNGEPVAGLGKVFLKYADTDGSTRARFGMNGRKFG 552
Query 135 GRTVKAKFYSEERFARGD 152
G V A +Y E++F +GD
Sbjct 553 GNEVVAVYYPEDKFEQGD 570
> pfa:PF14_0656 U2 snRNP auxiliary factor, putative; K12837 splicing
factor U2AF 65 kDa subunit
Length=833
Score = 47.4 bits (111), Expect = 2e-05, Method: Composition-based stats.
Identities = 34/90 (37%), Positives = 41/90 (45%), Gaps = 19/90 (21%)
Query 74 VDSDLKE---ETEEEAAKLGNLLQVRI---------FEAAGVPDDCAVRIFCEYERKEEA 121
VDS +E E +EEA K G L + I E G +IF Y + A
Sbjct 746 VDSQYEEILKEVKEEAEKYGTLQNIVIPKPNKDLSYTEGVG-------KIFLHYADEATA 798
Query 122 TRALLTFNGRAFGGRTVKAKFYSEERFARG 151
+A FNGR F R V A FYSEE F +G
Sbjct 799 RKAQYMFNGRLFEKRVVCAAFYSEEHFLKG 828
> cpv:cgd7_5220 splicing factor with 3 RRM domains ; K13091 RNA-binding
protein 39
Length=563
Score = 47.0 bits (110), Expect = 3e-05, Method: Composition-based stats.
Identities = 23/79 (29%), Positives = 42/79 (53%), Gaps = 5/79 (6%)
Query 73 QVDSDLKEETEEEAAKLGNLLQVRIFEAAGVPDDCAVRIFCEYERKEEATRALLTFNGRA 132
Q+ +++ + EEE K G LL+ + + + ++ +Y R EEA++A + F+GR
Sbjct 487 QILEEIQADVEEECGKYGTLLECFLDK-----EKMDGNVWVKYSRPEEASKAKMVFHGRF 541
Query 133 FGGRTVKAKFYSEERFARG 151
F GR + F +E F +
Sbjct 542 FAGRKLNVSFIKDEEFPKA 560
> tpv:TP03_0201 hypothetical protein
Length=509
Score = 44.7 bits (104), Expect = 2e-04, Method: Composition-based stats.
Identities = 35/114 (30%), Positives = 53/114 (46%), Gaps = 15/114 (13%)
Query 59 PTRILLLLNMVGRGQVDSD-----LKEETEEEAAKLGNLLQVRI--------FEAAGVPD 105
PTR+LLL N+V + +++ D + ++ E G +L+V + E D
Sbjct 358 PTRVLLLSNLVSKDELEDDEEYVDIIDDVRCECELYGVVLRVELPRVPKGLTEEEMKAFD 417
Query 106 DCAV-RIFCEYERKEEATRALLTFNGRAFGGRTVKAKFYSEERFARGDL-RPDP 157
+V F + E A++A +GR FG RTV A F+SE F G P P
Sbjct 418 PTSVGSAFVLFSTVESASKARKVLDGRKFGQRTVHAHFFSELYFLTGKFGNPKP 471
> tpv:TP04_0540 hypothetical protein
Length=486
Score = 41.2 bits (95), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 23/93 (24%), Positives = 43/93 (46%), Gaps = 3/93 (3%)
Query 60 TRILLLLNMVGRGQVDSDLKEETEEEAAKLGNLLQVRIFEAAGVPDDCAVRIFCEYERKE 119
+ ++++ NMV D +L E + E K G + V + +A ++ + +F + E
Sbjct 394 SNVIVIYNMVDPKLADENLANEVKVECNKYGTVTSVYLHFSA---NNDTLSVFVVFNTPE 450
Query 120 EATRALLTFNGRAFGGRTVKAKFYSEERFARGD 152
+A A+ N R F GR + K Y + G+
Sbjct 451 DADNAVRALNTRWFNGRQIMCKTYDASAYFSGN 483
> dre:402989 u2af2b, MGC77804, wu:fb73g02, zgc:77804; U2 small
nuclear RNA auxiliary factor 2b; K12837 splicing factor U2AF
65 kDa subunit
Length=475
Score = 40.8 bits (94), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 27/101 (26%), Positives = 47/101 (46%), Gaps = 6/101 (5%)
Query 59 PTRILLLLNMVGRGQVDSD-----LKEETEEEAAKLGNLLQVRIFEAA-GVPDDCAVRIF 112
PT +L L+NMV ++ D + E+ +EE +K G + + I G+ +IF
Sbjct 374 PTEVLCLMNMVAPEELIDDEEYEEIVEDVKEECSKYGQVKSIEIPRPVDGLDIPGTGKIF 433
Query 113 CEYERKEEATRALLTFNGRAFGGRTVKAKFYSEERFARGDL 153
E+ ++ +A+ GR F R V K+ + + R D
Sbjct 434 VEFTSVYDSQKAMQGLTGRKFANRVVVTKYCDPDAYHRRDF 474
> mmu:16589 Uhmk1, 4732477C12Rik, 4930500M09Rik, AA673513, AI449218,
AU021979, KIS, Kist; U2AF homology motif (UHM) kinase
1 (EC:2.7.11.1); K08877 U2AF homology motif (UHM) kinase 1
[EC:2.7.11.1]
Length=419
Score = 40.4 bits (93), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 32/111 (28%), Positives = 50/111 (45%), Gaps = 13/111 (11%)
Query 50 PRAVTFNMAPTRILLLLNMVGRGQVDS-----DLKEETEEEAAKLGNLLQVRIFEAAGVP 104
P M PT +L LLN++ +++ D+ E+ +EE K G ++ + VP
Sbjct 310 PHIEDLVMLPTPVLRLLNVLDDDYLENEDEYEDVVEDVKEECQKYGPVVSLL------VP 363
Query 105 DDCAVR--IFCEYERKEEATRALLTFNGRAFGGRTVKAKFYSEERFARGDL 153
+ R +F EY ++ A GR F G+ V A FY + RG L
Sbjct 364 KENPGRGQVFVEYANAGDSKAAQKLLTGRMFDGKFVVATFYPLSAYKRGYL 414
> cel:Y92C3B.2 uaf-1; U2AF splicing factor family member (uaf-1);
K12837 splicing factor U2AF 65 kDa subunit
Length=474
Score = 40.4 bits (93), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 28/97 (28%), Positives = 46/97 (47%), Gaps = 10/97 (10%)
Query 60 TRILLLLNMVGRGQVDSD-----LKEETEEEAAKLG---NLLQVRIFEAAGVPDDCAVRI 111
T IL L+NMV ++ +D + E+ +E +K G +L R +E VP ++
Sbjct 375 TEILCLMNMVTEDELKADDEYEEILEDVRDECSKYGIVRSLEIPRPYEDHPVP--GVGKV 432
Query 112 FCEYERKEEATRALLTFNGRAFGGRTVKAKFYSEERF 148
F E+ + RA GR F RTV +Y +++
Sbjct 433 FVEFASTSDCQRAQAALTGRKFANRTVVTSYYDVDKY 469
> dre:767721 uhmk1, MGC153241, zgc:153241; U2AF homology motif
(UHM) kinase 1 (EC:2.7.11.1); K08877 U2AF homology motif (UHM)
kinase 1 [EC:2.7.11.1]
Length=410
Score = 39.7 bits (91), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 29/109 (26%), Positives = 49/109 (44%), Gaps = 9/109 (8%)
Query 50 PRAVTFNMAPTRILLLLNMVGRGQVDS-----DLKEETEEEAAKLGNLLQVRIFEAAGVP 104
P + PT +L LLN++ + + D+ E+ +EE K G ++ + I +
Sbjct 301 PHIEDLVLLPTPVLRLLNVIDDSHLYNEDEYEDIIEDMKEECQKYGTVVSLLIPKE---- 356
Query 105 DDCAVRIFCEYERKEEATRALLTFNGRAFGGRTVKAKFYSEERFARGDL 153
+ ++F EY ++ A GR F G+ V A FY + RG L
Sbjct 357 NPGKGQVFVEYANAGDSKEAQRLLTGRTFDGKFVVATFYPLGAYKRGYL 405
> tpv:TP04_0035 U2 small nuclear ribonucleoprotein, auxiliary
factor, large subunit; K12837 splicing factor U2AF 65 kDa subunit
Length=380
Score = 38.9 bits (89), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 34/105 (32%), Positives = 48/105 (45%), Gaps = 16/105 (15%)
Query 59 PTRILLLLNMVGRGQVDSDLKEE-----TEEEAAKLGNLLQVRIFEAAGVPD------DC 107
P++++ LLNMV + SD +EEA K G L +V I PD +
Sbjct 275 PSKVIQLLNMVFHEDLISDYNYNEIVRLVKEEAQKYGPLQEVVIPR----PDKDLTFKEG 330
Query 108 AVRIFCEYERKEEATRALLTFNGRAFG-GRTVKAKFYSEERFARG 151
++F YE A +A FNGR F R V + F+ E+ F G
Sbjct 331 VGKVFIRYEDLLSARKAQYMFNGRVFDKNRIVCSAFFPEDLFITG 375
> bbo:BBOV_II004960 18.m06413; RNA recognition motif (RRM)-containing
protein; K12837 splicing factor U2AF 65 kDa subunit
Length=383
Score = 38.5 bits (88), Expect = 0.010, Method: Compositional matrix adjust.
Identities = 32/104 (30%), Positives = 51/104 (49%), Gaps = 14/104 (13%)
Query 59 PTRILLLLNMVGRGQVDSD-----LKEETEEEAAKLGNLLQVRIFEAAGVPDDCAVR--- 110
P+RI+ L+N+V + D +K+ EEA K G+L + I DD + +
Sbjct 278 PSRIVQLINIVFHEDLIQDKRYHEVKDAIMEEAKKYGHLEDIVIPRPN---DDLSYKEGV 334
Query 111 --IFCEYERKEEATRALLTFNGRAF-GGRTVKAKFYSEERFARG 151
+F ++ + + RA NGR F G R V A F+ +RF +G
Sbjct 335 GKVFLKFGDEISSRRAQYMLNGRVFDGNRIVCAAFFPLDRFLKG 378
> tpv:TP03_0691 splicing factor; K13091 RNA-binding protein 39
Length=644
Score = 38.5 bits (88), Expect = 0.011, Method: Composition-based stats.
Identities = 18/72 (25%), Positives = 41/72 (56%), Gaps = 5/72 (6%)
Query 77 DLKEETEEEAAKLGNLLQVRIFEAAGVPDDCAVRIFCEYERKEEATRALLTFNGRAFGGR 136
+++E+ +EE K G ++QV F PD +++ +++ ++A A + GR F G
Sbjct 571 EIEEDVKEECGKYGTVIQV--FVNKRNPDG---KVYVKFKNNDDAQAANKSLQGRYFAGN 625
Query 137 TVKAKFYSEERF 148
T++ + S++++
Sbjct 626 TIQVSYISDDQY 637
> bbo:BBOV_IV007070 23.m05938; hypothetical protein
Length=400
Score = 38.5 bits (88), Expect = 0.011, Method: Compositional matrix adjust.
Identities = 33/130 (25%), Positives = 57/130 (43%), Gaps = 28/130 (21%)
Query 59 PTRILLLLNMVGRGQVDSDLK-----EETEEEAAKLGNLLQVRI---------------- 97
PTR+LLL N+V + ++ D + ++ E + G +++V +
Sbjct 254 PTRVLLLANLVSKEDLEDDAEYYDIIDDVRCECEEYGPVVRVEMPRVPKGLTLDEIRNMD 313
Query 98 FEAAGVPDDCAVRIFCEYERKEEATRALLTFNGRAFGGRTVKAKFYSEERFARGDLRPDP 157
F A G CA +F E A++A +GR FG R V+ F+SE F G+
Sbjct 314 FSAVG----CAFVLFSNIEG---ASKARKVLDGRKFGHRIVECHFFSELLFHVGEFSNPA 366
Query 158 EQEIDQHVTV 167
+ +H ++
Sbjct 367 PNFLKEHSSM 376
> tgo:TGME49_108920 U2 small nuclear ribonucleoprotein auxiliary
factor U2AF
Length=704
Score = 38.5 bits (88), Expect = 0.011, Method: Composition-based stats.
Identities = 18/47 (38%), Positives = 30/47 (63%), Gaps = 1/47 (2%)
Query 112 FCEYERKEEATRALLTFNGRAFGGRTVKAKFYSEERFARGDLRPDPE 158
+ E+E E + +A NGR FGG+ V+A ++SE +F R ++ DP+
Sbjct 539 YVEFEDCEWSAKARKALNGRKFGGKIVEAHYFSEVKF-RKEMFYDPK 584
> cel:C07A4.1 hypothetical protein
Length=360
Score = 37.0 bits (84), Expect = 0.030, Method: Compositional matrix adjust.
Identities = 23/89 (25%), Positives = 41/89 (46%), Gaps = 11/89 (12%)
Query 69 VGRGQVDSDLKEETEEEAAKLGNLLQVRIFEAAGVPDDCAVRIFCEYERKEEATRALLTF 128
+ + D+DL++ + G++ +VRIF+ F YE+KE AT+A++
Sbjct 244 ISQQTTDADLRDL----FSTYGDIAEVRIFKTQRY-------AFVRYEKKECATKAIMEM 292
Query 129 NGRAFGGRTVKAKFYSEERFARGDLRPDP 157
NG+ G V+ + + L P P
Sbjct 293 NGKEMAGNQVRCSWGRTQAVPNQALNPLP 321
> ath:AT2G16940 RNA recognition motif (RRM)-containing protein
Length=561
Score = 37.0 bits (84), Expect = 0.034, Method: Compositional matrix adjust.
Identities = 33/122 (27%), Positives = 52/122 (42%), Gaps = 14/122 (11%)
Query 32 SVCGVARQGQDVTTLGQVPRAVTFNMAPTRILLLLNMVG-----RGQVDSDLKEETEEEA 86
+V G+A G G +P P+ LLL NM D D+KE+ +EE
Sbjct 439 AVAGLAGSG---IIPGVIPAGFDPIGVPSECLLLKNMFDPSTETEDDFDEDIKEDVKEEC 495
Query 87 AKLGNLLQVRIFEAAGVPDDCAVRIFCEYERKEEATRALLTFNGRAFGGRTVKAKFYSEE 146
+K G L IF V + ++ +E + A A +GR F G+ + A + + E
Sbjct 496 SKFGKLNH--IF----VDKNSVGFVYLRFENAQAAIGAQRALHGRWFAGKMITATYMTTE 549
Query 147 RF 148
+
Sbjct 550 AY 551
> tgo:TGME49_034520 U2 snRNP auxiliary factor or splicing factor,
putative ; K12837 splicing factor U2AF 65 kDa subunit
Length=553
Score = 36.6 bits (83), Expect = 0.039, Method: Compositional matrix adjust.
Identities = 37/116 (31%), Positives = 55/116 (47%), Gaps = 16/116 (13%)
Query 48 QVPRAVTFNMAPTRILLLLNMVGRGQVDSDLKE------ETEEEAAKLGNLLQVRIFEAA 101
QV A P++++ LLN V + + D KE + ++EA K G L +V + +
Sbjct 437 QVQAARKIGERPSKVVQLLNCVYQEDL-IDPKEYEAICDDIKQEAEKHGALEEVLVPKPN 495
Query 102 GVPDDCAVR-----IFCEYERKEEATRALLTFNGRAF-GGRTVKAKFYSEERFARG 151
+D + R +F Y A +A L NGR F R V A F+ EE+FA G
Sbjct 496 ---EDLSYREGVGKVFLRYSDVTAARKAQLMLNGRRFDSNRVVCAAFFPEEKFAAG 548
> dre:386806 cstf2, CstF-64, fb11e07, wu:fb11e07, zgc:56346, zgc:77730;
cleavage stimulation factor, 3' pre-RNA, subunit 2;
K14407 cleavage stimulation factor subunit 2
Length=488
Score = 36.6 bits (83), Expect = 0.040, Method: Compositional matrix adjust.
Identities = 14/36 (38%), Positives = 21/36 (58%), Gaps = 0/36 (0%)
Query 112 FCEYERKEEATRALLTFNGRAFGGRTVKAKFYSEER 147
FCEY+ +E A A+ NGR F GR ++ + E+
Sbjct 69 FCEYQDQETALSAMRNLNGREFSGRALRVDNAASEK 104
> mmu:83410 Cstf2t, 64kDa, C77975, tCstF-64, tauCstF-64; cleavage
stimulation factor, 3' pre-RNA subunit 2, tau; K14407 cleavage
stimulation factor subunit 2
Length=632
Score = 36.6 bits (83), Expect = 0.042, Method: Composition-based stats.
Identities = 22/74 (29%), Positives = 37/74 (50%), Gaps = 9/74 (12%)
Query 80 EETEEEA----AKLGNLLQVRIF--EAAGVPDDCAVRIFCEYERKEEATRALLTFNGRAF 133
E TEE+ +++G+++ R+ G P FCEY+ +E A A+ NGR F
Sbjct 26 EATEEQLKDIFSEVGSVVSFRLVYDRETGKPKGYG---FCEYQDQETALSAMRNLNGREF 82
Query 134 GGRTVKAKFYSEER 147
GR ++ + E+
Sbjct 83 SGRALRVDNAASEK 96
> pfa:PF07_0066 conserved Plasmodium protein, unknown function
Length=1125
Score = 36.6 bits (83), Expect = 0.043, Method: Composition-based stats.
Identities = 14/37 (37%), Positives = 23/37 (62%), Gaps = 0/37 (0%)
Query 112 FCEYERKEEATRALLTFNGRAFGGRTVKAKFYSEERF 148
F +E E AT+A +GR FG ++A ++SE++F
Sbjct 1051 FIHFENIESATKARKELSGRKFGANIIEANYFSEKKF 1087
> hsa:23283 CSTF2T, CstF-64T, DKFZp434C1013, KIAA0689; cleavage
stimulation factor, 3' pre-RNA, subunit 2, 64kDa, tau variant;
K14407 cleavage stimulation factor subunit 2
Length=616
Score = 36.6 bits (83), Expect = 0.044, Method: Composition-based stats.
Identities = 22/74 (29%), Positives = 37/74 (50%), Gaps = 9/74 (12%)
Query 80 EETEEEA----AKLGNLLQVRIF--EAAGVPDDCAVRIFCEYERKEEATRALLTFNGRAF 133
E TEE+ +++G+++ R+ G P FCEY+ +E A A+ NGR F
Sbjct 26 EATEEQLKDIFSEVGSVVSFRLVYDRETGKPKGYG---FCEYQDQETALSAMRNLNGREF 82
Query 134 GGRTVKAKFYSEER 147
GR ++ + E+
Sbjct 83 SGRALRVDNAASEK 96
> pfa:MAL13P1.120 splicing factor, putative; K13091 RNA-binding
protein 39
Length=864
Score = 36.2 bits (82), Expect = 0.050, Method: Composition-based stats.
Identities = 18/71 (25%), Positives = 37/71 (52%), Gaps = 5/71 (7%)
Query 76 SDLKEETEEEAAKLGNLLQVRIFEAAGVPDDCAVRIFCEYERKEEATRALLTFNGRAFGG 135
+D+ E+ +EE +K G ++ + + + +I+ +Y +E+ ++ NGR FGG
Sbjct 790 NDILEDVKEECSKYGKVVNIWL-----DTKNIDGKIYIKYSNNDESLKSFQFLNGRYFGG 844
Query 136 RTVKAKFYSEE 146
+ A F S +
Sbjct 845 SLINAYFISND 855
> mmu:72459 Htatsf1, 1600023H17Rik, 2600017A12Rik, 2700077B20Rik,
TAT-SF1; HIV TAT specific factor 1; K13093 HIV Tat-specific
factor 1
Length=757
Score = 36.2 bits (82), Expect = 0.051, Method: Composition-based stats.
Identities = 26/87 (29%), Positives = 43/87 (49%), Gaps = 11/87 (12%)
Query 61 RILLLLNMVGRGQVDSD------LKEETEEEAAKLGNLLQVRIFEAAGVPDDCAVRIFCE 114
R+++L NM + D ++E+ E +K G + ++ +F+ PD A F E
Sbjct 265 RVVILKNMFHPMDFEDDPLVLNEIREDLRVECSKFGQIRKLLLFDRH--PDGVASVSFRE 322
Query 115 YERKEEATRALLTFNGRAFGGRTVKAK 141
EEA + T +GR FGGR + A+
Sbjct 323 ---PEEADHCIQTLDGRWFGGRQITAQ 346
> xla:379871 cstf2, MGC53575, MGC83019, cstf-64; cleavage stimulation
factor, 3' pre-RNA, subunit 2, 64kDa; K14407 cleavage
stimulation factor subunit 2
Length=518
Score = 36.2 bits (82), Expect = 0.056, Method: Composition-based stats.
Identities = 14/36 (38%), Positives = 21/36 (58%), Gaps = 0/36 (0%)
Query 112 FCEYERKEEATRALLTFNGRAFGGRTVKAKFYSEER 147
FCEY+ +E A A+ NGR F GR ++ + E+
Sbjct 61 FCEYQDQETALSAMRNLNGREFSGRALRVDNAASEK 96
> hsa:1478 CSTF2, CstF-64; cleavage stimulation factor, 3' pre-RNA,
subunit 2, 64kDa; K14407 cleavage stimulation factor subunit
2
Length=577
Score = 35.8 bits (81), Expect = 0.063, Method: Composition-based stats.
Identities = 14/36 (38%), Positives = 21/36 (58%), Gaps = 0/36 (0%)
Query 112 FCEYERKEEATRALLTFNGRAFGGRTVKAKFYSEER 147
FCEY+ +E A A+ NGR F GR ++ + E+
Sbjct 61 FCEYQDQETALSAMRNLNGREFSGRALRVDNAASEK 96
> xla:380287 u2af2, MGC53441, u2af65; U2 small nuclear RNA auxiliary
factor 2; K12837 splicing factor U2AF 65 kDa subunit
Length=456
Score = 35.8 bits (81), Expect = 0.067, Method: Compositional matrix adjust.
Identities = 30/113 (26%), Positives = 48/113 (42%), Gaps = 6/113 (5%)
Query 47 GQVPRAVTFNMAPTRILLLLNMVGRGQ-VDSD----LKEETEEEAAKLGNLLQVRIFEAA 101
G + V PT +L L+NMV + +D D + E+ +E K G + + I
Sbjct 343 GLMSSQVQMGGHPTEVLCLMNMVVPEELIDDDEYEEIVEDVRDECGKYGAVKSIEIPRPV 402
Query 102 -GVPDDCAVRIFCEYERKEEATRALLTFNGRAFGGRTVKAKFYSEERFARGDL 153
GV +IF E+ + +A+ GR F R V K+ + + R D
Sbjct 403 DGVEVPGCGKIFVEFTSVFDCQKAMQGLTGRKFANRVVVTKYCDPDGYHRRDF 455
> mmu:108062 Cstf2, 64kDa, C630034J23Rik, Cstf64; cleavage stimulation
factor, 3' pre-RNA subunit 2; K14407 cleavage stimulation
factor subunit 2
Length=580
Score = 35.8 bits (81), Expect = 0.071, Method: Composition-based stats.
Identities = 14/36 (38%), Positives = 21/36 (58%), Gaps = 0/36 (0%)
Query 112 FCEYERKEEATRALLTFNGRAFGGRTVKAKFYSEER 147
FCEY+ +E A A+ NGR F GR ++ + E+
Sbjct 61 FCEYQDQETALSAMRNLNGREFSGRALRVDNAASEK 96
> tgo:TGME49_083740 mRNA processing protein, putative (EC:3.4.21.72)
Length=615
Score = 35.8 bits (81), Expect = 0.076, Method: Composition-based stats.
Identities = 22/69 (31%), Positives = 36/69 (52%), Gaps = 4/69 (5%)
Query 79 KEETEEEAAKLGNLLQVRI-FEAAGVPDDCAVRIFCEYERKEEATRALLTFNGRAFGGRT 137
+E+ + +++G +LQVRI ++ G A FCE+ E A +T N GGR
Sbjct 29 EEDLKTLLSRVGRVLQVRIKYDEGGQSKGFA---FCEFPDPETCYLAYVTLNNADLGGRK 85
Query 138 VKAKFYSEE 146
+K F ++E
Sbjct 86 LKIDFATDE 94
> hsa:27336 HTATSF1, TAT-SF1, dJ196E23.2; HIV-1 Tat specific factor
1; K13093 HIV Tat-specific factor 1
Length=755
Score = 34.7 bits (78), Expect = 0.16, Method: Composition-based stats.
Identities = 26/97 (26%), Positives = 45/97 (46%), Gaps = 11/97 (11%)
Query 51 RAVTFNMAPTRILLLLNMVGRGQVDSD------LKEETEEEAAKLGNLLQVRIFEAAGVP 104
RA M R++++ NM + D ++E+ E +K G + ++ +F+ P
Sbjct 254 RAGPSRMRHERVVIIKNMFHPMDFEDDPLVLNEIREDLRVECSKFGQIRKLLLFDRH--P 311
Query 105 DDCAVRIFCEYERKEEATRALLTFNGRAFGGRTVKAK 141
D A + EEA + T +GR FGGR + A+
Sbjct 312 DGVAS---VSFRDPEEADYCIQTLDGRWFGGRQITAQ 345
> cpv:cgd2_1480 splicing factor U2AF U2 SnRNP auxiliary factor
large subunit, RRM domain ; K12837 splicing factor U2AF 65
kDa subunit
Length=438
Score = 34.3 bits (77), Expect = 0.21, Method: Compositional matrix adjust.
Identities = 35/152 (23%), Positives = 65/152 (42%), Gaps = 34/152 (22%)
Query 32 SVCGVARQGQDVTTLGQVPRAVTFNM------------------APTRILLLLNM-VGRG 72
SV Q + + ++P ++T+N+ P++I+ LLN+ +
Sbjct 276 SVNSNIIQNSEYLHITEIPTSMTYNIFSNPVLGLMMKYSKQVGETPSQIIQLLNIFLPEE 335
Query 73 QVDSDLKEET----EEEAAKLGNLLQV-----RIFEAAGVPDDC--AVRIFCEYERKEEA 121
VD+++ T EA G +L++ ++ E C A ++F + A
Sbjct 336 LVDNEIYNSTLDSVRSEAEVYGTILEIFCPRPKVIEEF---HSCSGAGKVFIYFSDITAA 392
Query 122 TRALLTFNGRAFGG-RTVKAKFYSEERFARGD 152
RA FNGR F +TV A F+ E++ + +
Sbjct 393 RRAQYQFNGRVFDNIKTVSATFFPLEKYLKHE 424
Lambda K H
0.326 0.141 0.417
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4222647260
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40