bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_0682_orf1
Length=430
Score E
Sequences producing significant alignments: (Bits) Value
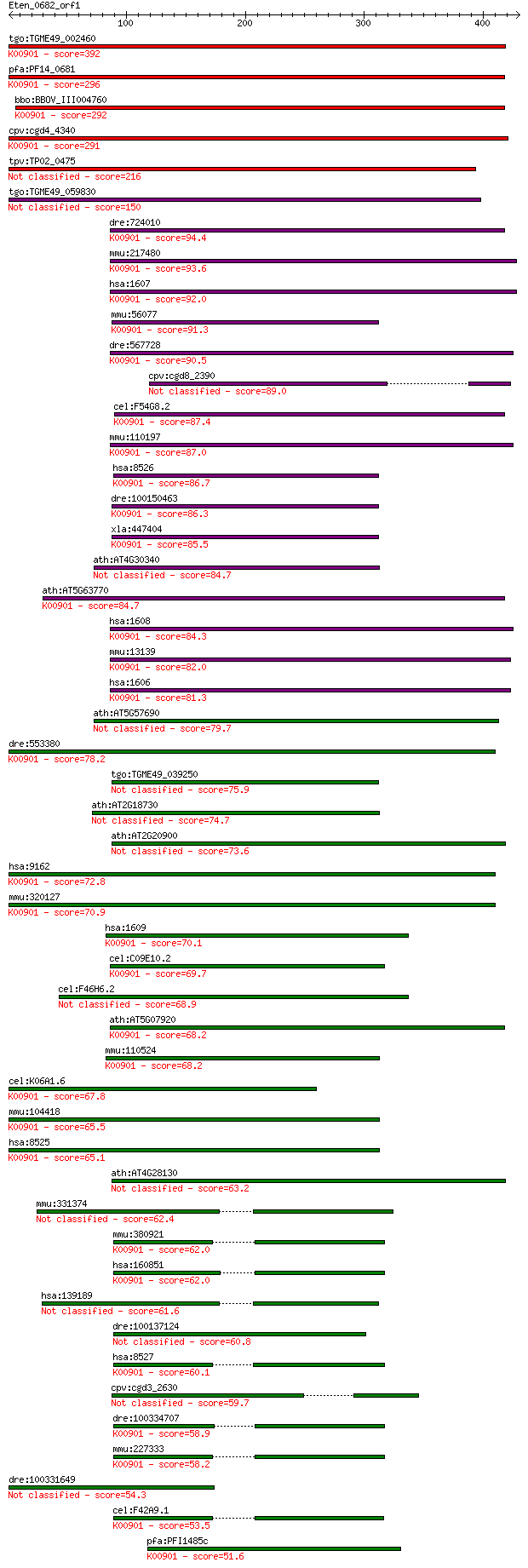
tgo:TGME49_002460 diacylglycerol kinase, putative (EC:4.1.1.70... 392 1e-108
pfa:PF14_0681 diacylglycerol kinase, putative; K00901 diacylgl... 296 9e-80
bbo:BBOV_III004760 17.m07427; diacylglycerol kinase (EC:2.7.1.... 292 2e-78
cpv:cgd4_4340 diacylglycerol kinase ; K00901 diacylglycerol ki... 291 4e-78
tpv:TP02_0475 diacylglycerol kinase 216 1e-55
tgo:TGME49_059830 diacylglycerol kinase, putative (EC:2.7.1.107) 150 9e-36
dre:724010 dgka; zgc:136759 (EC:2.7.1.107); K00901 diacylglyce... 94.4 9e-19
mmu:217480 Dgkb, 6430574F24, 90kda, C630029D13Rik, DAGK2, DGK,... 93.6 1e-18
hsa:1607 DGKB, DAGK2, DGK, DGK-BETA, KIAA0718; diacylglycerol ... 92.0 3e-18
mmu:56077 Dgke, C87606, DAGK6, DGK; diacylglycerol kinase, eps... 91.3 7e-18
dre:567728 diacylglycerol kinase beta-like; K00901 diacylglyce... 90.5 1e-17
cpv:cgd8_2390 diacylglycerol kinase 89.0 3e-17
cel:F54G8.2 dgk-3; DiacylGlycerol Kinase family member (dgk-3)... 87.4 1e-16
mmu:110197 Dgkg, 2900055E17Rik, 90kDa, AI854428, Dagk3, E43000... 87.0 1e-16
hsa:8526 DGKE, DAGK5, DAGK6, DGK; diacylglycerol kinase, epsil... 86.7 2e-16
dre:100150463 dgke; diacylglycerol kinase epsilon (EC:2.7.1.10... 86.3 2e-16
xla:447404 dgke, MGC81643; diacylglycerol kinase, epsilon 64kD... 85.5 4e-16
ath:AT4G30340 ATDGK7; ATDGK7 (Diacylglycerol kinase 7); diacyl... 84.7 6e-16
ath:AT5G63770 ATDGK2; ATDGK2 (Diacylglycerol kinase 2); diacyl... 84.7 6e-16
hsa:1608 DGKG, DAGK3, DGK-GAMMA, MGC104993, MGC133330; diacylg... 84.3 7e-16
mmu:13139 Dgka, 80kDa, AW146112, Dagk1; diacylglycerol kinase,... 82.0 4e-15
hsa:1606 DGKA, DAGK, DAGK1, DGK-alpha, MGC12821, MGC42356; dia... 81.3 8e-15
ath:AT5G57690 diacylglycerol kinase 79.7 2e-14
dre:553380 dgki; diacylglycerol kinase, zeta-like; K00901 diac... 78.2 6e-14
tgo:TGME49_039250 diacylglycerol kinase, putative (EC:2.7.1.10... 75.9 3e-13
ath:AT2G18730 diacylglycerol kinase, putative 74.7 7e-13
ath:AT2G20900 diacylglycerol kinase, putative 73.6 1e-12
hsa:9162 DGKI, DGK-IOTA; diacylglycerol kinase, iota (EC:2.7.1... 72.8 3e-12
mmu:320127 Dgki, C130010K08Rik; diacylglycerol kinase, iota (E... 70.9 1e-11
hsa:1609 DGKQ, DAGK, DAGK4, DAGK7; diacylglycerol kinase, thet... 70.1 2e-11
cel:C09E10.2 dgk-1; DiacylGlycerol Kinase family member (dgk-1... 69.7 2e-11
cel:F46H6.2 dgk-2; DiacylGlycerol Kinase family member (dgk-2) 68.9 4e-11
ath:AT5G07920 DGK1; DGK1 (DIACYLGLYCEROL KINASE1); calcium ion... 68.2 6e-11
mmu:110524 Dgkq, 110kDa, DAGK, DAGK7, Dagk4, Dgkd; diacylglyce... 68.2 6e-11
cel:K06A1.6 dgk-5; DiacylGlycerol Kinase family member (dgk-5)... 67.8 7e-11
mmu:104418 Dgkz, E130307B02Rik, F730209L11Rik, mDGK[z]; diacyl... 65.5 4e-10
hsa:8525 DGKZ, DAGK5, DAGK6, DGK-ZETA, hDGKzeta; diacylglycero... 65.1 5e-10
ath:AT4G28130 diacylglycerol kinase accessory domain-containin... 63.2 2e-09
mmu:331374 Dgkk, Gm360; diacylglycerol kinase kappa (EC:2.7.1.... 62.4 4e-09
mmu:380921 Dgkh, 5930402B05Rik, D130015C16, MGC156865; diacylg... 62.0 4e-09
hsa:160851 DGKH, DGKeta, DKFZp761I1510; diacylglycerol kinase,... 62.0 5e-09
hsa:139189 DGKK, MRX68; diacylglycerol kinase, kappa (EC:2.7.1... 61.6 6e-09
dre:100137124 si:ch211-93a19.1 60.8 1e-08
hsa:8527 DGKD, DGKdelta, FLJ26930, KIAA0145, dgkd-2; diacylgly... 60.1 2e-08
cpv:cgd3_2630 diacylglycerol kinase 59.7 2e-08
dre:100334707 diacylglycerol kinase, eta-like; K00901 diacylgl... 58.9 3e-08
mmu:227333 Dgkd, AI841987, D330025K09, DGKdelta, KIAA0145, dgk... 58.2 6e-08
dre:100331649 diacylglycerol kinase, iota-like 54.3 1e-06
cel:F42A9.1 dgk-4; DiacylGlycerol Kinase family member (dgk-4)... 53.5 1e-06
pfa:PFI1485c DGK1; diacylglycerol kinase, putative (EC:2.7.1.1... 51.6 6e-06
> tgo:TGME49_002460 diacylglycerol kinase, putative (EC:4.1.1.70
2.7.1.107); K00901 diacylglycerol kinase [EC:2.7.1.107]
Length=678
Score = 392 bits (1007), Expect = 1e-108, Method: Compositional matrix adjust.
Identities = 207/469 (44%), Positives = 280/469 (59%), Gaps = 54/469 (11%)
Query 1 LFSNPTSGGNKASAFTRTAVSQLTLTEPCPLKVFIHDIREGPSGSKPGFLLLRSVA---- 56
+F NPTSGGNKA+AFT A+ +LT+T P P ++I DIREG SG+KPGF LLR+
Sbjct 204 IFINPTSGGNKAAAFTE-AMCRLTMTSPFPCNIYIFDIREGISGNKPGFRLLRAATEVAF 262
Query 57 ---------------------AAAAAAAAAAAGE------PRAAAD---------GSSSS 80
A A A + A AG+ P A D G
Sbjct 263 SRESSLEAGASPASVSSASLHAEAGAGSKAEAGKTDPDGVPTAGEDQRKRGAGERGERED 322
Query 81 KKAQH----------PIVRVLVAGGDGTVMWCIDEMHKTGVQPHVCAVGVVPYGTGNDFA 130
K + ++RVLVAGGDGTVMWC E + P A GV+PYGTGNDFA
Sbjct 323 KGDREREEKGEIQDDNVIRVLVAGGDGTVMWCAAEADAHRIDPMKIAFGVIPYGTGNDFA 382
Query 131 NAFGWRPFNAANPFDASLHTLRNIVEHCMRATVVYHDLWSVKVTLKPGGHFNRINSTTRK 190
NAFGW+ + PFD ++ T+R ++ +A VV+HDLWSV V LK G F +INS TRK
Sbjct 383 NAFGWKEWRGLRPFDGAMKTMRRLLSRWSQARVVHHDLWSVNVVLKDDGEFTKINSETRK 442
Query 191 KEVV-EAEGEPVKELSFTMSNYFSMGVESRIGRGFDRYRTKSQVFNKMRYGIEGFKKTFV 249
K+V+ ++ G V++++F MSNYFSMGVESRIGRGFDR+R +SQ+ NKM YGIEG KK +
Sbjct 443 KQVLQDSTGRRVQKMTFVMSNYFSMGVESRIGRGFDRHRRQSQLLNKMTYGIEGVKKAWF 502
Query 250 TRTKSINEIILDLRIHPNTPEEQTLFTTDKAAAKAQQLPLLNKTASLVVLNIPSFSGGND 309
RT +I+ I+ L P P+E+ +F T + P+L K+ SL+ LNIPSFS GND
Sbjct 503 KRTLTIDNIVDGLLESPGEPDERVVFRTKDSTLP--DGPILKKSVSLIALNIPSFSAGND 560
Query 310 IWAPSKKLATRMFDKQKRKQNKELLKVPQQMGDEKLEFMTFKNIPSLGMEFMLHGRGCRL 369
IWA S + ++ + + + Q GD +LEF++F ++ S+G+EF HGR R+
Sbjct 561 IWATSHSVGILTKSTSLNRELRTIAQEKQLTGDRELEFLSFPSVTSMGVEFACHGRARRI 620
Query 370 HSGKGPWQISFRSLDEKTRVYFQTDGEYYQLTLPASVFVSHNRRVRVLS 418
+ G+GPW+I F+ L +VYFQ DGE++Q+ P V + HNR V+VL+
Sbjct 621 NQGRGPWKILFKDLSAHAKVYFQVDGEFFQMARPDFVTIEHNRTVQVLA 669
> pfa:PF14_0681 diacylglycerol kinase, putative; K00901 diacylglycerol
kinase [EC:2.7.1.107]
Length=499
Score = 296 bits (759), Expect = 9e-80, Method: Compositional matrix adjust.
Identities = 170/496 (34%), Positives = 250/496 (50%), Gaps = 85/496 (17%)
Query 1 LFSNPTSGGNKASAFTRTAVSQLTLTEPCPLKVFIHDIREGPSGSKPGFLLLRSVA---- 56
LF NPTSGGNKAS FT V+++ +P FI++I EG G KPGF+ L ++
Sbjct 6 LFVNPTSGGNKASIFTEFGVNEIIFHKPHKCHFFIYNILEGEKGKKPGFINLSNIMNNHN 65
Query 57 ---------------AAAAAAAAAAAGEPRAAADG----SSSSKKAQHP----------- 86
+ + G + +G + SSK +
Sbjct 66 KYESKGFSASSLNNKSFSMTNDKITCGNEKKEMEGEGILNGSSKVDEEDKNIYNSKNNNN 125
Query 87 ---------------------IVRVLVAGGDGTVMWCIDEMHKTGVQPHVCAVGVVPYGT 125
V VLVAGGDGTV W + E + + A+GV+P+GT
Sbjct 126 NMNYFNNKKIETKKTLGHKDIFVYVLVAGGDGTVNWVLKEAEYYNINDNNFAIGVIPFGT 185
Query 126 GNDFANAFGWRPFNAANPFDASLHTLRNIVEHCMRATVVYHDLWSVKVTLKPGGHFNRIN 185
GNDFA AFGW+ + L+ IV+ ++ V HD W++ V LK G+FN+IN
Sbjct 186 GNDFAKAFGWKKIDRMLNHSFLFDVLKKIVDQTFKSKVEKHDYWNIHVVLKEQGYFNKIN 245
Query 186 STTRKKE-VVEAEGEPVKELSFTMSNYFSMGVESRIGRGFDRYRTKSQVFNKMRYGIEGF 244
S T+KKE ++ + E + E+ MSNYFS+G++SRIGRGF+R+R KS V NK Y +EGF
Sbjct 246 SRTKKKETLLNDDKENLHEMKLCMSNYFSIGIDSRIGRGFERHRQKSAVCNKFIYAVEGF 305
Query 245 KKTFVTRTKSINEII--------------------LDLRIHPNTPEEQTLFTTDKAAAKA 284
KK + IN II + + +FTT+
Sbjct 306 KKVSFKKNTPINLIIDKMINMDNNNDNNDNDNNIDGNDNNNDGNDNNNVIFTTN----HN 361
Query 285 QQLPLLNKTASLVVLNIPSFSGGNDIWAPSKKLAT---RMFDKQKRKQNKELLKVPQQMG 341
+ PLL K S++ +NIPS+S GNDIW + K+ + +++Q + L K Q++G
Sbjct 362 ESYPLLKKAMSIICINIPSYSSGNDIWNYTHKIGLILPKNITNDQKEQYQILKKTKQEVG 421
Query 342 DEKLEFMTFKNIPSLGMEFMLHGRGCRLHSGKGPWQISFRSLDEKTRVYFQTDGEYYQLT 401
D LEF+ +++ LG+EF L GR R+H G GPW++ F+ ++K VYFQ DGEYY ++
Sbjct 422 DGILEFVIYQSGVDLGLEFTLKGRAYRVHQGFGPWKLLFK--EQKYNVYFQVDGEYYLMS 479
Query 402 LPASVFVSHNRRVRVL 417
LP + + H +++ VL
Sbjct 480 LPDYISIEHYKKINVL 495
> bbo:BBOV_III004760 17.m07427; diacylglycerol kinase (EC:2.7.1.107);
K00901 diacylglycerol kinase [EC:2.7.1.107]
Length=680
Score = 292 bits (747), Expect = 2e-78, Method: Compositional matrix adjust.
Identities = 155/415 (37%), Positives = 231/415 (55%), Gaps = 39/415 (9%)
Query 7 SGGNKASAFTRTAVSQLTLTEPCPLKVFIHDIREGPSGSKPGFLLLRSVAAAAAAAAAAA 66
SGG KA+ F V++L T+P P V+I+ I +GPSG KPGF LR AA
Sbjct 300 SGGQKAATFFEPNVNELHFTKPVPSVVYIYSITDGPSGHKPGFEHLRDCIAA-------- 351
Query 67 AGEPRAAADGSSSSKKAQHPIVRVLVAGGDGTVMWCIDEMHKTGVQPHVCAVGVVPYGTG 126
+ + +V++ GGDG+VMW I+EM G+ + +VPYGTG
Sbjct 352 ---------------RTEDDHFKVVICGGDGSVMWMIEEMDAHGIDCNSVVFSIVPYGTG 396
Query 127 NDFANAFGWRPFNAANPFDASLHTLRNIVEHCMRATVVYHDLWSVKVTLKPGGHFNRINS 186
NDFA A W F NPFD ++ LR ++E +T V HD W V +T++P G FNRIN
Sbjct 397 NDFARAVHWDNFTGLNPFDNNMSALRRVIERLFNSTEVLHDFWKVVLTVEPEGSFNRINQ 456
Query 187 TTRKKE-VVEAEGEPVKELSFTMSNYFSMGVESRIGRGFDRYRTKSQVFNKMRYGIEGFK 245
+R+KE VV+ G + F M NYFS+GV++RIGRGFDR R++S + NK+ Y +G K
Sbjct 457 KSRQKETVVDESGSDALRMEFVMGNYFSIGVDARIGRGFDRLRSQSSLVNKLIYLWQGTK 516
Query 246 KTF---VTRTKSINEIILDLRIHPNTPEEQTLFTTDKAAAKAQQLPLLNKTASLVVLNIP 302
TF + K I++++ +T+FTT+ P+L ++++L+ LNIP
Sbjct 517 NTFRRSIRVDKQIDKML------SGESYSKTVFTTEMGNLSN---PVLPRSSALIALNIP 567
Query 303 SFSGGNDIWAPSKKLATRMFDKQKRKQNKELLKVPQQMGDEKLEFMTFKNIPSLGMEFML 362
S+S G D +A S ++ + + EL Q+MGD +LEF+ ++ + + +F
Sbjct 568 SYSAGIDPFARSCRVG---LENLTDAECSELTHFSQKMGDHRLEFLAYRKVYHIAADFCG 624
Query 363 HGRGCRLHSGKGPWQISFRSLDEKTRVYFQTDGEYYQLTLPASVFVSHNRRVRVL 417
R+H+ GPW+I F+ L +VYFQ DGE+Y + P V +SH+R +R+L
Sbjct 625 TSLARRVHASGGPWKIVFKELHPSEKVYFQIDGEFYVMLQPKDVEISHSRTIRIL 679
> cpv:cgd4_4340 diacylglycerol kinase ; K00901 diacylglycerol
kinase [EC:2.7.1.107]
Length=503
Score = 291 bits (745), Expect = 4e-78, Method: Compositional matrix adjust.
Identities = 174/488 (35%), Positives = 244/488 (50%), Gaps = 93/488 (19%)
Query 1 LFSNPTSGGNKASAFTRTAVSQLTLTEPCPLKVFIHDIREGPSGSKPGFLLLRSVAAAAA 60
LF NPTSGGNKAS FT++ + ++ P V I+DIR+G +G K GFL ++ A +
Sbjct 24 LFVNPTSGGNKASVFTKSGMDMFRISNPVSASVHIYDIRQGETGKKEGFLKVKETANSMG 83
Query 61 AAAAAAAGEPRAAADGSSSSKKAQHPIVRVLVAGGDGTVMWCIDEMHKTGVQPHVCAVGV 120
+ + ++ AGGDGTVMWC+DE+ KT V +GV
Sbjct 84 DNSIP----------------------LYIIAAGGDGTVMWCVDELEKTSVDFDKLVIGV 121
Query 121 VPYGTGNDFANAFGWRPFNAANPFDASLHTLRNIVEHCMRATVVYHDLWSVKVTLKPGGH 180
+P+GTGNDF+ A W NPFD LH R ++ + A ++ HD+WSVK + G
Sbjct 122 IPFGTGNDFSRALNWMTI-LDNPFDNGLHRFRQLISDWINADIIEHDIWSVKAVVDSYGC 180
Query 181 FNRINSTTRKKEVVEAEGEP--------------------VKELSFTMSNYFSMGVESRI 220
F +++S +RKK+VV +G EL+F MSNYFS+GVESRI
Sbjct 181 FYKVDSYSRKKKVVNKQGRANARQSNHEFSRGIDRGSNTIATELTFRMSNYFSVGVESRI 240
Query 221 GRGFDRYRTKSQVFNKMRYGIEGFKKTFVTRTKSINEIILDLRIHPN---------TPEE 271
G GFDRYRTK+ V NKMRY +EG K TF T + +II ++ E
Sbjct 241 GIGFDRYRTKNAVLNKMRYALEGVKNTF-RHTTPVYDIIDKCTVNTTCNKEDSDLIVSSE 299
Query 272 QTLFTTDKAAA-KAQQL-----------------------------------PLLNKTAS 295
+ LF TD+ A K+ P L AS
Sbjct 300 KVLFVTDQNAKFKSNSCTCSKGGYEFVDTQNSISEHNRQSTGHYQNSVENPSPTLMPCAS 359
Query 296 LVVLNIPSFSGGNDIWAPSKKLATRMFDKQKRKQNKELLKVPQQMGDEKLEFMTFKNIPS 355
L+ +NIPSF+ GNDIW K F K +R + +L Q+MGDE +E ++F + +
Sbjct 360 LIFINIPSFASGNDIWRYCKGNCGIKF-KSRRDSIENMLFEQQKMGDEIIECVSFPSAVA 418
Query 356 LGMEFMLHGRGCRLHSGKGPWQISFR---SLDEKTRVYFQTDGEYYQLTLPASVFVSHNR 412
+G+E + G G R+ KGP ++ F+ SL +VYFQ DGE+ P VSH+R
Sbjct 419 IGLEIAIKGMGRRVFQNKGPVRLHFKQKNSLATGRKVYFQVDGEFLIAYHPLYFDVSHSR 478
Query 413 RVRVLSLK 420
+++VL K
Sbjct 479 KIKVLQGK 486
> tpv:TP02_0475 diacylglycerol kinase
Length=334
Score = 216 bits (551), Expect = 1e-55, Method: Compositional matrix adjust.
Identities = 134/406 (33%), Positives = 193/406 (47%), Gaps = 92/406 (22%)
Query 1 LFSNPTSGGNKASAFTRTAVSQLTLTEPCPLKVFIHDIREGPSGSKPGFLLLRSVAAAAA 60
++ NP SGG AS F + V L LT+P PL + I I EG SG+KPGF+ LR +
Sbjct 6 IYVNPESGGRNASHFIKQGVKSLELTKPEPLTLHICSIFEGESGNKPGFIQLRDFLSYE- 64
Query 61 AAAAAAAGEPRAAADGSSSSKKAQHPIVRVLVAGGDGTVMWCIDEMHKTGVQPHVCAVGV 120
EP+ +V V+ AGGDGT+ W + E+ K + P + G+
Sbjct 65 --------EPQE--------------LVFVITAGGDGTLTWVVSEVEKHSINPDLLCFGI 102
Query 121 VPYGTG-------------NDFANAFGWRPFNAANPFDASLHTLRNIVEHCMRATVVYHD 167
+PYGTG NDFA A W F PF+ + L + M++ + HD
Sbjct 103 IPYGTGWCRKMQNKLFILGNDFARALKWHKFRNLKPFEDNFKPLMTYLNKLMKSDPIKHD 162
Query 168 LWSVKVTLKPGGHFNRINSTTRKKEVVEAEGEPVKELSFTMSNYFSMGVESRIGRGFDRY 227
W V + L PGG FN+INS TR K
Sbjct 163 FWDVYLKLNPGGSFNKINSKTRLK------------------------------------ 186
Query 228 RTKSQVFNKMRYGIEGFKKTFVTRTKSINEIILDLRIHPNTPEEQTLFTTDKAAAKAQQL 287
RY +E FKKTF+ N++ + ++ E+ LF TD K + L
Sbjct 187 ----------RYILEAFKKTFMGLVYVNNQV--ERMVYGEN--EEILFVTD---PKEKNL 229
Query 288 PLLNKTASLVVLNIPSFSGGNDIWAPSKKLATRMFDKQKRKQNKELLKVPQQMGDEKLEF 347
P+L K+ASLV LNIPSFS G D ++ + +A + ++ +K ELL Q MGD+KLEF
Sbjct 230 PVLLKSASLVALNIPSFSSGIDAFSLASGIALKNVSEEYKK---ELLIADQVMGDKKLEF 286
Query 348 MTFKNIPSLGMEFMLHGRGCRLHSGKGPWQISFRSLDEKTRVYFQT 393
++++ +G++ + G+ RLH G GPW+I F+ L + + YFQ
Sbjct 287 FCYRDMKDVGLDILKFGKSKRLHVGGGPWKIIFKDLSKDEKCYFQV 332
> tgo:TGME49_059830 diacylglycerol kinase, putative (EC:2.7.1.107)
Length=628
Score = 150 bits (379), Expect = 9e-36, Method: Compositional matrix adjust.
Identities = 126/417 (30%), Positives = 190/417 (45%), Gaps = 49/417 (11%)
Query 1 LFSNPTSGGNKASAF----TRTAVSQLTLTEPCPLKVFIHDIREGPSGSKPGFLLLRSVA 56
LF N +GG + + R TL + C +V I+D+ SG F LR
Sbjct 211 LFVNRKAGGQAGARWLTGANRNGEDSHTLQDVCT-RVLIYDLLANHSG----FQFLRETV 265
Query 57 AAAAAAAAAAAGEPRAAADGSSSSKKAQHPIVRVLVAGGDGTVMWCIDEMHKTGVQPHVC 116
RA + +KK H +VRV+ GGDGTVMW E V
Sbjct 266 EVK-----------RALSHKQKPTKK--HELVRVIAVGGDGTVMWVNREAVAANVPIAWI 312
Query 117 AVGVVPYGTGNDFANAFGWRPFNAANPFDASLHTLRNIVEHCMRATVVYHDLWSVKVTLK 176
A G+V +GTGNDFA ++GW NA TL+ +V+ A V HD W + V+
Sbjct 313 AFGIVGFGTGNDFAQSYGWTHGNAQKLDIFRGDTLKRLVQMWQNAVVARHDTWRMSVSTY 372
Query 177 PGGHFNRINSTTRKKE-VVEAE-GEPVKELSFTMSNYFSMGVESRIGRGFDRYRTKSQVF 234
GG+F +N TR E + + E G + Y +G ++ +G FDR R+ S++
Sbjct 373 DGGYFEHVNCDTRTLERITDGETGLHARSFESPFILYLGIGFDALVGIEFDRLRSTSRLR 432
Query 235 NKMRYGIEGFKKTFVTRTKSINEIILDLRIHPNTPEEQTLFTTDKAAAKAQQLPLLNKTA 294
N++ YG+ F K K + E I D + +++ LFTT+ K L +
Sbjct 433 NRIMYGV-AFLKLLRRSWKRVTEQI-DTVYVVDRGQKKVLFTTN--TEKYPTAMRLESSI 488
Query 295 SLVVLNIPSFSGGNDIW---------APSKKLATR-MFDKQKRKQNKELLKVPQQMGDEK 344
S+ LN S GG +W P+K+ TR ++D +R+ L+ Q MGD +
Sbjct 489 SMTFLNNRSMIGGIPLWRHTTRVGVLPPAKEGETRTIYDTYRRR----LINAHQSMGDSR 544
Query 345 LEFMTFKN----IPSLGMEFMLHGRGCRLHSGKGPWQISFRSLDEKTRVYFQTDGEY 397
+E + F I S+ +++ G RL +GP+ +SF ++ V FQTDGE+
Sbjct 545 IELIAFHGRFDLIRSVLVDYDAAG---RLLQHRGPFLVSFYPEEQAGPVGFQTDGEF 598
> dre:724010 dgka; zgc:136759 (EC:2.7.1.107); K00901 diacylglycerol
kinase [EC:2.7.1.107]
Length=727
Score = 94.4 bits (233), Expect = 9e-19, Method: Compositional matrix adjust.
Identities = 95/342 (27%), Positives = 148/342 (43%), Gaps = 64/342 (18%)
Query 86 PIVRVLVAGGDGTVMWCIDEMHKTGVQPHVCAVGVVPYGTGNDFANAFGWRPFNAANPFD 145
P R+LV GGDGTV W +D + K + P V V+P GTGND A W +D
Sbjct 429 PNYRILVCGGDGTVGWILDAIDKANL-PVRPPVAVLPLGTGNDLARCLRW-----GGGYD 482
Query 146 ASLHTLRNIVEHCMRATVVYHDLWSVKVTLKPGGHFNRINSTTRKKEVVEAEGEPVKELS 205
L I++ + D WS++VTL E + G+PV
Sbjct 483 G--MDLGRILKDIEVSEEGPMDRWSIQVTL----------------EDSQERGDPVP--Y 522
Query 206 FTMSNYFSMGVESRIGRGFDRYRTK------SQVFNKMRYGIEGFKKTFVTRTKSINEII 259
++NYFS+GV++ I F R K S++ NK+ Y +T K + E +
Sbjct 523 EIINNYFSIGVDASIAHRFHTMREKHPQKFNSRMKNKLWYFEFATSETISASCKKLKECL 582
Query 260 LDLRIHPNTPEEQTLFTTDKAAAKAQQLPLLNKTASLVVLNIPSFSGGNDIWAPSKKLAT 319
T + L L+ + +LNIPS GG+++W +KK +
Sbjct 583 -----------------TIECCGTQLDLSSLS-LEGIAILNIPSMHGGSNLWGEAKK-SD 623
Query 320 RMFDKQKRKQ---NKELLKV-PQQMGDEKLEFMTFKNIPSLGMEFMLHGRGCRLHSGKGP 375
RM QK + + E+LKV PQ M D++LE + + +G + RL
Sbjct 624 RM--DQKLPEVIVDPEILKVSPQDMSDKRLEVVGLEGAMEMGQIYTGLKSAVRLAKTS-- 679
Query 376 WQISFRSLDEKTRVYFQTDGEYYQLTLPASVFVSHNRRVRVL 417
QI+ R+ K + Q DGE + + P ++ ++H + +L
Sbjct 680 -QITIRT---KKPLPMQIDGEPW-MQPPCTIHITHKNQASML 716
> mmu:217480 Dgkb, 6430574F24, 90kda, C630029D13Rik, DAGK2, DGK,
DGK-beta, mKIAA0718; diacylglycerol kinase, beta (EC:2.7.1.107);
K00901 diacylglycerol kinase [EC:2.7.1.107]
Length=795
Score = 93.6 bits (231), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 97/357 (27%), Positives = 150/357 (42%), Gaps = 68/357 (19%)
Query 86 PIVRVLVAGGDGTVMWCIDEMHKTGVQPHVCAVGVVPYGTGNDFANAFGWRPFNAANPFD 145
P RVL GGDGTV W +D + K V H V ++P GTGND A W ++
Sbjct 478 PDFRVLACGGDGTVGWILDCIEKANVVKHP-PVAILPLGTGNDLARCLRW-----GGGYE 531
Query 146 ASLHTLRNIVEHCMRATVVYHDLWSVKVTLKPGGHFNRINSTTRKKEVVEAEGEPVKELS 205
L I++ +T + D W +VT K E +G+PV
Sbjct 532 GE--NLMKILKDIESSTEIMLDRWKFEVT------------PNDKDE----KGDPVP--Y 571
Query 206 FTMSNYFSMGVESRIGRGFDRYRTK------SQVFNKMRYGIEGFKKTFVTRTKSINEII 259
++NYFS+GV++ I F R K S++ NK Y G +TF K ++E +
Sbjct 572 SIINNYFSIGVDASIAHRFHIMREKHPEKFNSRMKNKFWYFEFGTSETFSATCKKLHESV 631
Query 260 LDLRIHPNTPEEQTLFTTDKAAAKAQQLPLLNKT-ASLVVLNIPSFSGGNDIWAPS-KKL 317
+ Q+ L+N + + +LNIPS GG+++W S KK
Sbjct 632 -------------------EIECDGVQIDLINISLEGIAILNIPSMHGGSNLWGESKKKR 672
Query 318 ATRMFDKQKRKQN------KELLKVPQQMGDEKLEFMTFKNIPSLGMEFM-LHGRGCRLH 370
+ R +K+ + KEL Q + D+ LE + + +G + L G RL
Sbjct 673 SHRRIEKKGSDKRPTLTDAKELKFASQDLSDQLLEVVGLEGAMEMGQIYTGLKSAGRRLA 732
Query 371 SGKGPWQISFRSLDEKTRVYFQTDGEYYQLTLPASVFVSHNRRVRVLSLKPPGKMAL 427
Q S + + Q DGE + T P ++ ++H + +L + PP K L
Sbjct 733 ------QCSSVVIRTSKSLPMQIDGEPWMQT-PCTIKITHKNQAPML-MGPPPKTGL 781
> hsa:1607 DGKB, DAGK2, DGK, DGK-BETA, KIAA0718; diacylglycerol
kinase, beta 90kDa (EC:2.7.1.107); K00901 diacylglycerol kinase
[EC:2.7.1.107]
Length=804
Score = 92.0 bits (227), Expect = 3e-18, Method: Compositional matrix adjust.
Identities = 96/357 (26%), Positives = 149/357 (41%), Gaps = 68/357 (19%)
Query 86 PIVRVLVAGGDGTVMWCIDEMHKTGVQPHVCAVGVVPYGTGNDFANAFGWRPFNAANPFD 145
P RVL GGDGTV W +D + K V H V ++P GTGND A W ++
Sbjct 487 PDFRVLACGGDGTVGWVLDCIEKANVGKHP-PVAILPLGTGNDLARCLRW-----GGGYE 540
Query 146 ASLHTLRNIVEHCMRATVVYHDLWSVKVTLKPGGHFNRINSTTRKKEVVEAEGEPVKELS 205
L I++ +T + D W +V K E +G+PV
Sbjct 541 GE--NLMKILKDIENSTEIMLDRWKFEVI------------PNDKDE----KGDPVP--Y 580
Query 206 FTMSNYFSMGVESRIGRGFDRYRTK------SQVFNKMRYGIEGFKKTFVTRTKSINEII 259
++NYFS+GV++ I F R K S++ NK Y G +TF K ++E +
Sbjct 581 SIINNYFSIGVDASIAHRFHIMREKHPEKFNSRMKNKFWYFEFGTSETFSATCKKLHESV 640
Query 260 LDLRIHPNTPEEQTLFTTDKAAAKAQQLPLLNKT-ASLVVLNIPSFSGGNDIWAPSKKLA 318
+ Q+ L+N + + +LNIPS GG+++W SKK
Sbjct 641 -------------------EIECDGVQIDLINISLEGIAILNIPSMHGGSNLWGESKKRR 681
Query 319 T-RMFDKQKRKQN------KELLKVPQQMGDEKLEFMTFKNIPSLGMEFM-LHGRGCRLH 370
+ R +K+ + KEL Q + D+ LE + + +G + L G RL
Sbjct 682 SHRRIEKKGSDKRTTVTDAKELKFASQDLSDQLLEVVGLEGAMEMGQIYTGLKSAGRRLA 741
Query 371 SGKGPWQISFRSLDEKTRVYFQTDGEYYQLTLPASVFVSHNRRVRVLSLKPPGKMAL 427
Q S + + Q DGE + T P ++ ++H + +L + PP K L
Sbjct 742 ------QCSCVVIRTSKSLPMQIDGEPWMQT-PCTIKITHKNQAPML-MGPPPKTGL 790
> mmu:56077 Dgke, C87606, DAGK6, DGK; diacylglycerol kinase, epsilon
(EC:2.7.1.107); K00901 diacylglycerol kinase [EC:2.7.1.107]
Length=564
Score = 91.3 bits (225), Expect = 7e-18, Method: Compositional matrix adjust.
Identities = 77/233 (33%), Positives = 108/233 (46%), Gaps = 54/233 (23%)
Query 88 VRVLVAGGDGTVMWC---IDEMHKTGVQPHVCAVGVVPYGTGNDFANAFGWRPFNAANPF 144
VRVLV GGDGTV W IDEM G + ++ V V+P GTGND +N GW A
Sbjct 269 VRVLVCGGDGTVGWVLDAIDEMKIKGQEKYIPEVAVLPLGTGNDLSNTLGWGTGYAGEIP 328
Query 145 DASLHTLRNIVEHCMRATVVYHDLWSVKVTLKPGGHFNRINSTTRKKEVVEAEGEPVKEL 204
A + LRN++E A + D W V+VT K G++N RK +
Sbjct 329 VAQV--LRNVME----ADGIKLDRWKVQVTNK--GYYN-----LRKPK------------ 363
Query 205 SFTMSNYFSMGVESRIGRGFDRYRTK------SQVFNKMRYGIEGFKKTFVTRTKSINEI 258
FTM+NYFS+G ++ + F +R K S++ NK Y G K V K +N+
Sbjct 364 EFTMNNYFSVGPDALMALNFHAHREKAPSLFSSRILNKAVYLFYGTKDCLVQECKDLNKK 423
Query 259 ILDLRIHPNTPEEQTLFTTDKAAAKAQQLPLLNKTASLVVLNIPSFSGGNDIW 311
I +L + + +LP L ++VLNI + GG +W
Sbjct 424 I-ELELD----------------GERVELPNLE---GIIVLNIGYWGGGCRLW 456
> dre:567728 diacylglycerol kinase beta-like; K00901 diacylglycerol
kinase [EC:2.7.1.107]
Length=893
Score = 90.5 bits (223), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 95/357 (26%), Positives = 147/357 (41%), Gaps = 72/357 (20%)
Query 86 PIVRVLVAGGDGTVMWCIDEMHKTGVQPHVCAVGVVPYGTGNDFANAFGWRPFNAANPFD 145
P RVL GGDG+V W +D + K H V ++P GTGND A W
Sbjct 577 PDFRVLACGGDGSVGWILDCIDKASFARHP-PVAILPLGTGNDLARCLRW----GGGYEG 631
Query 146 ASLHTLRNIVEHCMRATVVYHDLWSVKVTLKPGGHFNRINSTTRKKEVVEAEGEPVKELS 205
SL +EH +T V D W++ + + E +G+PV
Sbjct 632 GSLVKFLRDIEH---STEVLLDRWNIDIV----------------PDDKEEKGDPVP--Y 670
Query 206 FTMSNYFSMGVESRIGRGFDRYRTK------SQVFNKMRYGIEGFKKTFVTRTKSINEII 259
++NYFS+GV++ I F R K S++ NK+ Y G +T K +NE I
Sbjct 671 SIVNNYFSIGVDASIAHRFHLMREKHPEKFNSRMKNKLWYFEFGTTETISATCKKLNETI 730
Query 260 LDLRIHPNTPEEQTLFTTDKAAAKAQQLPLLNKTASLVVLNIPSFSGGNDIWAPSKKLAT 319
E D ++ + + VLNIPS GG+++W +KK
Sbjct 731 ---------EVECDGIILDLSSTSLE---------GIAVLNIPSMHGGSNLWGETKK--R 770
Query 320 RMFDKQKRK-----------QNKELLKVPQQMGDEKLEFMTFKNIPSLGMEFM-LHGRGC 367
R +++ +K KEL Q + D+ LE + + +G + L G
Sbjct 771 RNYNRMSKKVPERMTGSTVTDAKELKFCVQDLSDQLLEVVGLEGAIEMGQIYTGLKSAGR 830
Query 368 RLHSGKGPWQISFRSLDEKTRVYFQTDGEYYQLTLPASVFVSHNRRVRVLSLKPPGK 424
RL Q S ++ + Q DGE + + P ++ ++H +V +L L PP K
Sbjct 831 RLA------QCSNVTIRTSRLLPMQIDGEPW-MQPPCTIRITHKNQVPML-LGPPQK 879
> cpv:cgd8_2390 diacylglycerol kinase
Length=616
Score = 89.0 bits (219), Expect = 3e-17, Method: Compositional matrix adjust.
Identities = 63/223 (28%), Positives = 101/223 (45%), Gaps = 24/223 (10%)
Query 119 GVVPYGTGNDFANAFGWRPFNAANPFDASLHTLRNIVEHCMRATVVYHDLWSVKVTL--K 176
V+P+GTGND+A +FGW + L ++ + ++ D W ++V +
Sbjct 144 AVLPFGTGNDWAKSFGWSSYGNMKFMKDDFSPLIDLARGIFTSNLINFDTWMIEVEIFDS 203
Query 177 PGGHFNRINSTTRKKEVV---EAEGEPVKELSFTMSNYFSMGVESRIGRGFDRYRTKSQV 233
F R+N +++ E V + EG+ + L NYFS G ESR+G FD YR ++ +
Sbjct 204 EDSSFQRVNPISQELERVNNNDDEGKKLLLLKRRCINYFSFGEESRVGITFDTYRKRNVL 263
Query 234 FNKMRYGIEGFKKTFVTRTK---------SINEIILDLRIHPNTPEEQ---------TLF 275
N+ YG+ G T K S +I L+ HPN T+
Sbjct 264 LNRALYGLAGSMFTVNMNNKHQSNTPLSHSTRKIYLEDPEHPNKETNNLECALCPNGTVH 323
Query 276 TTDKAAAKAQQLPLLNKTASLVVLNIPSFSGGNDIWAPSKKLA 318
+++ + + P L + SLV LNI +F GG +W SK +
Sbjct 324 SSEDSQENS-TCPTLLSSVSLVFLNIETFGGGVKLWKNSKNVG 365
Score = 34.3 bits (77), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 14/35 (40%), Positives = 20/35 (57%), Gaps = 0/35 (0%)
Query 388 RVYFQTDGEYYQLTLPASVFVSHNRRVRVLSLKPP 422
VYFQ DGEYY P V ++++++VL P
Sbjct 574 EVYFQIDGEYYLAKEPKQCSVKYDQQIKVLRCIIP 608
> cel:F54G8.2 dgk-3; DiacylGlycerol Kinase family member (dgk-3);
K00901 diacylglycerol kinase [EC:2.7.1.107]
Length=812
Score = 87.4 bits (215), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 84/336 (25%), Positives = 143/336 (42%), Gaps = 51/336 (15%)
Query 90 VLVAGGDGTVMWCIDEMHKTGVQPHVCAVGVVPYGTGNDFANAFGWRPFNAANPFDASLH 149
+LV GGDGT+ W ++ M K V V+P GTGND A W + +LH
Sbjct 480 ILVCGGDGTIGWVLESMDKMTFPHGRPPVAVLPLGTGNDLARCLRW----GGGYENENLH 535
Query 150 TLRNIVEHCMRATVVYHDLWSVKVTLKPGGHFNRINSTTRKKEVVEAEGEPVKELSFTMS 209
I+E +++++ D W +K+ + N + R+ E P + ++
Sbjct 536 ---KILEQIEKSSLIDMDRWQIKIEITE-------NKSARRAS--EKGDTPPYSI---IN 580
Query 210 NYFSMGVESRIGRGFDRYRTK------SQVFNKMRYGIEGFKKTFVTRTKSINEIILDLR 263
NYFS+GV++ I F R K S++ NK+ Y G +T + K+++E I
Sbjct 581 NYFSIGVDASIAHRFHVMREKFPEKFNSRMRNKLWYFELGTSETLSSSCKNLHEQI---- 636
Query 264 IHPNTPEEQTLFTTDKAAAKAQQLPLLNKTASLVVLNIPSFSGGNDIWAPSKKLATRM-- 321
+ ++ Q L + +LNIPS GG+++W S+K RM
Sbjct 637 ---------DILCDGESIDLGQDASL----EGIALLNIPSIYGGSNLWGRSRKSKGRMPG 683
Query 322 FDKQKRKQNKELLKVPQQMGDEKLEFMTFKNIPSLGMEFMLHGRGCRLHSGKGPWQISFR 381
K + +L Q +GD +E + ++ +G + RG R S Q S
Sbjct 684 LFPMKNAEKMQLQTRVQDIGDGLIELVGLESAMQMG-QIKAGVRGARRLS-----QCSTV 737
Query 382 SLDEKTRVYFQTDGEYYQLTLPASVFVSHNRRVRVL 417
+ Q DGE + + P + ++H + ++L
Sbjct 738 VIQTHKSFPMQIDGEPW-MQPPCIIQITHKNQAKML 772
> mmu:110197 Dgkg, 2900055E17Rik, 90kDa, AI854428, Dagk3, E430001K23Rik,
KIAA4131, mKIAA4131; diacylglycerol kinase, gamma
(EC:2.7.1.107); K00901 diacylglycerol kinase [EC:2.7.1.107]
Length=788
Score = 87.0 bits (214), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 90/349 (25%), Positives = 146/349 (41%), Gaps = 64/349 (18%)
Query 86 PIVRVLVAGGDGTVMWCIDEMHKTGVQPHVCAVGVVPYGTGNDFANAFGWRPFNAANPFD 145
P RVL GGDGTV W +D + K H V V+P GTGND A W ++
Sbjct 480 PDFRVLACGGDGTVGWILDCIDKANFTKHP-PVAVLPLGTGNDLARCLRW-----GGGYE 533
Query 146 ASLHTLRNIVEHCMRATVVYHDLWSVKVTLKPGGHFNRINSTTRKKEVVEAEGEPVKELS 205
+L I++ ++ +V D W ++V ++EV + P
Sbjct 534 GG--SLTKILKEIEQSPLVMLDRWYLEV--------------MPREEVENGDQVPYN--- 574
Query 206 FTMSNYFSMGVESRIGRGFDRYRTK------SQVFNKMRYGIEGFKKTFVTRTKSINEII 259
M+NYFS+GV++ I F R K S++ NK+ Y G +TF K +++ I
Sbjct 575 -IMNNYFSIGVDASIAHRFHMMREKHPEKFNSRMKNKLWYFEFGTSETFAATCKKLHDHI 633
Query 260 LDLRIHPNTPEEQTLFTTDKAAAKAQQLPLLNKTASLVVLNIPSFSGGNDIWAPSKKLAT 319
+L + +F + +LNIPS GG ++W +KK
Sbjct 634 -ELECDGVEVDLSNIF-----------------LEGIAILNIPSMYGGTNLWGETKK--N 673
Query 320 RMFDKQKRK---QNKELLKVPQQMGDEKLEFMTFKNIPSLGMEFM-LHGRGCRLHSGKGP 375
R ++ RK KEL Q + D+ LE + + +G + L G RL
Sbjct 674 RAVIRESRKSVTDPKELKCCVQDLSDQLLEVVGLEGAMEMGQIYTGLKSAGRRLA----- 728
Query 376 WQISFRSLDEKTRVYFQTDGEYYQLTLPASVFVSHNRRVRVLSLKPPGK 424
Q S ++ + Q DGE + + ++ ++H + ++ + PP K
Sbjct 729 -QCSSVTIRTNKLLPMQVDGEPW-MQPQCTIKITHKNQAPMM-MGPPQK 774
> hsa:8526 DGKE, DAGK5, DAGK6, DGK; diacylglycerol kinase, epsilon
64kDa (EC:2.7.1.107); K00901 diacylglycerol kinase [EC:2.7.1.107]
Length=567
Score = 86.7 bits (213), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 73/232 (31%), Positives = 106/232 (45%), Gaps = 54/232 (23%)
Query 89 RVLVAGGDGTVMWC---IDEMHKTGVQPHVCAVGVVPYGTGNDFANAFGWRPFNAANPFD 145
RVLV GGDGTV W +D+M G + ++ V V+P GTGND +N GW A
Sbjct 273 RVLVCGGDGTVGWVLDAVDDMKIKGQEKYIPQVAVLPLGTGNDLSNTLGWGTGYAGEIPV 332
Query 146 ASLHTLRNIVEHCMRATVVYHDLWSVKVTLKPGGHFNRINSTTRKKEVVEAEGEPVKELS 205
A + LRN++E A + D W V+VT K G++N RK +
Sbjct 333 AQV--LRNVME----ADGIKLDRWKVQVTNK--GYYN-----LRKPK------------E 367
Query 206 FTMSNYFSMGVESRIGRGFDRYRTK------SQVFNKMRYGIEGFKKTFVTRTKSINEII 259
FTM+NYFS+G ++ + F +R K S++ NK Y G K V K +N+ +
Sbjct 368 FTMNNYFSVGPDALMALNFHAHREKAPSLFSSRILNKAVYLFYGTKDCLVQECKDLNKKV 427
Query 260 LDLRIHPNTPEEQTLFTTDKAAAKAQQLPLLNKTASLVVLNIPSFSGGNDIW 311
+ ++ A LP L ++VLNI + GG +W
Sbjct 428 ------------ELELDGERVA-----LPSLE---GIIVLNIGYWGGGCRLW 459
> dre:100150463 dgke; diacylglycerol kinase epsilon (EC:2.7.1.107);
K00901 diacylglycerol kinase [EC:2.7.1.107]
Length=564
Score = 86.3 bits (212), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 77/234 (32%), Positives = 103/234 (44%), Gaps = 57/234 (24%)
Query 88 VRVLVAGGDGTVMWC---IDEMHKTGVQPHVCAVGVVPYGTGNDFANAFGWRPFNAAN-P 143
VRVLV GGDGTV W ID M G + V ++P GTGND +N+ GW A P
Sbjct 261 VRVLVCGGDGTVGWVLDAIDTMKLKGQDQFIPLVTILPLGTGNDLSNSLGWGAGYAGEIP 320
Query 144 FDASLHTLRNIVEHCMRATVVYHDLWSVKVTLKPGGHFNRINSTTRKKEVVEAEGEPVKE 203
+ LRN++E A VV D W V+V K G +F RK +V
Sbjct 321 VE---QVLRNVLE----AEVVKMDRWKVQVASK-GNYF-------RKPKV---------- 355
Query 204 LSFTMSNYFSMGVESRIGRGFDRYRTK------SQVFNKMRYGIEGFKKTFVTRTKSINE 257
+M+NYFS+G ++ + F +R K S++ NK Y + G K V K
Sbjct 356 --LSMNNYFSVGPDALMALNFHVHREKTPSFFSSRIINKAVYFLYGTKDCLVQECKD--- 410
Query 258 IILDLRIHPNTPEEQTLFTTDKAAAKAQQLPLLNKTASLVVLNIPSFSGGNDIW 311
LD RI EQ LP L ++V NI ++ GG +W
Sbjct 411 --LDKRIELELDGEQV------------TLPNLE---GIIVCNIGNWGGGCRLW 447
> xla:447404 dgke, MGC81643; diacylglycerol kinase, epsilon 64kDa;
K00901 diacylglycerol kinase [EC:2.7.1.107]
Length=552
Score = 85.5 bits (210), Expect = 4e-16, Method: Compositional matrix adjust.
Identities = 73/234 (31%), Positives = 103/234 (44%), Gaps = 57/234 (24%)
Query 88 VRVLVAGGDGTVMWC---IDEMHKTGVQPHVCAVGVVPYGTGNDFANAFGWRPFNAAN-P 143
V+VLV GGDGTV W +DEM G++ V V V+P GTGND AN GW A + P
Sbjct 252 VKVLVCGGDGTVGWVLDAVDEMKIKGLEGCVPQVAVLPLGTGNDLANTLGWGAGYAGDVP 311
Query 144 FDASLHTLRNIVEHCMRATVVYHDLWSVKVTLKPGGHFNRINSTTRKKEVVEAEGEPVKE 203
+ LRNI M A + D W V+VT K + RK +V
Sbjct 312 VE---QILRNI----MDADSIKLDRWKVQVTNK--------GYSLRKPKV---------- 346
Query 204 LSFTMSNYFSMGVESRIGRGFDRYRTK------SQVFNKMRYGIEGFKKTFVTRTKSINE 257
+M+NYFS+G ++ + F +R K S++ NK Y G K V K +N+
Sbjct 347 --LSMNNYFSVGPDALMALNFHTHREKTPSLFSSRLVNKAVYLFYGTKDCLVQECKDLNK 404
Query 258 IILDLRIHPNTPEEQTLFTTDKAAAKAQQLPLLNKTASLVVLNIPSFSGGNDIW 311
+ + +++ L N +VVLNI + GG +W
Sbjct 405 KV-------------------ELELDGERIDLPN-LEGIVVLNIGYWGGGCRLW 438
> ath:AT4G30340 ATDGK7; ATDGK7 (Diacylglycerol kinase 7); diacylglycerol
kinase
Length=492
Score = 84.7 bits (208), Expect = 6e-16, Method: Compositional matrix adjust.
Identities = 68/262 (25%), Positives = 122/262 (46%), Gaps = 38/262 (14%)
Query 73 AADGSSSSKKAQHPIVRVLVAGGDGTVMW---CIDEMHKTGVQPHVCAVGVVPYGTGNDF 129
AA G +++ + I R++VAGGDGTV W C+ E+HK G + H+ VGV+P GTGND
Sbjct 147 AAKGDECARECREKI-RIMVAGGDGTVGWVLGCLGELHKDG-KSHIPPVGVIPLGTGNDL 204
Query 130 ANAFGWRPFNAANPFDASLHTLRNIVEHCMRATVVYHDLWSVKVTLKPGGHFNRINS--- 186
+ +F W + PF A ++ + ++ D W + V++ G + S
Sbjct 205 SRSFSW---GGSFPF-AWRSAMKRTLHRATLGSIARLDSWKIVVSMPSGEVVDPPYSLKP 260
Query 187 ---TTRKKEVVEAEGE-PVKELSF--TMSNYFSMGVESRIGRGFDR------YRTKSQVF 234
T + ++A+G+ P K S+ NYFS+G+++++ GF Y + V
Sbjct 261 TIEETALDQALDADGDVPPKAKSYEGVFYNYFSIGMDAQVAYGFHHLRNEKPYLAQGPVT 320
Query 235 NKMRYG----IEGFKKTFVTRTKSINEIILDLRIHPNTPEEQTLFTTDKAAAKAQQLPLL 290
NK+ Y +G+ T ++ + ++IH ++ +++ +
Sbjct 321 NKIIYSSYSCTQGWFCTPCVNNPALRGLRNIMKIH----------IKKANCSEWEEIHVP 370
Query 291 NKTASLVVLNIPSFSGGNDIWA 312
S+VVLN+ ++ G W
Sbjct 371 KSVRSIVVLNLYNYGSGRHPWG 392
> ath:AT5G63770 ATDGK2; ATDGK2 (Diacylglycerol kinase 2); diacylglycerol
kinase; K00901 diacylglycerol kinase [EC:2.7.1.107]
Length=558
Score = 84.7 bits (208), Expect = 6e-16, Method: Compositional matrix adjust.
Identities = 100/394 (25%), Positives = 156/394 (39%), Gaps = 74/394 (18%)
Query 30 PLKVFIHDIREGPSGSKPGFLLLRSVAAAAAAAAAAAAGEPRAAADGSSSSKKAQHPIVR 89
PL VFI+ SG + G L R + G + G K ++ R
Sbjct 188 PLLVFIN----AKSGGQLGPFLHRRLNMLLNPVQVFELGSCQGPDAGLDLCSKVKY--FR 241
Query 90 VLVAGGDGTVMWCIDEMHKTGVQPHVCAVGVVPYGTGNDFANAFGWRPFNAANPFDASLH 149
VLV GGDGTV W +D + K + V ++P GTGND + W + SL
Sbjct 242 VLVCGGDGTVAWVLDAIEKRNFESPP-PVAILPLGTGNDLSRVLQWGRGISVVDGQGSLR 300
Query 150 TLRNIVEHCMRATVVYHDLWSVKVTLKPGGHFNRINSTTRKKEVVEAEGEPVKELSFTMS 209
T ++H A V D WSVK+ + +T K P +E M
Sbjct 301 TFLQDIDH---AAVTMLDRWSVKI----------VEESTEKF--------PAREGHKFMM 339
Query 210 NYFSMGVESRIGRGFDRYRTK------SQVFNKMRYGIEGFKKTFVTRTKSINEIILDLR 263
NY +G ++++ F R + SQ NK+RY EG + I+D R
Sbjct 340 NYLGIGCDAKVAYEFHMMRQEKPEKFCSQFVNKLRYAKEGARD------------IMD-R 386
Query 264 IHPNTPEEQTLFTTDKAAAKAQQLPLLNKTASLVVLNIPSFSGGNDIWAPSKKLATRMFD 323
+ P + L + K ++P + L+VLNI S+ GG D+W
Sbjct 387 ACADLPWQVWL----EVDGKDIEIP--KDSEGLIVLNIGSYMGGVDLW------------ 428
Query 324 KQKRKQNKELLKVPQQMGDEKLEFMTFKNIPSLGMEFMLHGRGCRLHSGKGPWQISFRSL 383
Q ++ + + Q M D+ LE + + LG + + RL GK +
Sbjct 429 -QNDYEHDDNFSI-QCMHDKTLEVVCVRGAWHLGKLQVGLSQARRLAQGK------VIRI 480
Query 384 DEKTRVYFQTDGEYYQLTLPASVFVSHNRRVRVL 417
+ Q DGE + + P + ++H+ +V +L
Sbjct 481 HVSSPFPVQIDGEPF-IQQPGCLEITHHGQVFML 513
> hsa:1608 DGKG, DAGK3, DGK-GAMMA, MGC104993, MGC133330; diacylglycerol
kinase, gamma 90kDa (EC:2.7.1.107); K00901 diacylglycerol
kinase [EC:2.7.1.107]
Length=766
Score = 84.3 bits (207), Expect = 7e-16, Method: Compositional matrix adjust.
Identities = 89/349 (25%), Positives = 144/349 (41%), Gaps = 64/349 (18%)
Query 86 PIVRVLVAGGDGTVMWCIDEMHKTGVQPHVCAVGVVPYGTGNDFANAFGWRPFNAANPFD 145
P RVL GGDGTV W +D + K H V V+P GTGND A W ++
Sbjct 458 PDFRVLACGGDGTVGWILDCIDKANFAKHP-PVAVLPLGTGNDLARCLRW-----GGGYE 511
Query 146 ASLHTLRNIVEHCMRATVVYHDLWSVKVTLKPGGHFNRINSTTRKKEVVEAEGEPVKELS 205
+L I++ ++ +V D W ++V ++EV + P
Sbjct 512 GG--SLTKILKDIEQSPLVMLDRWHLEV--------------IPREEVENGDQVPYS--- 552
Query 206 FTMSNYFSMGVESRIGRGFDRYRTK------SQVFNKMRYGIEGFKKTFVTRTKSINEII 259
M+NYFS+GV++ I F R K S++ NK+ Y G +TF K +++ I
Sbjct 553 -IMNNYFSIGVDASIAHRFHVMREKHPEKFNSRMKNKLWYFEFGTSETFAATCKKLHDHI 611
Query 260 LDLRIHPNTPEEQTLFTTDKAAAKAQQLPLLNKTASLVVLNIPSFSGGNDIWAPSKKLAT 319
D + L + +LNIPS GG ++W +KK
Sbjct 612 --------------ELECDGVGVDLSNIFL----EGIAILNIPSMYGGTNLWGENKK--N 651
Query 320 RMFDKQKRK---QNKELLKVPQQMGDEKLEFMTFKNIPSLGMEFM-LHGRGCRLHSGKGP 375
R ++ RK KEL Q + D+ LE + + +G + L G RL
Sbjct 652 RAVIRESRKGVTDPKELKFCVQDLSDQLLEVVGLEGAMEMGQIYTGLKSAGRRLA----- 706
Query 376 WQISFRSLDEKTRVYFQTDGEYYQLTLPASVFVSHNRRVRVLSLKPPGK 424
Q + ++ + Q DGE + + ++ ++H + ++ + PP K
Sbjct 707 -QCASVTIRTNKLLPMQVDGEPW-MQPCCTIKITHKNQAPMM-MGPPQK 752
> mmu:13139 Dgka, 80kDa, AW146112, Dagk1; diacylglycerol kinase,
alpha (EC:2.7.1.107); K00901 diacylglycerol kinase [EC:2.7.1.107]
Length=730
Score = 82.0 bits (201), Expect = 4e-15, Method: Compositional matrix adjust.
Identities = 91/356 (25%), Positives = 156/356 (43%), Gaps = 75/356 (21%)
Query 86 PIVRVLVAGGDGTVMWCIDEMHKTGVQPHVCAVGVVPYGTGNDFANAFGW-RPFNAANPF 144
P R+LV GGDGTV W ++ + K V V V+P GTGND A W R + N
Sbjct 420 PQFRILVCGGDGTVGWVLETIDKANFAT-VPPVAVLPLGTGNDLARCLRWGRGYEGEN-- 476
Query 145 DASLHTLRNIVEHCMRATVVYHDLWSVKVTLKPGGHFNRINSTTRKKEVVEAEGEPVKEL 204
LR I++ + VVY D W ++V + G + +PV
Sbjct 477 ------LRKILKDIELSKVVYLDRWFLEVIPQQNGE----------------KSDPVP-- 512
Query 205 SFTMSNYFSMGVESRIGRGFDRYRTKSQVFNKMRYGIEGFKKTFVTRTKSINEIILDLRI 264
S ++NYFS+GV++ I F R K + + F +R K N++
Sbjct 513 SQIINNYFSIGVDASIAHRFHLMREK-------------YPEKFNSRMK--NKL-----W 552
Query 265 HPNTPEEQTLFTTDKAAAKAQQLPLLNKTA--------SLVVLNIPSFSGGNDIWAPSKK 316
+ +++F+T K ++ + + K + VLNIPS GG+++W +K+
Sbjct 553 YFEFATSESIFSTCKKLEESVTVEICGKLLDLSDLSLEGIAVLNIPSTHGGSNLWGDTKR 612
Query 317 -------LATRMFDKQKRKQNKELLK--VPQQMGDEKLEFMTFKNIPSLGMEFM-LHGRG 366
+ + K + ++LK VP M D++LE + + +G + L G
Sbjct 613 PHGDTCEINQALGSAAKIITDPDILKTCVP-DMSDKRLEVVGIEGAIEMGQIYTRLKSAG 671
Query 367 CRLHSGKGPWQISFRSLDEKTRVYFQTDGEYYQLTLPASVFVSHNRRVRVLSLKPP 422
RL +I+F++ + Q DGE + + P ++ ++H ++ +L + PP
Sbjct 672 HRLAKCS---EITFQT---TKTLPMQIDGEPW-MQAPCTIKITHKNQMPML-MGPP 719
> hsa:1606 DGKA, DAGK, DAGK1, DGK-alpha, MGC12821, MGC42356; diacylglycerol
kinase, alpha 80kDa (EC:2.7.1.107); K00901 diacylglycerol
kinase [EC:2.7.1.107]
Length=735
Score = 81.3 bits (199), Expect = 8e-15, Method: Compositional matrix adjust.
Identities = 89/353 (25%), Positives = 152/353 (43%), Gaps = 68/353 (19%)
Query 86 PIVRVLVAGGDGTVMWCIDEMHKTGVQPHVCAVGVVPYGTGNDFANAFGWRPFNAANPFD 145
P R+LV GGDGTV W ++ + K + P + V V+P GTGND A W ++
Sbjct 425 PDSRILVCGGDGTVGWILETIDKANL-PVLPPVAVLPLGTGNDLARCLRW-----GGGYE 478
Query 146 ASLHTLRNIVEHCMRATVVYHDLWSVKVTLKPGGHFNRINSTTRKKEVVEAEGEPVKELS 205
L I++ + VV+ D WSV+V + E + +PV
Sbjct 479 G--QNLAKILKDLEMSKVVHMDRWSVEVI----------------PQQTEEKSDPVP--F 518
Query 206 FTMSNYFSMGVESRIGRGFDRYRTK------SQVFNKMRYGIEGFKKTFVTRTKSINEII 259
++NYFS+GV++ I F R K S++ NK+ Y ++ + K + E +
Sbjct 519 QIINNYFSIGVDASIAHRFHIMREKYPEKFNSRMKNKLWYFEFATSESIFSTCKKLEESL 578
Query 260 LDLRIHPNTPEEQTLFTTDKAAAKAQQLPLLNKTASLVVLNIPSFSGGNDIWAPSKKLAT 319
T + K L L+ + VLNIPS GG+++W +++
Sbjct 579 -----------------TVEICGKPLDLSNLS-LEGIAVLNIPSMHGGSNLWGDTRRPHG 620
Query 320 RMFD-------KQKRKQNKELLK--VPQQMGDEKLEFMTFKNIPSLGMEFM-LHGRGCRL 369
++ K + ++LK VP + D++LE + + +G + L G RL
Sbjct 621 DIYGINQALGATAKVITDPDILKTCVP-DLSDKRLEVVGLEGAIEMGQIYTKLKNAGRRL 679
Query 370 HSGKGPWQISFRSLDEKTRVYFQTDGEYYQLTLPASVFVSHNRRVRVLSLKPP 422
+I+F + + Q DGE + T P ++ ++H ++ +L PP
Sbjct 680 AKCS---EITFHT---TKTLPMQIDGEPWMQT-PCTIKITHKNQMPMLMGPPP 725
> ath:AT5G57690 diacylglycerol kinase
Length=487
Score = 79.7 bits (195), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 90/361 (24%), Positives = 158/361 (43%), Gaps = 48/361 (13%)
Query 73 AADGSSSSKKAQHPIVRVLVAGGDGTVMW---CIDEMHKTGVQPHVCAVGVVPYGTGNDF 129
A+ G +K+ + + R++VAGGDGTV W C+ E++ P V V ++P GTGND
Sbjct 143 ASRGDECAKEIREKM-RIVVAGGDGTVGWVLGCLGELNLQNRLP-VPPVSIMPLGTGNDL 200
Query 130 ANAFGWRPFNAANPFDASLHTLRNIVEHCMRATVVYHDLWSVKVTLKPGGHFNRINSTTR 189
+ +FGW + PF A ++ + A + D W++ +T+ G + S
Sbjct 201 SRSFGW---GGSFPF-AWKSAIKRTLHRASVAPISRLDSWNILITMPSGEIVDPPYSLKA 256
Query 190 KKEV-----VEAEGE---PVKELSFTMSNYFSMGVESRIGRGFDRYRTK------SQVFN 235
+E +E EGE NYFS+G+++++ GF R + + N
Sbjct 257 TQECYIDQNLEIEGEIPPSTNGYEGVFYNYFSIGMDAQVAYGFHHLRNEKPYLANGPIAN 316
Query 236 KMRYGIEGFKKTFVTRTKSINEIILDLRIHPNTPEEQTLFTTDKAAAKAQQLPLLNKTAS 295
K+ Y G + + T IN+ LR N TL +++ +++P+ +
Sbjct 317 KIIYSGYGCSQGWFL-THCINDP--GLRGLKNI---MTLHIKKLDSSEWEKVPVPKSVRA 370
Query 296 LVVLNIPSFSGGNDIWAPSKK--LATRMFDKQKRKQNKELLKVPQQMGDEKLEFMTFKNI 353
+V LN+ S+ G + W K+ L R F + + + LL++ G ++ +F +
Sbjct 371 VVALNLHSYGSGRNPWGNLKQDYLEKRGF--VEAQADDGLLEI---FGLKQGWHASFVMV 425
Query 354 PSLGMEFMLHGRGCRLHSGKGPWQISFRSLDEKTRVYFQTDGEYYQ--LTLPASVFVSHN 411
+ + + RL G W+ +F Q DGE ++ +T S FV
Sbjct 426 ELISAKHIAQAAAIRLEIRGGDWKDAF----------MQMDGEPWKQPMTRDYSTFVDIK 475
Query 412 R 412
R
Sbjct 476 R 476
> dre:553380 dgki; diacylglycerol kinase, zeta-like; K00901 diacylglycerol
kinase [EC:2.7.1.107]
Length=1086
Score = 78.2 bits (191), Expect = 6e-14, Method: Compositional matrix adjust.
Identities = 107/417 (25%), Positives = 156/417 (37%), Gaps = 108/417 (25%)
Query 1 LFSNPTSGGNKASAFTRTAVSQLTLTEPCPLKVFIHDIREGPSGSKPGFLLLRSVAAAAA 60
+F NP SGGN+ + +T + L P +VF D+ +G G + G + R V
Sbjct 366 VFVNPKSGGNQGAKIIQTFLWYLN-----PRQVF--DLSQG--GPQEGLEMYRKVHN--- 413
Query 61 AAAAAAAGEPRAAADGSSSSKKAQHPIVRVLVAGGDGTVMWCIDEMHKTGVQPHVCAVGV 120
+R+L GGDGTV W + + + + P AV V
Sbjct 414 ---------------------------LRILACGGDGTVGWILSALDQLQLNPSP-AVAV 445
Query 121 VPYGTGNDFANAFGWRPFNAANPFDASLHTLRNIVEHCMRATVVYHDLWSVKVTLKP-GG 179
+P GTGND A W P L I+ H +V D W++ V P G
Sbjct 446 LPLGTGNDLARTLNWGGGYTDEP-------LSKILSHVEDGNIVQLDRWNLVVKPNPEAG 498
Query 180 HFNRINSTTRKKEVVEAEGEPVKELSFTMSNYFSMGVESRIGRGFDRYRT------KSQV 233
R T K P+ +NYFS+G ++ + F R S+
Sbjct 499 PEERDEQVTDKL--------PLD----VFNNYFSLGFDAHVTLEFHESREANPEKFNSRF 546
Query 234 FNKMRYGIEGFKKTFVTRTKSINEIILDLRIHPNTPEEQTLFTTDKAAAKAQQLPLLNKT 293
NKM Y F + +K DL H + T T+ K Q L K
Sbjct 547 RNKMFYAGTAFSDFLMGSSK-------DLAKHIKVVCDGTDLTS-----KVQDL----KL 590
Query 294 ASLVVLNIPSFSGGNDIWA-PSKKLATRMFDKQKRKQNKELLKVPQQMGDEKLEFMTFKN 352
LV LNIP + G W PS+ F+ PQ+ D +E + F
Sbjct 591 QCLVFLNIPRYCAGTMPWGNPSEH---HDFE-------------PQRHDDGCIEVIGF-T 633
Query 353 IPSLGMEFMLHGRGCRLHSGKGPWQISFRSLDEKTRVYFQTDGEYYQLTLPASVFVS 409
+ SL + G G RL+ + +F+S + Q DGE +L P+ + +S
Sbjct 634 MTSLAT-LQVGGHGERLNQCREVTLTTFKS------IPMQVDGEPCKLA-PSVIHIS 682
> tgo:TGME49_039250 diacylglycerol kinase, putative (EC:2.7.1.107
2.7.11.13 3.4.21.72)
Length=1841
Score = 75.9 bits (185), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 79/286 (27%), Positives = 108/286 (37%), Gaps = 115/286 (40%)
Query 88 VRVLVAGGDGTVMWCIDEMHKT-------------------------------------- 109
+RVLV GGDGTV W ID +HK
Sbjct 864 LRVLVCGGDGTVGWIIDSIHKVYGAEAAEEERGSEAQTGDEVDSGEAAGKVGVEGEGRRG 923
Query 110 ---------GVQPHVC------AVGVVPYGTGNDFANAFGWRPFNAANPFDASLHTLRNI 154
G + C VG+ P GTGND +N GW FD +I
Sbjct 924 RRESEGVAWGSRDRACDLRSLVPVGICPLGTGNDLSNVLGW-----GFSFDG------DI 972
Query 155 VEHCMR---ATVVYHDLWSVKVTLKPGGHFNRINSTTRKKEVVEAEGEPVKELSFTMSNY 211
++H ++ A DLW VKV ++ N+T +VE T SNY
Sbjct 973 MKHLLKIQSAVSSTLDLWKVKVI------SDKTNAT-----LVET----------TFSNY 1011
Query 212 FSMGVESRIGRGFDRYRT------KSQVFNKMRYGIEGFKKTFVTRTKSINEIILDLRIH 265
+GV +RI F + R +S++ NK YG GF+ VT N + L+I
Sbjct 1012 LDVGVAARIVLKFHKLREENPELFQSRLGNKFLYGEVGFRDFLVT----PNIALRGLKI- 1066
Query 266 PNTPEEQTLFTTDKAAAKAQQLPLLNKTASLVVLNIPSFSGGNDIW 311
F + A LP L + V+NIPSF+GG ++W
Sbjct 1067 ---------FCDGQEIA----LPYLE---GICVVNIPSFAGGVELW 1096
> ath:AT2G18730 diacylglycerol kinase, putative
Length=488
Score = 74.7 bits (182), Expect = 7e-13, Method: Compositional matrix adjust.
Identities = 66/264 (25%), Positives = 118/264 (44%), Gaps = 39/264 (14%)
Query 71 RAAADGSSSSKKAQHPIVRVLVAGGDGTVMW---CIDEMHKTGVQPHVCAVGVVPYGTGN 127
+ AA+G +K+ + + R++VAGGDGTV W C+ E++K G + + VGV+P GTGN
Sbjct 142 KVAAEGDECAKECRARL-RIMVAGGDGTVGWVLGCLGELNKDG-KSQIPPVGVIPLGTGN 199
Query 128 DFANAFGWRPFNAANPFDASLHTLRNIVEHCMRATVVYHDLWSVKVTLKPGGHFNRINST 187
D + +FGW + PF A ++ + V D W + V++ G + S
Sbjct 200 DLSRSFGW---GGSFPF-AWRSAVKRTLHRASMGPVARLDSWKILVSMPSGEVVDPPYSL 255
Query 188 TRKKEVVEAEG-------EPV-KELSFTMSNYFSMGVESRIGRGFDR------YRTKSQV 233
+E +G P+ K NY S+G+++++ GF Y + +
Sbjct 256 KPAEENELDQGLDAGIDAPPLAKAYEGVFYNYLSIGMDAQVAYGFHHLRNTKPYLAQGPI 315
Query 234 FNKMRYGIEGFKKTFVTRTKSINEIILD-----LRIHPNTPEEQTLFTTDKAAAKAQQLP 288
NK+ Y G + + T +N+ L ++IH ++ +++
Sbjct 316 SNKIIYSSFGCSQGWFC-TPCVNDPGLRGLRNIMKIH----------IKKVNCSQWEEIA 364
Query 289 LLNKTASLVVLNIPSFSGGNDIWA 312
+ S+V LN+ S+ G+ W
Sbjct 365 VPKNVRSIVALNLHSYGSGSHPWG 388
> ath:AT2G20900 diacylglycerol kinase, putative
Length=509
Score = 73.6 bits (179), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 89/357 (24%), Positives = 143/357 (40%), Gaps = 55/357 (15%)
Query 88 VRVLVAGGDGTVMWCIDEMHKTGVQPHVCAVGVVPYGTGNDFANAFGWRPFNAANPFDAS 147
++++VAGGDGT W + + + H + VP GTGN+ AFGW N A
Sbjct 105 LKIIVAGGDGTAGWLLGVVCDLKLS-HPPPIATVPLGTGNNLPFAFGWGKKNPGTDRTA- 162
Query 148 LHTLRNIVEHCMRATVVYHDLWSVKV---TLKPGGHFN---------RINSTTRKKEVVE 195
+ + +E ++A V+ D W + + T K GG + +++ R E
Sbjct 163 ---VESFLEQVLKAKVMKIDNWHILMRMKTPKEGGSCDPVAPLELPHSLHAFHRVSPTDE 219
Query 196 AEGEPVKELSFTMSNYFSMGVESRIGRGFDRYRT------KSQVFNKMRYGIEGFKKTFV 249
E NYFS+G++++I F R K+Q+ N+ Y G + +
Sbjct 220 LNKEGCHTFRGGFWNYFSLGMDAQISYAFHSERKLHPEKFKNQLVNQSTYVKLGCTQGWF 279
Query 250 TRTKSINEIILDLRIHPNTPEEQTLFTTDKAA--AKAQQLPLLNKTASLVVLNIPSFSGG 307
+ HP + L A + Q L + + S+V LN+PSFSGG
Sbjct 280 CAS----------LFHPASRNIAQLAKVKIATRNGQWQDLHIPHSIRSIVCLNLPSFSGG 329
Query 308 NDIWAPSKKLATRMFDKQKRKQNKELLKVPQQMGDEKLEFMTFKNIPSLGMEFMLHGRGC 367
+ W RKQ L P + D +E + F+N + +G G
Sbjct 330 LNPWGTP----------NPRKQRDRGL-TPPFVDDGLIEVVGFRNAWHGLVLLAPNGHGT 378
Query 368 RLHSGKGPWQISFRSLDEKT-RVYFQTDGEYYQLTLPAS-----VFVSHNRRVRVLS 418
RL +I F T + + DGE ++ LP V +SH +V +L+
Sbjct 379 RLAQAN---RIRFEFHKGATDHTFMRMDGEPWKQPLPLDDETVMVEISHLGQVNMLA 432
> hsa:9162 DGKI, DGK-IOTA; diacylglycerol kinase, iota (EC:2.7.1.107);
K00901 diacylglycerol kinase [EC:2.7.1.107]
Length=1065
Score = 72.8 bits (177), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 108/416 (25%), Positives = 153/416 (36%), Gaps = 107/416 (25%)
Query 1 LFSNPTSGGNKASAFTRTAVSQLTLTEPCPLKVFIHDI-REGPSGSKPGFLLLRSVAAAA 59
+F NP SGGN+ T V Q+ + P +VF D+ +EGP K L R V
Sbjct 379 VFVNPKSGGNQG-----TKVLQMFMWYLNPRQVF--DLSQEGP---KDALELYRKV---- 424
Query 60 AAAAAAAAGEPRAAADGSSSSKKAQHPIVRVLVAGGDGTVMWCIDEMHKTGVQPHVCAVG 119
P +R+L GGDGTV W + + + + P VG
Sbjct 425 --------------------------PNLRILACGGDGTVGWILSILDELQLSPQP-PVG 457
Query 120 VVPYGTGNDFANAFGWRPFNAANPFDASLHTLRNIVEHCMRATVVYHDLWSVKVTLKPGG 179
V+P GTGND A W P L + + TVV D W++ V P
Sbjct 458 VLPLGTGNDLARTLNWGGGYTDEPVSKILCQVED-------GTVVQLDRWNLHVERNP-- 508
Query 180 HFNRINSTTRKKEVVEAEGEPVKELSFTMSNYFSMGVESRIGRGFDRYRT------KSQV 233
E E K +NYFS+G ++ + F R S+
Sbjct 509 ----------DLPPEELEDGVCKLPLNVFNNYFSLGFDAHVTLEFHESREANPEKFNSRF 558
Query 234 FNKMRYGIEGFKKTFVTRTKSINEIILDLRIHPNTPEEQTLFTTDKAAAKAQQLPLLNKT 293
NKM Y F F+ R+ DL H + T T K Q+L K
Sbjct 559 RNKMFYAGAAF-SDFLQRSSR------DLSKHVKVVCDGTDLT-----PKIQEL----KF 602
Query 294 ASLVVLNIPSFSGGNDIWAPSKKLATRMFDKQKRKQNKELLKVPQQMGDEKLEFMTFKNI 353
+V LNIP + G W F+ PQ+ D +E + F +
Sbjct 603 QCIVFLNIPRYCAGTMPWGNPGD--HHDFE-------------PQRHDDGYIEVIGF-TM 646
Query 354 PSLGMEFMLHGRGCRLHSGKGPWQISFRSLDEKTRVYFQTDGEYYQLTLPASVFVS 409
SL + G G RLH + ++++S + Q DGE +L PA + +S
Sbjct 647 ASLAA-LQVGGHGERLHQCREVMLLTYKS------IPMQVDGEPCRLA-PAMIRIS 694
> mmu:320127 Dgki, C130010K08Rik; diacylglycerol kinase, iota
(EC:2.7.1.107); K00901 diacylglycerol kinase [EC:2.7.1.107]
Length=1071
Score = 70.9 bits (172), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 107/417 (25%), Positives = 153/417 (36%), Gaps = 109/417 (26%)
Query 1 LFSNPTSGGNKASAFTRTAVSQLTLTEPCPLKVFIHDI-REGPSGSKPGFLLLRSVAAAA 59
+F NP SGGN+ T V Q+ + P +VF D+ +EGP K + R V
Sbjct 374 VFVNPKSGGNQG-----TKVLQMFMWYLNPRQVF--DLSQEGP---KDALEMYRKV---- 419
Query 60 AAAAAAAAGEPRAAADGSSSSKKAQHPIVRVLVAGGDGTVMWCIDEMHKTGVQPHVCAVG 119
P +R+L GGDGTV W + + + + P VG
Sbjct 420 --------------------------PNLRILACGGDGTVGWILSILDELQLSPQP-PVG 452
Query 120 VVPYGTGNDFANAFGWRPFNAANPFDASLHTLRNIVEHCMRATVVYHDLWSVKVTLKPGG 179
V+P GTGND A W P L + + T+V D W++ V P
Sbjct 453 VLPLGTGNDLARTLNWGGGYTDEPVSKILCQVED-------GTIVQLDRWNLHVERNP-- 503
Query 180 HFNRINSTTRKKEVVEAEGEPVKELSFTMSNYFSMGVESRIGRGFDRYRT------KSQV 233
E E K +NYFS+G ++ + F R S+
Sbjct 504 ----------DLPPEELEDGVCKLPLNVFNNYFSLGFDAHVTLEFHESREANPEKFNSRF 553
Query 234 FNKMRYGIEGFKKTFVTRTKSINEIILDLRIHPNTPEEQTLFTTDKAAAKAQQLPLLNKT 293
NKM Y F F+ R+ DL H + T T K Q L K
Sbjct 554 RNKMFYAGAAFSD-FLQRSSR------DLSKHVKVVCDGTDLT-----PKIQDL----KF 597
Query 294 ASLVVLNIPSFSGGNDIWA-PSKKLATRMFDKQKRKQNKELLKVPQQMGDEKLEFMTFKN 352
+V LNIP + G W P F+ PQ+ D +E + F
Sbjct 598 QCIVFLNIPRYCAGTMPWGNPGDH---HDFE-------------PQRHDDGYIEVIGF-T 640
Query 353 IPSLGMEFMLHGRGCRLHSGKGPWQISFRSLDEKTRVYFQTDGEYYQLTLPASVFVS 409
+ SL + G G RLH + ++++S + Q DGE +L PA + +S
Sbjct 641 MASLAA-LQVGGHGERLHQCREVMLLTYKS------IPMQVDGEPCRLA-PAMIRIS 689
> hsa:1609 DGKQ, DAGK, DAGK4, DAGK7; diacylglycerol kinase, theta
110kDa (EC:2.7.1.107); K00901 diacylglycerol kinase [EC:2.7.1.107]
Length=942
Score = 70.1 bits (170), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 72/264 (27%), Positives = 120/264 (45%), Gaps = 58/264 (21%)
Query 83 AQHPIVRVLVAGGDGTVMWCIDEMHKTGVQPHVC---AVGVVPYGTGNDFANAFGWRP-F 138
+Q P RVLV GGDGTV W + + +T + C +V ++P GTGND W +
Sbjct 634 SQVPCFRVLVCGGDGTVGWVLGALEETRYR-LACPEPSVAILPLGTGNDLGRVLRWGAGY 692
Query 139 NAANPFDASLHTLRNIVEHCMRATVVYHDLWSVKVTLKPGGHFNRINSTTRKKEVVEAEG 198
+ +PF L ++ E A V D W++ + G N T A+
Sbjct 693 SGEDPFS----VLLSVDE----ADAVLMDRWTILLDAHEAGSAE--NDT--------ADA 734
Query 199 EPVKELSFTMSNYFSMGVESRIGRGFDRYRTK------SQVFNKMRYGIEGFKKTFVTRT 252
EP K + MSNY +G+++ + F + R + S++ NK Y G +K ++ +
Sbjct 735 EPPKIVQ--MSNYCGIGIDAELSLDFHQAREEEPGKFTSRLHNKGVYVRVGLQK--ISHS 790
Query 253 KSINEIILDLRIHPNTPEEQTLFTTDKAAAKAQQLPLLNKTASLVVLNIPSFSGGNDIWA 312
+S+++ I L++ E Q + +LP + L+ +NIPS+ G D+W
Sbjct 791 RSLHKQI-RLQV-----ERQEV-----------ELPSIE---GLIFINIPSWGSGADLWG 830
Query 313 PSKKLATRMFDKQKRKQNKELLKV 336
TR +K + + LL+V
Sbjct 831 SDSD--TRF---EKPRMDDGLLEV 849
> cel:C09E10.2 dgk-1; DiacylGlycerol Kinase family member (dgk-1);
K00901 diacylglycerol kinase [EC:2.7.1.107]
Length=794
Score = 69.7 bits (169), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 72/252 (28%), Positives = 107/252 (42%), Gaps = 60/252 (23%)
Query 86 PIVRVLVAGGDGTVMW---CIDEMHKTGVQPHVC---AVGVVPYGTGNDFANAFGWRP-- 137
P ++L GGDGT+ W C+D Q C G+VP GTGND A W
Sbjct 439 PKYKILACGGDGTIGWVLQCLD----IAKQDAACFSPPCGIVPLGTGNDLARVLRWGGGY 494
Query 138 FNAANPFDASLHTLRNIVEHCMRATVVYHDLWSVKVTLKPGGHFNRINSTTRKKEVVEAE 197
NP D L++++E A V D W+V + N+ S+ + E+ E
Sbjct 495 TGEENPMD----ILKDVIE----ADTVKLDRWAVVFHEE---ERNQPTSSGNQTEMNEQT 543
Query 198 -GEPVKELS-FTMSNYFSMGVESRIGRGFDRYRT------KSQVFNKMRYGIEGFKKTFV 249
P + S M+NYF +G+++ + F R +S++FNK +Y G +K F
Sbjct 544 MNNPEDQTSMIIMNNYFGIGIDADVCLKFHNKRDANPEKFQSRLFNKTQYAKIGLQKMFF 603
Query 250 TRT-----KSINEIILDLRIHPNTPEEQTLFTTDKAAAKAQQLPLLNKTASLVVLNIPSF 304
RT K I E+ +D RI +LP +VVLN+ S+
Sbjct 604 ERTCKDLWKRI-ELEVDGRI--------------------IELP---NIEGIVVLNLLSW 639
Query 305 SGGNDIWAPSKK 316
G + W SK+
Sbjct 640 GSGANPWGTSKE 651
> cel:F46H6.2 dgk-2; DiacylGlycerol Kinase family member (dgk-2)
Length=536
Score = 68.9 bits (167), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 77/305 (25%), Positives = 123/305 (40%), Gaps = 70/305 (22%)
Query 43 SGSKPGFLLLRSVAAAAAAAAAAAAGEPRAAADGSSSSKKAQHP--IVRVLVAGGDGTVM 100
SGS G LLR+ A A + AA S +HP VR+L+AGGDGT+
Sbjct 212 SGSGAGKQLLRNFRAHLHPAQVVDVLKSNIAA---SLRWIDEHPNVDVRILIAGGDGTIC 268
Query 101 WCIDEMHKTGVQPHVCAVGVVPYGTGNDFANAFGWRPFNAANPFDASLHTLRNIVEHCMR 160
+D++ + V V+P GTGND + W D + ++ ++E
Sbjct 269 SALDQIDTLSRRI---PVAVLPLGTGNDLSRLLKW-----GKKCDGDIDVIK-LMEDIQE 319
Query 161 ATVVYHDLWSVKVTLKPGGHFNRINSTTRKKEVVEAEGEPVKELSFTMSNYFSMGVESRI 220
A V D W+ I++ ++KK V + + +M+NY S+GV++ +
Sbjct 320 AEVTLVDRWT-------------IDAESQKKLGVRLQSNK----TLSMTNYVSVGVDACV 362
Query 221 GRGFDRYR------TKSQVFNKMRYGIEGFKKTFVTRTKSINEII---LDLRIHPNTPEE 271
G R S++ NK + G K F K +NE I LD +H N P+
Sbjct 363 TLGMQNTRESIPRAMSSRLLNKFLFFTFGTKDVFERVCKGLNERIDLYLD-DVHVNLPDI 421
Query 272 QTLFTTDKAAAKAQQLPLLNKTASLVVLNIPSFSGGNDIWAPSKKLATRMFDKQKRKQNK 331
+ L+ LNIP + G WA D +++ +
Sbjct 422 E----------------------GLIFLNIPYWGAGVKPWATYN-------DSHRQECDD 452
Query 332 ELLKV 336
E+++V
Sbjct 453 EMIEV 457
> ath:AT5G07920 DGK1; DGK1 (DIACYLGLYCEROL KINASE1); calcium ion
binding / diacylglycerol kinase; K00901 diacylglycerol kinase
[EC:2.7.1.107]
Length=728
Score = 68.2 bits (165), Expect = 6e-11, Method: Compositional matrix adjust.
Identities = 81/339 (23%), Positives = 127/339 (37%), Gaps = 73/339 (21%)
Query 86 PIVRVLVAGGDGTVMWCIDEMHKTG-VQPHVCAVGVVPYGTGNDFANAFGWRPFNAANPF 144
P RVLV GGDGT W +D + K + P AV ++P GTGND + W +
Sbjct 411 PHFRVLVCGGDGTAGWVLDAIEKQNFISPP--AVAILPAGTGNDLSRVLNWGGGLGSVER 468
Query 145 DASLHTLRNIVEHCMRATVVYHDLWSVKVTLKPGGHFNRINSTTRKKEVVEAEGEPVKEL 204
L T+ +EH A V D W V + + G +P K
Sbjct 469 QGGLSTVLQNIEH---AAVTVLDRWKVSILNQQGKQL-----------------QPPK-- 506
Query 205 SFTMSNYFSMGVESRIGRGFDRYRTK------SQVFNKMRYGIEGFKKTFVTRTKSINEI 258
M+NY +G ++++ R + SQ NK+ Y EG +
Sbjct 507 --YMNNYIGVGCDAKVALEIHNLREENPERFYSQFMNKVLYAREGARS------------ 552
Query 259 ILDLRIHPNTPEEQTLFTTDKAAAKAQQLPLLNKTASLVVLNIPSFSGGNDIWAPSKKLA 318
I+D R + P + + + + ++V NI S+ GG D+W
Sbjct 553 IMD-RTFEDFPWQV------RVEVDGVDIEVPEDAEGILVANIGSYMGGVDLW------- 598
Query 319 TRMFDKQKRKQNKELLKVPQQMGDEKLEFMTFKNIPSLGMEFMLHGRGCRLHSGKGPWQI 378
Q + E PQ M D+ +E ++ LG + R RL G
Sbjct 599 ------QNEDETYENFD-PQSMHDKIVEVVSISGTWHLGKLQVGLSRARRLAQG------ 645
Query 379 SFRSLDEKTRVYFQTDGEYYQLTLPASVFVSHNRRVRVL 417
S + + Q DGE + P ++ +SH+ + +L
Sbjct 646 SAVKIQLCAPLPVQIDGEPWN-QQPCTLTISHHGQAFML 683
> mmu:110524 Dgkq, 110kDa, DAGK, DAGK7, Dagk4, Dgkd; diacylglycerol
kinase, theta (EC:2.7.1.107); K00901 diacylglycerol kinase
[EC:2.7.1.107]
Length=934
Score = 68.2 bits (165), Expect = 6e-11, Method: Compositional matrix adjust.
Identities = 64/242 (26%), Positives = 112/242 (46%), Gaps = 57/242 (23%)
Query 83 AQHPIVRVLVAGGDGTVMWCIDEMHKTGVQPHVC----AVGVVPYGTGNDFANAFGWRP- 137
+Q P RVLV GGDGTV W + + +T + H+ +V ++P GTGND W
Sbjct 626 SQVPSFRVLVCGGDGTVGWVLAALEET--RRHLACPEPSVAILPLGTGNDLGRVLRWGAG 683
Query 138 FNAANPFDASLHTLRNIVEHCMRATVVYHDLWSVKVTLKPGGHFNRINSTTRKKEVVEAE 197
++ +PF +++ A V D W++ + + I+ST + VVE E
Sbjct 684 YSGEDPF--------SVLVSVDEADAVLMDRWTILLDA------HEIDST--ENNVVETE 727
Query 198 GEPVKELSFTMSNYFSMGVESRIGRGFDRYRTK------SQVFNKMRYGIEGFKKTFVTR 251
+ + M+NY +G+++ + F + R + S+ NK Y G +K ++
Sbjct 728 PPKIVQ----MNNYCGIGIDAELSLDFHQAREEEPGKFTSRFHNKGVYVRVGLQK--ISH 781
Query 252 TKSIN-EIILDLRIHPNTPEEQTLFTTDKAAAKAQQLPLLNKTASLVVLNIPSFSGGNDI 310
++S++ EI L + E+Q + +LP + L+ +NIPS+ G D+
Sbjct 782 SRSLHKEIRLQV-------EQQEV-----------ELPSIE---GLIFINIPSWGSGADL 820
Query 311 WA 312
W
Sbjct 821 WG 822
> cel:K06A1.6 dgk-5; DiacylGlycerol Kinase family member (dgk-5);
K00901 diacylglycerol kinase [EC:2.7.1.107]
Length=937
Score = 67.8 bits (164), Expect = 7e-11, Method: Compositional matrix adjust.
Identities = 70/268 (26%), Positives = 105/268 (39%), Gaps = 67/268 (25%)
Query 1 LFSNPTSGGNKASAFTRTAVSQLTLTEPCPLKVFIHDIREGPSGSKPGFLLLRSVAAAAA 60
+F NP SGGNK S T L P +VF +GP K G + R V
Sbjct 421 VFVNPKSGGNKGSKALHTLCWLLN-----PRQVFDITSLKGP---KFGLEMFRKVVTQ-- 470
Query 61 AAAAAAAGEPRAAADGSSSSKKAQHPIVRVLVAGGDGTVMWCIDEMHKTGVQPHVCAVGV 120
+R+LV GGDGTV W + + P + +
Sbjct 471 ---------------------------LRILVCGGDGTVGWVLSTLDNLN-WPAYPPMAI 502
Query 121 VPYGTGNDFANAFGWRPFNAANPFDASLHTLRNIVEHCMRATVVYH-DLWSVKVTLKPGG 179
+P GTGND A GW + P + +++ + T+V H D W + V +P
Sbjct 503 MPLGTGNDLARCMGWGGVFSDEP-------ISQLMQAILHETIVTHLDRWRIDV--EPNT 553
Query 180 HFNRINSTTRKKEVVEAEGEPVKE-LSFT-MSNYFSMGVESRIGRGFDRYRT------KS 231
N +E E + ++ L T M+NYFS+G ++ + F R+ S
Sbjct 554 SCN-----------LEEEDDGMQSALPLTVMTNYFSIGADAHVALQFHHSRSANPQMLNS 602
Query 232 QVFNKMRYGIEGFKKTFVTRTKSINEII 259
++ N++ YG G F K + E I
Sbjct 603 RLKNRIAYGGLGTIDLFKRSWKDLCEYI 630
> mmu:104418 Dgkz, E130307B02Rik, F730209L11Rik, mDGK[z]; diacylglycerol
kinase zeta (EC:2.7.1.107); K00901 diacylglycerol
kinase [EC:2.7.1.107]
Length=1123
Score = 65.5 bits (158), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 75/325 (23%), Positives = 116/325 (35%), Gaps = 94/325 (28%)
Query 1 LFSNPTSGGNKASAFTRTAVSQLTLTEPCPLKVFIHDIREGPSGSKPGFLLLRSVAAAAA 60
+F NP SGGN+ + ++ + L P +VF D+ +G G + + R V
Sbjct 493 VFVNPKSGGNQGAKIIQSFLWYLN-----PRQVF--DLSQG--GPREALEMYRKVHN--- 540
Query 61 AAAAAAAGEPRAAADGSSSSKKAQHPIVRVLVAGGDGTVMWCIDEMHKTGVQPHVCAVGV 120
+R+L GGDGTV W + + + ++P V +
Sbjct 541 ---------------------------LRILACGGDGTVGWILSTLDQLRLKPPP-PVAI 572
Query 121 VPYGTGNDFANAFGWRPFNAANPFDASLHTLRNIVEHCMRATVVYHDLWSVKVTLKP-GG 179
+P GTGND A W P + I+ H VV D W ++ P G
Sbjct 573 LPLGTGNDLARTLNWGGGYTDEP-------VSKILSHVEEGNVVQLDRWDLRAEPNPEAG 625
Query 180 HFNRINSTTRKKEVVEAEGEPVKELSFTMSNYFSMGVESRIGRGFDRYRT------KSQV 233
R + T + + +NYFS+G ++ + F R S+
Sbjct 626 PEERDDGATDRLPLD------------VFNNYFSLGFDAHVTLEFHESREANPEKFNSRF 673
Query 234 FNKMRYGIEGFKKTFVTRTKSINEII------LDLRIHPNTPEEQTLFTTDKAAAKAQQL 287
NKM Y F + +K + + I +DL TP+ Q L
Sbjct 674 RNKMFYAGTAFSDFLMGSSKDLAKHIRVVCDGMDL-----TPKIQDL------------- 715
Query 288 PLLNKTASLVVLNIPSFSGGNDIWA 312
K +V LNIP + G W
Sbjct 716 ----KPQCIVFLNIPRYCAGTMPWG 736
> hsa:8525 DGKZ, DAGK5, DAGK6, DGK-ZETA, hDGKzeta; diacylglycerol
kinase, zeta (EC:2.7.1.107); K00901 diacylglycerol kinase
[EC:2.7.1.107]
Length=1117
Score = 65.1 bits (157), Expect = 5e-10, Method: Compositional matrix adjust.
Identities = 76/325 (23%), Positives = 115/325 (35%), Gaps = 94/325 (28%)
Query 1 LFSNPTSGGNKASAFTRTAVSQLTLTEPCPLKVFIHDIREGPSGSKPGFLLLRSVAAAAA 60
+F NP SGGN+ + ++ + L P +VF D+ +G G K + R V
Sbjct 487 VFVNPKSGGNQGAKIIQSFLWYLN-----PRQVF--DLSQG--GPKEALEMYRKVHN--- 534
Query 61 AAAAAAAGEPRAAADGSSSSKKAQHPIVRVLVAGGDGTVMWCIDEMHKTGVQPHVCAVGV 120
+R+L GGDGTV W + + + ++P V +
Sbjct 535 ---------------------------LRILACGGDGTVGWILSTLDQLRLKPPP-PVAI 566
Query 121 VPYGTGNDFANAFGWRPFNAANPFDASLHTLRNIVEHCMRATVVYHDLWSVKVTLKP-GG 179
+P GTGND A W P + I+ H VV D W + P G
Sbjct 567 LPLGTGNDLARTLNWGGGYTDEP-------VSKILSHVEEGNVVQLDRWDLHAEPNPEAG 619
Query 180 HFNRINSTTRKKEVVEAEGEPVKELSFTMSNYFSMGVESRIGRGFDRYRT------KSQV 233
+R T + + +NYFS+G ++ + F R S+
Sbjct 620 PEDRDEGATDRLPL------------DVFNNYFSLGFDAHVTLEFHESREANPEKFNSRF 667
Query 234 FNKMRYGIEGFKKTFVTRTKSINEII------LDLRIHPNTPEEQTLFTTDKAAAKAQQL 287
NKM Y F + +K + + I +DL TP+ Q L
Sbjct 668 RNKMFYAGTAFSDFLMGSSKDLAKHIRVVCDGMDL-----TPKIQDL------------- 709
Query 288 PLLNKTASLVVLNIPSFSGGNDIWA 312
K +V LNIP + G W
Sbjct 710 ----KPQCVVFLNIPRYCAGTMPWG 730
> ath:AT4G28130 diacylglycerol kinase accessory domain-containing
protein
Length=466
Score = 63.2 bits (152), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 88/360 (24%), Positives = 145/360 (40%), Gaps = 67/360 (18%)
Query 88 VRVLVAGGDGTVMW---CIDEMHKTGVQPHVCAVGVVPYGTGNDFANAFGWRPFNAANPF 144
++++VAGGDGT W + +++ + P + VP GTGN+ AFGW N
Sbjct 112 LKIIVAGGDGTAGWLLGVVSDLNLSNPPP----IATVPLGTGNNLPFAFGWGKKNPGT-- 165
Query 145 DASLHTLRNIVEHCMRATVVYHDLW------------SVKVTLKPGGHFNRINSTTRKKE 192
D S ++ + + + A + D W S +TLK RI + ++
Sbjct 166 DRS--SVESFLGKVINAKEMKIDNWKILMRMKHPKEGSCDITLKLPHSLPRIFPSDQEN- 222
Query 193 VVEAEGEPVKELSFTMSNYFSMGVESRIGRGFDRYRT------KSQVFNKMRYGIEGFKK 246
EG F NYFS+G+++++ F R K+Q+ N+ Y
Sbjct 223 ---MEGYHTYRGGFW--NYFSLGMDAQVSYAFHSQRKLHPERFKNQLVNQSTY------- 270
Query 247 TFVTRTKSINEIILDLRIHPNTPEEQTLFTTDKAAAKAQ--QLPLLNKTASLVVLNIPSF 304
+ HP++ L Q L + S+V LN+PSF
Sbjct 271 ---LKLSCTQGWFFASLFHPSSQNIAKLAKIQICDRNGQWNDLHIPQSIRSIVCLNLPSF 327
Query 305 SGGNDIWA-PSKKLATRMFDKQKRKQNKELLKVPQQMGDEKLEFMTFKNIPSLGMEFMLH 363
SGG + W P+ K KQ L P + D +E + F+N + +
Sbjct 328 SGGLNPWGTPNPK-----------KQRDRSLTAP-FVDDGLIEIVGFRNAWHGLILLSPN 375
Query 364 GRGCRLHSGKGPWQISFRSLDEKTRVYFQTDGEYYQLTLPAS-----VFVSHNRRVRVLS 418
G G RL ++ F+ K Y + DGE ++ LP++ V +SH+ +V +L+
Sbjct 376 GHGTRLAQANRV-RLEFKKGAAK-HAYMRIDGEPWKQPLPSNDETVMVEISHHGQVNMLA 433
> mmu:331374 Dgkk, Gm360; diacylglycerol kinase kappa (EC:2.7.1.107)
Length=1118
Score = 62.4 bits (150), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 47/157 (29%), Positives = 64/157 (40%), Gaps = 22/157 (14%)
Query 25 LTEPCPLKVFIHDIREGPSGSKPGFLLLRSVAAAAAAAAA--AAAGEPRAAADGSSSSKK 82
LT CPL +FI+ SG G + LR + A G P A + +
Sbjct 329 LTCSCPLLIFIN----SKSGDHQGIIFLRKFKQYLNPSQVFDLAKGGPEAGIAMFKNFAR 384
Query 83 AQHPIVRVLVAGGDGTVMWCIDEMHKTGVQPHVCAVGVVPYGTGNDFANAFGWRPF--NA 140
RVLV GGDG+V W + + G+ C + ++P GTGND A GW
Sbjct 385 -----FRVLVCGGDGSVSWVLSTIDAYGLHDR-CQLAIIPLGTGNDLARVLGWGAVWSKG 438
Query 141 ANPFDASLHTLRNIVEHCMRATVVYHDLWSVKVTLKP 177
+P D I+ +A V D WSV + P
Sbjct 439 TSPLD--------ILSRVEQAHVRILDRWSVMIRETP 467
Score = 44.3 bits (103), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 36/123 (29%), Positives = 58/123 (47%), Gaps = 28/123 (22%)
Query 207 TMSNYFSMGVESRIGRGFDRYRTK------SQVFNKMRYGIEGFKKTFVTRTKSINEIIL 260
M+NYF +G++++I F+ R + S++ NK+ YG+ G K+ + + E
Sbjct 703 VMNNYFGIGLDAKISLEFNSRREEHPEQYNSRLKNKIWYGLLGSKELLQRSYRKLEE--- 759
Query 261 DLRIHPNTPEEQTLFTTDKAAAKAQQLPLLNKTASLVVLNIPSFSGGNDIWAPSKKLATR 320
RIH E A LP L +VVLNI S++GG + W ++ AT
Sbjct 760 --RIHLECDGE------------AVSLPNLQ---GIVVLNITSYAGGVNFWGRNR--ATT 800
Query 321 MFD 323
+D
Sbjct 801 EYD 803
> mmu:380921 Dgkh, 5930402B05Rik, D130015C16, MGC156865; diacylglycerol
kinase, eta (EC:2.7.1.107); K00901 diacylglycerol
kinase [EC:2.7.1.107]
Length=1156
Score = 62.0 bits (149), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 32/83 (38%), Positives = 43/83 (51%), Gaps = 7/83 (8%)
Query 89 RVLVAGGDGTVMWCIDEMHKTGVQPHVCAVGVVPYGTGNDFANAFGWRPFNAANPFDASL 148
R+LV GGDG+V W + E+ K + C +GV+P GTGND A GW +D
Sbjct 381 RILVCGGDGSVGWVLSEIDKLNLHKQ-CQLGVLPLGTGNDLARVLGW-----GGSYDDDT 434
Query 149 HTLRNIVEHCMRATVVYHDLWSV 171
L I+E RA+ D WS+
Sbjct 435 Q-LPQILEKLERASTKMLDRWSI 456
Score = 43.9 bits (102), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 30/115 (26%), Positives = 54/115 (46%), Gaps = 26/115 (22%)
Query 208 MSNYFSMGVESRIGRGFDRYR------TKSQVFNKMRYGIEGFKKTFVTRTKSINEIILD 261
M+NYF +G++++I F+ R +S+ N M YG+ G ++ K++
Sbjct 763 MNNYFGIGLDAKISLEFNNKREEHPEKCRSRTKNLMWYGVLGTRELLQRSYKNL------ 816
Query 262 LRIHPNTPEEQTLFTTDKAAAKAQQLPLLNKTASLVVLNIPSFSGGNDIWAPSKK 316
E++ D Q +PL + + VLNIPS++GG + W +K+
Sbjct 817 --------EQRVQLECD-----GQYIPLPS-LQGIAVLNIPSYAGGTNFWGGTKE 857
> hsa:160851 DGKH, DGKeta, DKFZp761I1510; diacylglycerol kinase,
eta (EC:2.7.1.107); K00901 diacylglycerol kinase [EC:2.7.1.107]
Length=1164
Score = 62.0 bits (149), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 36/99 (36%), Positives = 48/99 (48%), Gaps = 16/99 (16%)
Query 89 RVLVAGGDGTVMWCIDEMHKTGVQPHVCAVGVVPYGTGNDFANAFGWRPFNAANPFDASL 148
R+LV GGDG+V W + E+ K + C +GV+P GTGND A GW +D
Sbjct 384 RILVCGGDGSVGWVLSEIDKLNLNKQ-CQLGVLPLGTGNDLARVLGW-----GGSYDDDT 437
Query 149 HTLRNIVEHCMRATVVYHDLWSV---------KVTLKPG 178
L I+E RA+ D WS+ K +L PG
Sbjct 438 Q-LPQILEKLERASTKMLDRWSIMTYELKLPPKASLLPG 475
Score = 44.3 bits (103), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 30/115 (26%), Positives = 54/115 (46%), Gaps = 26/115 (22%)
Query 208 MSNYFSMGVESRIGRGFDRYR------TKSQVFNKMRYGIEGFKKTFVTRTKSINEIILD 261
M+NYF +G++++I F+ R +S+ N M YG+ G ++ K++
Sbjct 771 MNNYFGIGLDAKISLEFNNKREEHPEKCRSRTKNLMWYGVLGTRELLQRSYKNL------ 824
Query 262 LRIHPNTPEEQTLFTTDKAAAKAQQLPLLNKTASLVVLNIPSFSGGNDIWAPSKK 316
E++ D Q +PL + + VLNIPS++GG + W +K+
Sbjct 825 --------EQRVQLECD-----GQYIPLPS-LQGIAVLNIPSYAGGTNFWGGTKE 865
> hsa:139189 DGKK, MRX68; diacylglycerol kinase, kappa (EC:2.7.1.107)
Length=1271
Score = 61.6 bits (148), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 48/153 (31%), Positives = 65/153 (42%), Gaps = 22/153 (14%)
Query 29 CPLKVFIHDIREGPSGSKPGFLLLRSVAAAAAAAAA--AAAGEPRAAADGSSSSKKAQHP 86
CPL +FI+ SG G + LR + G P A G S K
Sbjct 490 CPLLIFIN----SKSGDHQGIVFLRKFKQYLNPSQVFDLLKGGPEA---GLSMFKNFAR- 541
Query 87 IVRVLVAGGDGTVMWCIDEMHKTGVQPHVCAVGVVPYGTGNDFANAFGWRPF--NAANPF 144
R+LV GGDG+V W + + G+ C + V+P GTGND A GW F + +P
Sbjct 542 -FRILVCGGDGSVSWVLSLIDAFGLH-EKCQLAVIPLGTGNDLARVLGWGAFWNKSKSPL 599
Query 145 DASLHTLRNIVEHCMRATVVYHDLWSVKVTLKP 177
D I+ +A+V D WSV + P
Sbjct 600 D--------ILNRVEQASVRILDRWSVMIRETP 624
Score = 43.1 bits (100), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 32/111 (28%), Positives = 51/111 (45%), Gaps = 26/111 (23%)
Query 207 TMSNYFSMGVESRIGRGFDRYRTK------SQVFNKMRYGIEGFKKTFVTRTKSINEIIL 260
M+NYF +G++++I F+ R + S++ NKM YG+ G K+ + + E
Sbjct 857 VMNNYFGIGLDAKISLDFNTRRDEHPGQYNSRLKNKMWYGLLGTKELLQRSYRKLEE--- 913
Query 261 DLRIHPNTPEEQTLFTTDKAAAKAQQLPLLNKTASLVVLNIPSFSGGNDIW 311
R+H E LP L +VVLNI S++GG + W
Sbjct 914 --RVHLECDGETI------------SLPNLQ---GIVVLNITSYAGGINFW 947
> dre:100137124 si:ch211-93a19.1
Length=586
Score = 60.8 bits (146), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 58/240 (24%), Positives = 93/240 (38%), Gaps = 54/240 (22%)
Query 89 RVLVAGGDGTVMWCIDEMHKTGVQPHVCAVGVVPYGTGNDFANAFGWRPFNAANPFDASL 148
R+LV GGDG+V W + E+ K + C +GV+P GTGND A GW P +
Sbjct 240 RILVCGGDGSVGWVLSEIDKLNLHKQ-CQLGVLPLGTGNDLARVLGWGPSCDDDT----- 293
Query 149 HTLRNIVEHCMRATVVYHDLWSV---KVTLKP-------------------------GGH 180
L I+E RA+ D WS+ ++ + P H
Sbjct 294 -QLPQILEKLERASTKMLDRWSIMTYEIKIPPKHSCPATPEEAEDGQLQISAYEDSVAAH 352
Query 181 FNRINSTTRKKEVVEAEGEPVKELSFTMSNYFSMGVESRIGRGFDRYRTKSQVFNKMRYG 240
+I ++ + V+ + K L T+ + SR+G+ ++R S+ NK
Sbjct 353 LTKILNSDQHSVVISSS----KVLCETVKEFV-----SRVGKCYERGSDSSEEENKDNEK 403
Query 241 IEGFKKTFVTRTKSINEIILDLRIHPNTPEEQTLFTTDKAAAKAQQLPLLNKTASLVVLN 300
E K+ L L P +P E+ + + ++ P +L VLN
Sbjct 404 DEDTKELET----------LPLAKSPCSPPERRISRSTQSCGSFSITPFTTSKENLPVLN 453
> hsa:8527 DGKD, DGKdelta, FLJ26930, KIAA0145, dgkd-2; diacylglycerol
kinase, delta 130kDa (EC:2.7.1.107); K00901 diacylglycerol
kinase [EC:2.7.1.107]
Length=1170
Score = 60.1 bits (144), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 33/83 (39%), Positives = 43/83 (51%), Gaps = 7/83 (8%)
Query 89 RVLVAGGDGTVMWCIDEMHKTGVQPHVCAVGVVPYGTGNDFANAFGWRPFNAANPFDASL 148
R+LV GGDG+V W + E+ + C +GV+P GTGND A GW +A D L
Sbjct 329 RILVCGGDGSVGWVLSEIDSLNLHKQ-CQLGVLPLGTGNDLARVLGW---GSACDDDTQL 384
Query 149 HTLRNIVEHCMRATVVYHDLWSV 171
I+E RA+ D WSV
Sbjct 385 P---QILEKLERASTKMLDRWSV 404
Score = 47.0 bits (110), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 33/116 (28%), Positives = 54/116 (46%), Gaps = 26/116 (22%)
Query 207 TMSNYFSMGVESRIGRGFDRYR------TKSQVFNKMRYGIEGFKKTFVTRTKSINEIIL 260
M+NYF +G++++I F+ R +S+ N M YG+ G K+ K++
Sbjct 719 VMNNYFGIGLDAKISLDFNNKRDEHPEKCRSRTKNMMWYGVLGTKELLHRTYKNL----- 773
Query 261 DLRIHPNTPEEQTLFTTDKAAAKAQQLPLLNKTASLVVLNIPSFSGGNDIWAPSKK 316
E++ L D + LP L A VLNIPS++GG + W +K+
Sbjct 774 ---------EQKVLLECD---GRPIPLPSLQGIA---VLNIPSYAGGTNFWGGTKE 814
> cpv:cgd3_2630 diacylglycerol kinase
Length=919
Score = 59.7 bits (143), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 48/171 (28%), Positives = 74/171 (43%), Gaps = 32/171 (18%)
Query 88 VRVLVAGGDGTVMWCIDEMHKT-GVQPH-VCAVGVVPYGTGNDFANAFGWR-PFNAANPF 144
+ +L+ GGDGTV W ID + GV + + + V+P GTGND + GW FN
Sbjct 406 LMILICGGDGTVRWVIDRCREIYGVNSNSLPPIAVLPLGTGNDLSRTLGWDVTFNG---- 461
Query 145 DASLHTLRNIVEHCMRATVVYHDLWSVKV-TLKPGGHFNRINSTTRKKEVVEAEGEPVKE 203
+ N ++ + + D+W LK G N+T ++
Sbjct 462 -----DILNFLKRICTSNIKQMDIWKCTAWDLKNGDS----NNTHDNHNML--------- 503
Query 204 LSFTMSNYFSMGVESRIGRGFDRYRT------KSQVFNKMRYGIEGFKKTF 248
S T NY +G+ +RI F R KS++ N++ YG GF+ F
Sbjct 504 FSSTFINYLDIGIAARIALKFHNLREAYPQHFKSRLGNQLVYGEVGFRDFF 554
Score = 35.0 bits (79), Expect = 0.59, Method: Compositional matrix adjust.
Identities = 15/55 (27%), Positives = 31/55 (56%), Gaps = 4/55 (7%)
Query 291 NKTASLVVLNIPSFSGGNDIWAPSKKLATRMFDKQKRKQNKELLKVPQQMGDEKL 345
N+ L++ NIPSFSGG ++W K++ + F K ++ ++ ++ + K+
Sbjct 659 NRLEGLIICNIPSFSGGVNLW----KISNQFFKKPRKNRDTNTIRRTTTLTSRKM 709
> dre:100334707 diacylglycerol kinase, eta-like; K00901 diacylglycerol
kinase [EC:2.7.1.107]
Length=924
Score = 58.9 bits (141), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 33/85 (38%), Positives = 44/85 (51%), Gaps = 7/85 (8%)
Query 89 RVLVAGGDGTVMWCIDEMHKTGVQPHVCAVGVVPYGTGNDFANAFGWRPFNAANPFDASL 148
R+LV GGDG+V W + E+ + C +GV+P GTGND A GW +A D
Sbjct 222 RILVCGGDGSVGWVLSEIDALTLHKQ-CQLGVLPLGTGNDLARVLGW---GSACDDDTQ- 276
Query 149 HTLRNIVEHCMRATVVYHDLWSVKV 173
L I+E RA+ D WS+ V
Sbjct 277 --LPQILEKLERASTKMLDRWSIMV 299
Score = 47.0 bits (110), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 33/115 (28%), Positives = 54/115 (46%), Gaps = 26/115 (22%)
Query 208 MSNYFSMGVESRIGRGFDRYR------TKSQVFNKMRYGIEGFKKTFVTRTKSINEIILD 261
M+NYF +G++++I F+ R +S+ N M YG+ G K+ K++
Sbjct 473 MNNYFGIGLDAKISLDFNNKRDEHPEKCRSRTKNMMWYGVLGTKELLHRTYKNL------ 526
Query 262 LRIHPNTPEEQTLFTTDKAAAKAQQLPLLNKTASLVVLNIPSFSGGNDIWAPSKK 316
E++ L D + LP L A VLNIPS++GG + W +K+
Sbjct 527 --------EQRVLLECD---GRPIPLPSLQGIA---VLNIPSYAGGTNFWGGTKE 567
> mmu:227333 Dgkd, AI841987, D330025K09, DGKdelta, KIAA0145, dgkd-2;
diacylglycerol kinase, delta; K00901 diacylglycerol kinase
[EC:2.7.1.107]
Length=1220
Score = 58.2 bits (139), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 32/83 (38%), Positives = 43/83 (51%), Gaps = 7/83 (8%)
Query 89 RVLVAGGDGTVMWCIDEMHKTGVQPHVCAVGVVPYGTGNDFANAFGWRPFNAANPFDASL 148
R+LV GGDG+V W + E+ + C +GV+P GTGND A GW +A D L
Sbjct 373 RILVCGGDGSVGWVLSEIDSLNLHKQ-CQLGVLPLGTGNDLARVLGW---GSACDDDTQL 428
Query 149 HTLRNIVEHCMRATVVYHDLWSV 171
+ +E RA+ D WSV
Sbjct 429 PQILAKLE---RASTKMLDRWSV 448
Score = 45.8 bits (107), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 32/115 (27%), Positives = 54/115 (46%), Gaps = 26/115 (22%)
Query 208 MSNYFSMGVESRIGRGFDRYR------TKSQVFNKMRYGIEGFKKTFVTRTKSINEIILD 261
M+NYF +G++++I F+ R +S+ N M YG+ G K+ +++
Sbjct 766 MNNYFGIGLDAKISLDFNNKRDEHPEKCRSRTKNMMWYGVLGTKELLHRTYRNL------ 819
Query 262 LRIHPNTPEEQTLFTTDKAAAKAQQLPLLNKTASLVVLNIPSFSGGNDIWAPSKK 316
E++ L D + LP L A VLNIPS++GG + W +K+
Sbjct 820 --------EQKVLLECD---GRPIPLPSLQGIA---VLNIPSYAGGTNFWGGTKE 860
> dre:100331649 diacylglycerol kinase, iota-like
Length=948
Score = 54.3 bits (129), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 45/173 (26%), Positives = 68/173 (39%), Gaps = 47/173 (27%)
Query 1 LFSNPTSGGNKASAFTRTAVSQLTLTEPCPLKVFIHDIREGPSGSKPGFLLLRSVAAAAA 60
+F NP SGGN+ + + + L P +VF D+ +G G + L R V
Sbjct 297 VFVNPKSGGNQGTKLLQMFMWILN-----PRQVF--DLSQG--GPRDALELYRKV----- 342
Query 61 AAAAAAAGEPRAAADGSSSSKKAQHPIVRVLVAGGDGTVMWCIDEMHKTGVQPHVCAVGV 120
P +R+L GGDGTV W + + + + P V V
Sbjct 343 -------------------------PNLRILACGGDGTVGWILSALDELQMNPQP-PVAV 376
Query 121 VPYGTGNDFANAFGWRPFNAANPFDASLHTLRNIVEHCMRATVVYHDLWSVKV 173
+P GTGND A W P + ++ H +VV D W+++V
Sbjct 377 LPLGTGNDLARTLNWGGGYTDEP-------VSKVLCHVEDGSVVQLDRWNLQV 422
> cel:F42A9.1 dgk-4; DiacylGlycerol Kinase family member (dgk-4);
K00901 diacylglycerol kinase [EC:2.7.1.107]
Length=1372
Score = 53.5 bits (127), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 31/83 (37%), Positives = 40/83 (48%), Gaps = 7/83 (8%)
Query 89 RVLVAGGDGTVMWCIDEMHKTGVQPHVCAVGVVPYGTGNDFANAFGWRPFNAANPFDASL 148
RVLV GGDGTV W + + + C + ++P GTGND A GW +D +L
Sbjct 296 RVLVCGGDGTVGWVLSAFDRLNLHSK-CQLAILPLGTGNDLARVLGW----GHAFYDDTL 350
Query 149 HTLRNIVEHCMRATVVYHDLWSV 171
L +V RA D WSV
Sbjct 351 --LPQVVRTMERAHTKMLDRWSV 371
Score = 42.0 bits (97), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 32/115 (27%), Positives = 55/115 (47%), Gaps = 29/115 (25%)
Query 208 MSNYFSMGVESRIGRGF-------DRYRTKSQVFNKMRYGIEGFKKTFVTRTKSINEIIL 260
M+NYF +G++++I F ++ R++S++F M YGI G K+ +++
Sbjct 840 MNNYFGIGLDAKIALEFHNKREESEKTRSRSKLF--MWYGILGGKELMHRTYRNL----- 892
Query 261 DLRIHPNTPEEQTLFTTDKAAAKAQQLPLLNKTASLVVLNIPSFSGGNDIWAPSK 315
E++ D LP L +V+LNIPS+SGG + W +K
Sbjct 893 ---------EQRIKLECDGVPI---DLPSLQ---GIVILNIPSYSGGANFWGRNK 932
> pfa:PFI1485c DGK1; diacylglycerol kinase, putative (EC:2.7.1.107);
K00901 diacylglycerol kinase [EC:2.7.1.107]
Length=1360
Score = 51.6 bits (122), Expect = 6e-06, Method: Composition-based stats.
Identities = 51/222 (22%), Positives = 89/222 (40%), Gaps = 58/222 (26%)
Query 118 VGVVPYGTGNDFANAFGWRPFNAANPFDASLHTLRNIVEHCMRATVVYHDLWSVKVTLKP 177
+ ++P GTGND + + GW + ++ + N ++HC V D+W++K
Sbjct 956 ISILPLGTGNDLSISLGW-----GSEYNNDIFFYLNKLKHCKNELV---DVWNMK----- 1002
Query 178 GGHFNRINSTTRKKEVVEAEGEPVKELSFTMSNYFSMGVESRIGRGFDRYRTK------S 231
++ N+ + NYF +G+ +R+ FD R K S
Sbjct 1003 --GYDSNNNLILNNSFI---------------NYFDIGIIARLALHFDNIRKKYPHFFNS 1045
Query 232 QVFNKMRYGIEGFKK-TFVTRTKSINEIILDLRIHPNTPEEQTLFTTDKAAAKAQQLPLL 290
++ NK+ YG GF+ F T +N+ I K +++ +
Sbjct 1046 RIGNKILYGEVGFRDFCFNTYKYKLNKNI-------------------KIYCDGKKVNIE 1086
Query 291 NKTASLVVLNIPSFSGGNDIWAPS--KKLATRMFDKQKRKQN 330
S+ ++NIP F GG IW +K F+K+ K N
Sbjct 1087 ENLESVCIINIPYFLGGIKIWKDDELEKNYYSDFEKENNKMN 1128
Lambda K H
0.319 0.133 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 19893619620
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40