bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_0598_orf1
Length=143
Score E
Sequences producing significant alignments: (Bits) Value
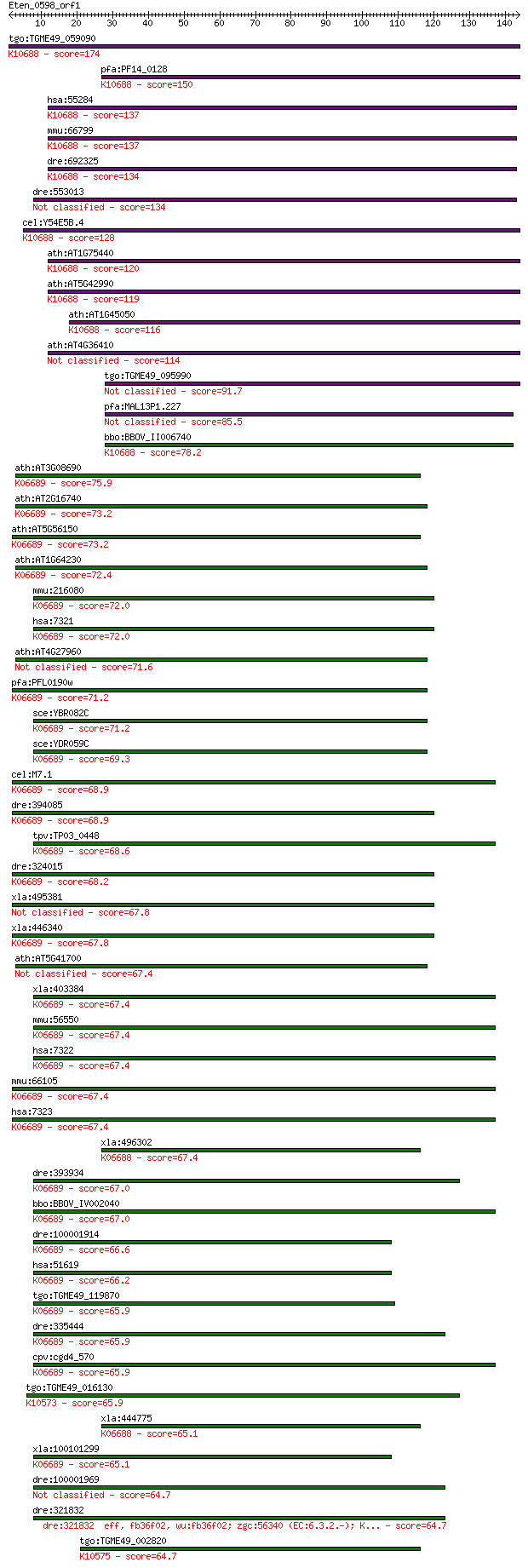
tgo:TGME49_059090 ubiquitin-conjugating enzyme domain-containi... 174 1e-43
pfa:PF14_0128 ubiquitin conjugating enzyme, putative; K10688 u... 150 2e-36
hsa:55284 UBE2W, FLJ11011, hUBC-16; ubiquitin-conjugating enzy... 137 8e-33
mmu:66799 Ube2w, 6130401J04Rik; ubiquitin-conjugating enzyme E... 137 9e-33
dre:692325 flj11011l, ube2w, zgc:136503; hypothetical protein ... 134 7e-32
dre:553013 ube2w, im:6905459, si:ch211-193n21.4; ubiquitin-con... 134 9e-32
cel:Y54E5B.4 ubc-16; UBiquitin Conjugating enzyme family membe... 128 6e-30
ath:AT1G75440 UBC16; UBC16 (ubiquitin-conjugating enzyme 16); ... 120 1e-27
ath:AT5G42990 UBC18; UBC18 (ubiquitin-conjugating enzyme 18); ... 119 3e-27
ath:AT1G45050 ATUBC2-1; ubiquitin-protein ligase; K10688 ubiqu... 116 2e-26
ath:AT4G36410 UBC17; UBC17 (UBIQUITIN-CONJUGATING ENZYME 17); ... 114 1e-25
tgo:TGME49_095990 ubiquitin-conjugating enzyme E2, putative (E... 91.7 7e-19
pfa:MAL13P1.227 ubiquitin conjugating enzyme, putative 85.5 5e-17
bbo:BBOV_II006740 18.m06553; ubiquitin-conjugating enzyme E2; ... 78.2 9e-15
ath:AT3G08690 UBC11; UBC11 (UBIQUITIN-CONJUGATING ENZYME 11); ... 75.9 5e-14
ath:AT2G16740 UBC29; UBC29 (ubiquitin-conjugating enzyme 29); ... 73.2 3e-13
ath:AT5G56150 UBC30; UBC30 (ubiquitin-conjugating enzyme 30); ... 73.2 3e-13
ath:AT1G64230 UBC28; ubiquitin-conjugating enzyme, putative; K... 72.4 4e-13
mmu:216080 Ube2d1, MGC28550, UBCH5; ubiquitin-conjugating enzy... 72.0 5e-13
hsa:7321 UBE2D1, E2(17)KB1, SFT, UBC4/5, UBCH5, UBCH5A; ubiqui... 72.0 5e-13
ath:AT4G27960 UBC9; UBC9 (UBIQUITIN CONJUGATING ENZYME 9); ubi... 71.6 8e-13
pfa:PFL0190w ubiquitin conjugating enzyme E2, putative (EC:6.3... 71.2 9e-13
sce:YBR082C UBC4; Ubc4p (EC:6.3.2.19); K06689 ubiquitin-conjug... 71.2 9e-13
sce:YDR059C UBC5; Ubc5p (EC:6.3.2.19); K06689 ubiquitin-conjug... 69.3 4e-12
cel:M7.1 let-70; LEThal family member (let-70); K06689 ubiquit... 68.9 4e-12
dre:394085 MGC66323, let-70; zgc:66323 (EC:6.3.2.-); K06689 ub... 68.9 5e-12
tpv:TP03_0448 ubiquitin-conjugating enzyme (EC:6.3.2.19); K066... 68.6 6e-12
dre:324015 ube2d1, wu:fc16h06, zgc:73096; ubiquitin-conjugatin... 68.2 8e-12
xla:495381 ube2d1, e2(17)kb1, sft, ubc4/5, ubch5, ubch5a; ubiq... 67.8 1e-11
xla:446340 ube2d2, MGC81261; ubiquitin-conjugating enzyme E2D ... 67.8 1e-11
ath:AT5G41700 UBC8; UBC8 (UBIQUITIN CONJUGATING ENZYME 8); pro... 67.4 1e-11
xla:403384 ube2d3, ube2d2, ube2d3.1, xubc4; ubiquitin-conjugat... 67.4 1e-11
mmu:56550 Ube2d2, 1500034D03Rik, Ubc2e, ubc4; ubiquitin-conjug... 67.4 1e-11
hsa:7322 UBE2D2, E2(17)KB2, PUBC1, UBC4, UBC4/5, UBCH5B; ubiqu... 67.4 1e-11
mmu:66105 Ube2d3, 1100001F19Rik, 9430029A22Rik, AA414951; ubiq... 67.4 1e-11
hsa:7323 UBE2D3, E2(17)KB3, MGC43926, MGC5416, UBC4/5, UBCH5C;... 67.4 1e-11
xla:496302 ube2c; ubiquitin-conjugating enzyme E2C (EC:6.3.2.1... 67.4 2e-11
dre:393934 MGC55886, Ube2d2, zgc:77149; zgc:55886 (EC:6.3.2.19... 67.0 2e-11
bbo:BBOV_IV002040 21.m02810; ubiquitin-conjugating enzyme E2 (... 67.0 2e-11
dre:100001914 ube2d4, MGC162263, zgc:162263; ubiquitin-conjuga... 66.6 2e-11
hsa:51619 UBE2D4, FLJ32004, HBUCE1; ubiquitin-conjugating enzy... 66.2 3e-11
tgo:TGME49_119870 ubiquitin-conjugating enzyme domain-containi... 65.9 4e-11
dre:335444 ube2d2, wu:fj13d01, zgc:73200; ubiquitin-conjugatin... 65.9 4e-11
cpv:cgd4_570 ubiquitin-conjugating enzyme ; K06689 ubiquitin-c... 65.9 4e-11
tgo:TGME49_016130 ubiquitin-conjugating enzyme E2, putative (E... 65.9 4e-11
xla:444775 ube2c, MGC81948; ubiquitin-conjugating enzyme E2 C ... 65.1 6e-11
xla:100101299 ube2d4, HBUCE1, ube2d3.2; ubiquitin-conjugating ... 65.1 7e-11
dre:100001969 ubiquitin-conjugating enzyme E2D 2-like 64.7 9e-11
dre:321832 eff, fb36f02, wu:fb36f02; zgc:56340 (EC:6.3.2.-); K... 64.7 9e-11
tgo:TGME49_002820 ubiquitin-conjugating enzyme E2, putative (E... 64.7 9e-11
> tgo:TGME49_059090 ubiquitin-conjugating enzyme domain-containing
protein (EC:6.3.2.19); K10688 ubiquitin-conjugating enzyme
E2 W [EC:6.3.2.19]
Length=286
Score = 174 bits (440), Expect = 1e-43, Method: Compositional matrix adjust.
Identities = 83/145 (57%), Positives = 101/145 (69%), Gaps = 2/145 (1%)
Query 1 NSKAARLLSRFPPSLLQYRAAAVD--GVRWLVAIEGAPDTAYANDPFLLSFKFFPKYPID 58
+S R+L+ PP L Y D G W +A+EGA T Y + FL+ F+F PKYPI+
Sbjct 142 SSARQRVLAVLPPHLTDYSMMDTDHDGFVWYIALEGAAGTLYEKEVFLVRFRFSPKYPIE 201
Query 59 SPELVFVAPFVPVHPHVYSNGHICLSILYDQWSPALGVKACCISLLSMLSSNREKQPPVD 118
+PE+ FV PF+PVHPHVYSNGHICLSILYD WSPALGV +C +SLLSM+SS R+KQ P D
Sbjct 202 APEVTFVPPFLPVHPHVYSNGHICLSILYDSWSPALGVSSCGMSLLSMVSSCRQKQKPAD 261
Query 119 DRVYCDSRGLRGPKGVKWHFHDDTV 143
D YC G + PK VKW FHDD +
Sbjct 262 DDAYCKVWGSKSPKNVKWVFHDDRI 286
> pfa:PF14_0128 ubiquitin conjugating enzyme, putative; K10688
ubiquitin-conjugating enzyme E2 W [EC:6.3.2.19]
Length=299
Score = 150 bits (378), Expect = 2e-36, Method: Compositional matrix adjust.
Identities = 63/117 (53%), Positives = 87/117 (74%), Gaps = 1/117 (0%)
Query 27 RWLVAIEGAPDTAYANDPFLLSFKFFPKYPIDSPELVFVAPFVPVHPHVYSNGHICLSIL 86
+W++ I GA +T Y+N+ F + FKF KYPI+SPE++F+ P+HPH+YSNGHICLSIL
Sbjct 184 KWIIQITGAENTLYSNETFQMQFKFTEKYPIESPEVIFLGQ-PPIHPHIYSNGHICLSIL 242
Query 87 YDQWSPALGVKACCISLLSMLSSNREKQPPVDDRVYCDSRGLRGPKGVKWHFHDDTV 143
YD WSP L V + C+S++SMLSS ++K+ P+DD +YC + PK +KW FHDD V
Sbjct 243 YDHWSPVLSVNSICLSIISMLSSCKKKRKPLDDILYCSTGPRISPKNMKWMFHDDKV 299
> hsa:55284 UBE2W, FLJ11011, hUBC-16; ubiquitin-conjugating enzyme
E2W (putative) (EC:6.3.2.19); K10688 ubiquitin-conjugating
enzyme E2 W [EC:6.3.2.19]
Length=162
Score = 137 bits (346), Expect = 8e-33, Method: Compositional matrix adjust.
Identities = 65/132 (49%), Positives = 89/132 (67%), Gaps = 2/132 (1%)
Query 12 PPSL-LQYRAAAVDGVRWLVAIEGAPDTAYANDPFLLSFKFFPKYPIDSPELVFVAPFVP 70
PP + L ++ +W+V +EGAP T Y + F L FKF +YP DSP+++F +P
Sbjct 31 PPGMTLNEKSVQNSITQWIVDMEGAPGTLYEGEKFQLLFKFSSRYPFDSPQVMFTGENIP 90
Query 71 VHPHVYSNGHICLSILYDQWSPALGVKACCISLLSMLSSNREKQPPVDDRVYCDSRGLRG 130
VHPHVYSNGHICLSIL + WSPAL V++ C+S++SMLSS +EK+ P D+ Y + +
Sbjct 91 VHPHVYSNGHICLSILTEDWSPALSVQSVCLSIISMLSSCKEKRRPPDNSFYVRTCN-KN 149
Query 131 PKGVKWHFHDDT 142
PK KW +HDDT
Sbjct 150 PKKTKWWYHDDT 161
> mmu:66799 Ube2w, 6130401J04Rik; ubiquitin-conjugating enzyme
E2W (putative) (EC:6.3.2.19); K10688 ubiquitin-conjugating
enzyme E2 W [EC:6.3.2.19]
Length=151
Score = 137 bits (346), Expect = 9e-33, Method: Compositional matrix adjust.
Identities = 64/132 (48%), Positives = 89/132 (67%), Gaps = 2/132 (1%)
Query 12 PPSL-LQYRAAAVDGVRWLVAIEGAPDTAYANDPFLLSFKFFPKYPIDSPELVFVAPFVP 70
PP + L ++ +W+V +EGAP T Y + F L FKF +YP DSP+++F +P
Sbjct 20 PPGMTLNEKSVQNSITQWIVDMEGAPGTLYEGEKFQLLFKFSSRYPFDSPQVMFTGENIP 79
Query 71 VHPHVYSNGHICLSILYDQWSPALGVKACCISLLSMLSSNREKQPPVDDRVYCDSRGLRG 130
+HPHVYSNGHICLSIL + WSPAL V++ C+S++SMLSS +EK+ P D+ Y + +
Sbjct 80 IHPHVYSNGHICLSILTEDWSPALSVQSVCLSIISMLSSCKEKRRPPDNSFYVRTCN-KN 138
Query 131 PKGVKWHFHDDT 142
PK KW +HDDT
Sbjct 139 PKKTKWWYHDDT 150
> dre:692325 flj11011l, ube2w, zgc:136503; hypothetical protein
FLJ11011-like (H. sapiens) (EC:6.3.2.19); K10688 ubiquitin-conjugating
enzyme E2 W [EC:6.3.2.19]
Length=151
Score = 134 bits (338), Expect = 7e-32, Method: Compositional matrix adjust.
Identities = 64/132 (48%), Positives = 88/132 (66%), Gaps = 2/132 (1%)
Query 12 PPSL-LQYRAAAVDGVRWLVAIEGAPDTAYANDPFLLSFKFFPKYPIDSPELVFVAPFVP 70
PP + L ++ +W+V +EGA T Y + F L FKF +YP DSP+++F +P
Sbjct 20 PPGMTLNEKSVQNTITQWIVDMEGASGTVYEGEKFQLLFKFSSRYPSDSPQVMFTGDNIP 79
Query 71 VHPHVYSNGHICLSILYDQWSPALGVKACCISLLSMLSSNREKQPPVDDRVYCDSRGLRG 130
VHPHVYSNGHICLSIL + WSPAL V++ C+S++SMLSS +EK+ P D+ Y + +
Sbjct 80 VHPHVYSNGHICLSILTEDWSPALSVQSVCLSIISMLSSCKEKRRPPDNSFYVRTCN-KN 138
Query 131 PKGVKWHFHDDT 142
PK KW +HDDT
Sbjct 139 PKKTKWWYHDDT 150
> dre:553013 ube2w, im:6905459, si:ch211-193n21.4; ubiquitin-conjugating
enzyme E2W (putative) (EC:6.3.2.19)
Length=151
Score = 134 bits (337), Expect = 9e-32, Method: Compositional matrix adjust.
Identities = 64/137 (46%), Positives = 87/137 (63%), Gaps = 3/137 (2%)
Query 8 LSRFPPS--LLQYRAAAVDGVRWLVAIEGAPDTAYANDPFLLSFKFFPKYPIDSPELVFV 65
L PP+ L R+ W + +EGA T Y + F L FKF +YP +SP+++F
Sbjct 15 LQNDPPAGMTLNERSVQNTITEWFIDMEGAQGTVYEGEKFQLLFKFSSRYPFESPQVMFT 74
Query 66 APFVPVHPHVYSNGHICLSILYDQWSPALGVKACCISLLSMLSSNREKQPPVDDRVYCDS 125
+PVHPHVYSNGHICLSIL + WSPAL V++ C+S++SMLSS +EK+ P D+ Y +
Sbjct 75 GENIPVHPHVYSNGHICLSILTEDWSPALSVQSVCLSIISMLSSCKEKRRPPDNSFYVKT 134
Query 126 RGLRGPKGVKWHFHDDT 142
+ PK KW +HDDT
Sbjct 135 CN-KNPKKTKWWYHDDT 150
> cel:Y54E5B.4 ubc-16; UBiquitin Conjugating enzyme family member
(ubc-16); K10688 ubiquitin-conjugating enzyme E2 W [EC:6.3.2.19]
Length=152
Score = 128 bits (322), Expect = 6e-30, Method: Compositional matrix adjust.
Identities = 67/140 (47%), Positives = 95/140 (67%), Gaps = 2/140 (1%)
Query 5 ARLLSRFPPSLL-QYRAAAVDGVRWLVAIEGAPDTAYANDPFLLSFKFFPKYPIDSPELV 63
A+L S P LL + + D +W + + GA T YA + F+L F F P+YP +SPE++
Sbjct 14 AQLKSEAPEGLLVDNTSTSNDLKQWKIGVVGAEGTLYAGEVFMLQFTFGPQYPFNSPEVM 73
Query 64 FVAPFVPVHPHVYSNGHICLSILYDQWSPALGVKACCISLLSMLSSNREKQPPVDDRVYC 123
FV +P HPH+YSNGHICLSIL D W+PAL V++ C+S+LSMLSS++EK+ P+DD +Y
Sbjct 74 FVGETIPAHPHIYSNGHICLSILSDDWTPALSVQSVCLSILSMLSSSKEKKHPIDDAIYV 133
Query 124 DSRGLRGPKGVKWHFHDDTV 143
+ + P +W FHDD+V
Sbjct 134 RTCS-KNPSKTRWWFHDDSV 152
> ath:AT1G75440 UBC16; UBC16 (ubiquitin-conjugating enzyme 16);
small conjugating protein ligase/ ubiquitin-protein ligase;
K10688 ubiquitin-conjugating enzyme E2 W [EC:6.3.2.19]
Length=161
Score = 120 bits (302), Expect = 1e-27, Method: Compositional matrix adjust.
Identities = 66/133 (49%), Positives = 90/133 (67%), Gaps = 3/133 (2%)
Query 12 PPSLLQYRAAAVDGVRWLVAIEGAPDTAYANDPFLLSFKFFPKYPIDSPELVFVAPFVPV 71
PP+ +++ + RW++ + GAP T YAND + L F YP++SP+++F+ P P+
Sbjct 31 PPTGFKHKVTD-NLQRWIIEVIGAPGTLYANDTYQLQVDFPEHYPMESPQVIFLHP-APL 88
Query 72 HPHVYSNGHICLSILYDQWSPALGVKACCISLLSMLSSNREKQPPVD-DRVYCDSRGLRG 130
HPH+YSNGHICL ILYD WSPA+ V + CIS+LSMLSS+ EKQ P D DR + + R
Sbjct 89 HPHIYSNGHICLDILYDSWSPAMTVSSICISILSMLSSSTEKQRPTDNDRYVKNCKNGRS 148
Query 131 PKGVKWHFHDDTV 143
PK +W FHDD V
Sbjct 149 PKETRWWFHDDKV 161
> ath:AT5G42990 UBC18; UBC18 (ubiquitin-conjugating enzyme 18);
small conjugating protein ligase/ ubiquitin-protein ligase;
K10688 ubiquitin-conjugating enzyme E2 W [EC:6.3.2.19]
Length=161
Score = 119 bits (298), Expect = 3e-27, Method: Compositional matrix adjust.
Identities = 65/133 (48%), Positives = 91/133 (68%), Gaps = 3/133 (2%)
Query 12 PPSLLQYRAAAVDGVRWLVAIEGAPDTAYANDPFLLSFKFFPKYPIDSPELVFVAPFVPV 71
PP+ ++R + +W++ + GAP T YAN+ + L +F YP+++P+++FV P P+
Sbjct 31 PPTGFKHRVTD-NLQKWVIEVTGAPGTLYANETYNLQVEFPQHYPMEAPQVIFVPP-APL 88
Query 72 HPHVYSNGHICLSILYDQWSPALGVKACCISLLSMLSSNREKQPPVD-DRVYCDSRGLRG 130
HPH+YSNGHICL ILYD WSPA+ V + CIS+LSMLSS+ EKQ P D DR + + R
Sbjct 89 HPHIYSNGHICLDILYDSWSPAMTVSSVCISILSMLSSSPEKQRPTDNDRYVKNCKNGRS 148
Query 131 PKGVKWHFHDDTV 143
PK +W FHDD V
Sbjct 149 PKETRWWFHDDKV 161
> ath:AT1G45050 ATUBC2-1; ubiquitin-protein ligase; K10688 ubiquitin-conjugating
enzyme E2 W [EC:6.3.2.19]
Length=161
Score = 116 bits (291), Expect = 2e-26, Method: Compositional matrix adjust.
Identities = 64/128 (50%), Positives = 86/128 (67%), Gaps = 3/128 (2%)
Query 18 YRAAAVDGV-RWLVAIEGAPDTAYANDPFLLSFKFFPKYPIDSPELVFVAPFVPVHPHVY 76
+R D + +W + + GAP T YAN+ + L +F YP+++P++VFV+P P HPH+Y
Sbjct 35 FRHKVTDNLQKWTIDVTGAPGTLYANETYQLQVEFPEHYPMEAPQVVFVSP-APSHPHIY 93
Query 77 SNGHICLSILYDQWSPALGVKACCISLLSMLSSNREKQPPVD-DRVYCDSRGLRGPKGVK 135
SNGHICL ILYD WSPA+ V + CIS+LSMLSS+ KQ P D DR + + R PK +
Sbjct 94 SNGHICLDILYDSWSPAMTVNSVCISILSMLSSSPAKQRPADNDRYVKNCKNGRSPKETR 153
Query 136 WHFHDDTV 143
W FHDD V
Sbjct 154 WWFHDDKV 161
> ath:AT4G36410 UBC17; UBC17 (UBIQUITIN-CONJUGATING ENZYME 17);
small conjugating protein ligase/ ubiquitin-protein ligase
Length=161
Score = 114 bits (284), Expect = 1e-25, Method: Compositional matrix adjust.
Identities = 60/133 (45%), Positives = 89/133 (66%), Gaps = 3/133 (2%)
Query 12 PPSLLQYRAAAVDGVRWLVAIEGAPDTAYANDPFLLSFKFFPKYPIDSPELVFVAPFVPV 71
PPS ++R + + RW++ + G P T YAN+ + L +F YP+++P+++F P P+
Sbjct 31 PPSGFKHRVSD-NLQRWIIEVHGVPGTLYANETYQLQVEFPEHYPMEAPQVIFQHP-APL 88
Query 72 HPHVYSNGHICLSILYDQWSPALGVKACCISLLSMLSSNREKQPPVDDRVYCDS-RGLRG 130
HPH+YSNGHICL +LYD WSPA+ + + C+S+LSMLSS+ KQ P D+ Y + + R
Sbjct 89 HPHIYSNGHICLDVLYDSWSPAMRLSSICLSILSMLSSSSVKQKPKDNDHYLKNCKHGRS 148
Query 131 PKGVKWHFHDDTV 143
PK +W FHDD V
Sbjct 149 PKETRWRFHDDKV 161
> tgo:TGME49_095990 ubiquitin-conjugating enzyme E2, putative
(EC:6.3.2.19)
Length=264
Score = 91.7 bits (226), Expect = 7e-19, Method: Compositional matrix adjust.
Identities = 45/116 (38%), Positives = 67/116 (57%), Gaps = 3/116 (2%)
Query 28 WLVAIEGAPDTAYANDPFLLSFKFFPKYPIDSPELVFVAPFVPVHPHVYSNGHICLSILY 87
WL+ + G + YAN+ + L P YP P F+ P PVH HVYSNG +CL++L
Sbjct 152 WLIEMTGMEGSPYANEMYRLKVVIPPDYPFSPPTCFFLQP-APVHVHVYSNGDVCLNLLG 210
Query 88 DQWSPALGVKACCISLLSMLSSNREKQPPVDDRVYCDSRGLRGPKGVKWHFHDDTV 143
W P+L + A +++LSML+S ++KQ P D+ + D G G ++ +HDD V
Sbjct 211 SDWRPSLSISAIAVAILSMLTSAKQKQLPTDNAAHMDVPA--GHHGTQFLYHDDKV 264
> pfa:MAL13P1.227 ubiquitin conjugating enzyme, putative
Length=278
Score = 85.5 bits (210), Expect = 5e-17, Method: Compositional matrix adjust.
Identities = 43/114 (37%), Positives = 68/114 (59%), Gaps = 3/114 (2%)
Query 28 WLVAIEGAPDTAYANDPFLLSFKFFPKYPIDSPELVFVAPFVPVHPHVYSNGHICLSILY 87
W+V G +T YAN+ + + F YP+ P +V+ P H HVYSNG ICLS+L
Sbjct 166 WIVQYVGLENTIYANEVYKIKIIFPDNYPL-KPPIVYFLQKPPKHTHVYSNGDICLSVLG 224
Query 88 DQWSPALGVKACCISLLSMLSSNREKQPPVDDRVYCDSRGLRGPKGVKWHFHDD 141
D ++P+L + +S++SMLSS +EK+ P+D+ + D++ G + +HDD
Sbjct 225 DDYNPSLSISGLILSIISMLSSAKEKKLPIDNYTHADAKP--GSSQNNFLYHDD 276
> bbo:BBOV_II006740 18.m06553; ubiquitin-conjugating enzyme E2;
K10688 ubiquitin-conjugating enzyme E2 W [EC:6.3.2.19]
Length=188
Score = 78.2 bits (191), Expect = 9e-15, Method: Compositional matrix adjust.
Identities = 41/114 (35%), Positives = 64/114 (56%), Gaps = 3/114 (2%)
Query 28 WLVAIEGAPDTAYANDPFLLSFKFFPKYPIDSPELVFVAPFVPVHPHVYSNGHICLSILY 87
W++ G P T Y + + L F +YP+ +P + F+ P PVH HVYSNG ICLS L
Sbjct 76 WIITWVGLPGTIYEGETYRLKILFPKEYPLKAPTVYFLQP-APVHAHVYSNGDICLSSLG 134
Query 88 DQWSPALGVKACCISLLSMLSSNREKQPPVDDRVYCDSRGLRGPKGVKWHFHDD 141
+ P V + +S++SMLS+ EK+ P D+ ++ + G ++ +HDD
Sbjct 135 SDYLPTATVTSFVLSIISMLSNATEKRLPADNHLHAEMPP--GSADARYLYHDD 186
> ath:AT3G08690 UBC11; UBC11 (UBIQUITIN-CONJUGATING ENZYME 11);
ubiquitin-protein ligase; K06689 ubiquitin-conjugating enzyme
E2 D/E [EC:6.3.2.19]
Length=148
Score = 75.9 bits (185), Expect = 5e-14, Method: Compositional matrix adjust.
Identities = 42/113 (37%), Positives = 57/113 (50%), Gaps = 2/113 (1%)
Query 3 KAARLLSRFPPSLLQYRAAAVDGVRWLVAIEGAPDTAYANDPFLLSFKFFPKYPIDSPEL 62
K + L + PPS A D W I G P++ YA FL+S F P YP P++
Sbjct 8 KELKDLQKDPPSNCSAGPVAEDMFHWQATIMGPPESPYAGGVFLVSIHFPPDYPFKPPKV 67
Query 63 VFVAPFVPVHPHVYSNGHICLSILYDQWSPALGVKACCISLLSMLSSNREKQP 115
F HP++ SNG ICL IL +QWSPAL + +S+ S+L+ P
Sbjct 68 SFKTKVY--HPNINSNGSICLDILKEQWSPALTISKVLLSICSLLTDPNPDDP 118
> ath:AT2G16740 UBC29; UBC29 (ubiquitin-conjugating enzyme 29);
ubiquitin-protein ligase; K06689 ubiquitin-conjugating enzyme
E2 D/E [EC:6.3.2.19]
Length=148
Score = 73.2 bits (178), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 41/115 (35%), Positives = 57/115 (49%), Gaps = 2/115 (1%)
Query 3 KAARLLSRFPPSLLQYRAAAVDGVRWLVAIEGAPDTAYANDPFLLSFKFFPKYPIDSPEL 62
K + L R PP D W I G ++ Y+ FL++ F P YP P++
Sbjct 8 KELKELQRDPPVSCSAGPTGEDMFHWQATIMGPNESPYSGGVFLVNIHFPPDYPFKPPKV 67
Query 63 VFVAPFVPVHPHVYSNGHICLSILYDQWSPALGVKACCISLLSMLSSNREKQPPV 117
VF HP++ SNG+ICL IL DQWSPAL + +S+ S+L+ P V
Sbjct 68 VFRTKVF--HPNINSNGNICLDILKDQWSPALTISKVLLSICSLLTDPNPDDPLV 120
> ath:AT5G56150 UBC30; UBC30 (ubiquitin-conjugating enzyme 30);
ubiquitin-protein ligase; K06689 ubiquitin-conjugating enzyme
E2 D/E [EC:6.3.2.19]
Length=148
Score = 73.2 bits (178), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 41/114 (35%), Positives = 56/114 (49%), Gaps = 2/114 (1%)
Query 2 SKAARLLSRFPPSLLQYRAAAVDGVRWLVAIEGAPDTAYANDPFLLSFKFFPKYPIDSPE 61
+K R L R PP D +W I G D+ +A FL++ F P YP P+
Sbjct 7 NKELRDLQRDPPVSCSAGPTGDDMFQWQATIMGPADSPFAGGVFLVTIHFPPDYPFKPPK 66
Query 62 LVFVAPFVPVHPHVYSNGHICLSILYDQWSPALGVKACCISLLSMLSSNREKQP 115
+ F HP++ SNG ICL IL +QWSPAL V +S+ S+L+ P
Sbjct 67 VAFRTKVY--HPNINSNGSICLDILKEQWSPALTVSKVLLSICSLLTDPNPDDP 118
> ath:AT1G64230 UBC28; ubiquitin-conjugating enzyme, putative;
K06689 ubiquitin-conjugating enzyme E2 D/E [EC:6.3.2.19]
Length=148
Score = 72.4 bits (176), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 41/115 (35%), Positives = 57/115 (49%), Gaps = 2/115 (1%)
Query 3 KAARLLSRFPPSLLQYRAAAVDGVRWLVAIEGAPDTAYANDPFLLSFKFFPKYPIDSPEL 62
K + L + PP+ A D W I G D+ Y+ FL++ F P YP P++
Sbjct 8 KELKDLQKDPPTSCSAGPVAEDMFHWQATIMGPSDSPYSGGVFLVTIHFPPDYPFKPPKV 67
Query 63 VFVAPFVPVHPHVYSNGHICLSILYDQWSPALGVKACCISLLSMLSSNREKQPPV 117
F HP+V SNG ICL IL +QWSPAL + +S+ S+L+ P V
Sbjct 68 AFRTKVF--HPNVNSNGSICLDILKEQWSPALTISKVLLSICSLLTDPNPDDPLV 120
> mmu:216080 Ube2d1, MGC28550, UBCH5; ubiquitin-conjugating enzyme
E2D 1, UBC4/5 homolog (yeast) (EC:6.3.2.19); K06689 ubiquitin-conjugating
enzyme E2 D/E [EC:6.3.2.19]
Length=147
Score = 72.0 bits (175), Expect = 5e-13, Method: Compositional matrix adjust.
Identities = 42/112 (37%), Positives = 52/112 (46%), Gaps = 2/112 (1%)
Query 8 LSRFPPSLLQYRAAAVDGVRWLVAIEGAPDTAYANDPFLLSFKFFPKYPIDSPELVFVAP 67
L R PP+ D W I G PD+AY F L+ F YP P++ F
Sbjct 13 LQRDPPAHCSAGPVGDDLFHWQATIMGPPDSAYQGGVFFLTVHFPTDYPFKPPKIAFTTK 72
Query 68 FVPVHPHVYSNGHICLSILYDQWSPALGVKACCISLLSMLSSNREKQPPVDD 119
HP++ SNG ICL IL QWSPAL V +S+ S+L P V D
Sbjct 73 IY--HPNINSNGSICLDILRSQWSPALTVSKVLLSICSLLCDPNPDDPLVPD 122
> hsa:7321 UBE2D1, E2(17)KB1, SFT, UBC4/5, UBCH5, UBCH5A; ubiquitin-conjugating
enzyme E2D 1 (UBC4/5 homolog, yeast) (EC:6.3.2.19);
K06689 ubiquitin-conjugating enzyme E2 D/E [EC:6.3.2.19]
Length=147
Score = 72.0 bits (175), Expect = 5e-13, Method: Compositional matrix adjust.
Identities = 42/112 (37%), Positives = 52/112 (46%), Gaps = 2/112 (1%)
Query 8 LSRFPPSLLQYRAAAVDGVRWLVAIEGAPDTAYANDPFLLSFKFFPKYPIDSPELVFVAP 67
L R PP+ D W I G PD+AY F L+ F YP P++ F
Sbjct 13 LQRDPPAHCSAGPVGDDLFHWQATIMGPPDSAYQGGVFFLTVHFPTDYPFKPPKIAFTTK 72
Query 68 FVPVHPHVYSNGHICLSILYDQWSPALGVKACCISLLSMLSSNREKQPPVDD 119
HP++ SNG ICL IL QWSPAL V +S+ S+L P V D
Sbjct 73 IY--HPNINSNGSICLDILRSQWSPALTVSKVLLSICSLLCDPNPDDPLVPD 122
> ath:AT4G27960 UBC9; UBC9 (UBIQUITIN CONJUGATING ENZYME 9); ubiquitin-protein
ligase
Length=178
Score = 71.6 bits (174), Expect = 8e-13, Method: Compositional matrix adjust.
Identities = 40/115 (34%), Positives = 57/115 (49%), Gaps = 2/115 (1%)
Query 3 KAARLLSRFPPSLLQYRAAAVDGVRWLVAIEGAPDTAYANDPFLLSFKFFPKYPIDSPEL 62
K + L + PP+ A D W I G D+ Y+ FL++ F P YP P++
Sbjct 38 KELKDLQKDPPTSCSAGPVAEDMFHWQATIMGPSDSPYSGGVFLVTIHFPPDYPFKPPKV 97
Query 63 VFVAPFVPVHPHVYSNGHICLSILYDQWSPALGVKACCISLLSMLSSNREKQPPV 117
F HP++ SNG ICL IL +QWSPAL + +S+ S+L+ P V
Sbjct 98 AFRTKVF--HPNINSNGSICLDILKEQWSPALTISKVLLSICSLLTDPNPDDPLV 150
> pfa:PFL0190w ubiquitin conjugating enzyme E2, putative (EC:6.3.2.19);
K06689 ubiquitin-conjugating enzyme E2 D/E [EC:6.3.2.19]
Length=147
Score = 71.2 bits (173), Expect = 9e-13, Method: Compositional matrix adjust.
Identities = 39/116 (33%), Positives = 58/116 (50%), Gaps = 2/116 (1%)
Query 2 SKAARLLSRFPPSLLQYRAAAVDGVRWLVAIEGAPDTAYANDPFLLSFKFFPKYPIDSPE 61
+K + L++ PP+ D W I G D+ Y N + L+ KF P YP P+
Sbjct 7 TKELQDLNKDPPTNCSAGPIGDDLFFWQATIMGPGDSPYENGVYFLNIKFPPDYPFKPPK 66
Query 62 LVFVAPFVPVHPHVYSNGHICLSILYDQWSPALGVKACCISLLSMLSSNREKQPPV 117
++F HP++ + G ICL IL DQWSPAL + +S+ S+L+ P V
Sbjct 67 IIFTTKIY--HPNINTAGAICLDILKDQWSPALTISKVLLSISSLLTDPNADDPLV 120
> sce:YBR082C UBC4; Ubc4p (EC:6.3.2.19); K06689 ubiquitin-conjugating
enzyme E2 D/E [EC:6.3.2.19]
Length=148
Score = 71.2 bits (173), Expect = 9e-13, Method: Compositional matrix adjust.
Identities = 40/110 (36%), Positives = 54/110 (49%), Gaps = 2/110 (1%)
Query 8 LSRFPPSLLQYRAAAVDGVRWLVAIEGAPDTAYANDPFLLSFKFFPKYPIDSPELVFVAP 67
L R PP+ D W +I G D+ YA F LS F YP P++ F
Sbjct 14 LERDPPTSCSAGPVGDDLYHWQASIMGPADSPYAGGVFFLSIHFPTDYPFKPPKISFTTK 73
Query 68 FVPVHPHVYSNGHICLSILYDQWSPALGVKACCISLLSMLSSNREKQPPV 117
HP++ +NG+ICL IL DQWSPAL + +S+ S+L+ P V
Sbjct 74 IY--HPNINANGNICLDILKDQWSPALTLSKVLLSICSLLTDANPDDPLV 121
> sce:YDR059C UBC5; Ubc5p (EC:6.3.2.19); K06689 ubiquitin-conjugating
enzyme E2 D/E [EC:6.3.2.19]
Length=148
Score = 69.3 bits (168), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 40/110 (36%), Positives = 54/110 (49%), Gaps = 2/110 (1%)
Query 8 LSRFPPSLLQYRAAAVDGVRWLVAIEGAPDTAYANDPFLLSFKFFPKYPIDSPELVFVAP 67
L R PP+ D W +I G D+ YA F LS F YP P++ F
Sbjct 14 LGRDPPASCSAGPVGDDLYHWQASIMGPSDSPYAGGVFFLSIHFPTDYPFKPPKVNFTTK 73
Query 68 FVPVHPHVYSNGHICLSILYDQWSPALGVKACCISLLSMLSSNREKQPPV 117
HP++ S+G+ICL IL DQWSPAL + +S+ S+L+ P V
Sbjct 74 IY--HPNINSSGNICLDILKDQWSPALTLSKVLLSICSLLTDANPDDPLV 121
> cel:M7.1 let-70; LEThal family member (let-70); K06689 ubiquitin-conjugating
enzyme E2 D/E [EC:6.3.2.19]
Length=147
Score = 68.9 bits (167), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 43/137 (31%), Positives = 59/137 (43%), Gaps = 4/137 (2%)
Query 2 SKAARLLSRFPPSLLQYRAAAVDGVRWLVAIEGAPDTAYANDPFLLSFKFFPKYPIDSPE 61
K + L R PP+ D W I G P++ Y F L+ F YP P+
Sbjct 7 QKELQDLGRDPPAQCSAGPVGDDLFHWQATIMGPPESPYQGGVFFLTIHFPTDYPFKPPK 66
Query 62 LVFVAPFVPVHPHVYSNGHICLSILYDQWSPALGVKACCISLLSMLSSNREKQPPVDD-- 119
+ F HP++ SNG ICL IL QWSPAL + +S+ S+L P V +
Sbjct 67 VAFTTRIY--HPNINSNGSICLDILRSQWSPALTISKVLLSICSLLCDPNPDDPLVPEIA 124
Query 120 RVYCDSRGLRGPKGVKW 136
R+Y R +W
Sbjct 125 RIYKTDRERYNQLAREW 141
> dre:394085 MGC66323, let-70; zgc:66323 (EC:6.3.2.-); K06689
ubiquitin-conjugating enzyme E2 D/E [EC:6.3.2.19]
Length=147
Score = 68.9 bits (167), Expect = 5e-12, Method: Compositional matrix adjust.
Identities = 42/118 (35%), Positives = 52/118 (44%), Gaps = 2/118 (1%)
Query 2 SKAARLLSRFPPSLLQYRAAAVDGVRWLVAIEGAPDTAYANDPFLLSFKFFPKYPIDSPE 61
K + L R PPS D W I G D+ Y F L+ F YP P+
Sbjct 7 QKELQDLQRDPPSQCSAGPLGEDLFHWQATIMGPGDSPYQGGVFFLTIHFPTDYPFKPPK 66
Query 62 LVFVAPFVPVHPHVYSNGHICLSILYDQWSPALGVKACCISLLSMLSSNREKQPPVDD 119
+ F HP++ SNG ICL IL QWSPAL V +S+ S+L P V D
Sbjct 67 VAFTTKIY--HPNINSNGSICLDILRSQWSPALTVSKVLLSICSLLCDPNPDDPLVPD 122
> tpv:TP03_0448 ubiquitin-conjugating enzyme (EC:6.3.2.19); K06689
ubiquitin-conjugating enzyme E2 D/E [EC:6.3.2.19]
Length=147
Score = 68.6 bits (166), Expect = 6e-12, Method: Compositional matrix adjust.
Identities = 40/131 (30%), Positives = 62/131 (47%), Gaps = 4/131 (3%)
Query 8 LSRFPPSLLQYRAAAVDGVRWLVAIEGAPDTAYANDPFLLSFKFFPKYPIDSPELVFVAP 67
L++ PP+ D W I G ++ Y N + L+ F YP P++ F
Sbjct 13 LTKDPPTNCSAGPVGDDMFHWQATIMGPHNSLYQNGVYFLNIHFPSDYPFKPPKVAFTTK 72
Query 68 FVPVHPHVYSNGHICLSILYDQWSPALGVKACCISLLSMLSSNREKQPPVDD--RVYCDS 125
HP++ +NG ICL IL DQWSPAL + +S+ S+L+ P V + ++Y +
Sbjct 73 VY--HPNINNNGAICLDILKDQWSPALTISKVLLSISSLLTDPNPDDPLVPEIAQIYKQN 130
Query 126 RGLRGPKGVKW 136
R L +W
Sbjct 131 RKLYESTVREW 141
> dre:324015 ube2d1, wu:fc16h06, zgc:73096; ubiquitin-conjugating
enzyme E2D 1 (UBC4/5 homolog, yeast) (EC:6.3.2.-); K06689
ubiquitin-conjugating enzyme E2 D/E [EC:6.3.2.19]
Length=147
Score = 68.2 bits (165), Expect = 8e-12, Method: Compositional matrix adjust.
Identities = 41/118 (34%), Positives = 52/118 (44%), Gaps = 2/118 (1%)
Query 2 SKAARLLSRFPPSLLQYRAAAVDGVRWLVAIEGAPDTAYANDPFLLSFKFFPKYPIDSPE 61
K + L R PP+ D W I G D+ Y F L+ F YP P+
Sbjct 7 QKELQDLQRDPPAQCSAGPVGDDLFHWQATIMGPSDSPYQGGVFFLTIHFPTDYPFKPPK 66
Query 62 LVFVAPFVPVHPHVYSNGHICLSILYDQWSPALGVKACCISLLSMLSSNREKQPPVDD 119
+ F HP++ SNG ICL IL QWSPAL V +S+ S+L P V D
Sbjct 67 VAFTTKIY--HPNINSNGSICLDILRSQWSPALTVSKVLLSICSLLCDPNPDDPLVPD 122
> xla:495381 ube2d1, e2(17)kb1, sft, ubc4/5, ubch5, ubch5a; ubiquitin-conjugating
enzyme E2D 1 (UBC4/5 homolog)
Length=147
Score = 67.8 bits (164), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 41/118 (34%), Positives = 51/118 (43%), Gaps = 2/118 (1%)
Query 2 SKAARLLSRFPPSLLQYRAAAVDGVRWLVAIEGAPDTAYANDPFLLSFKFFPKYPIDSPE 61
K L R PP+ D W I G D+ Y F L+ F YP P+
Sbjct 7 QKELNDLQRDPPAQCSAGPVGDDLFHWQATIMGPTDSPYQGGVFFLTIHFPTDYPFKPPK 66
Query 62 LVFVAPFVPVHPHVYSNGHICLSILYDQWSPALGVKACCISLLSMLSSNREKQPPVDD 119
+ F HP++ SNG ICL IL QWSPAL V +S+ S+L P V D
Sbjct 67 VAFTTKIY--HPNINSNGSICLDILRSQWSPALTVSKVLLSICSLLCDPNPDDPLVPD 122
> xla:446340 ube2d2, MGC81261; ubiquitin-conjugating enzyme E2D
2 (UBC4/5 homolog, yeast) (EC:6.3.2.-); K06689 ubiquitin-conjugating
enzyme E2 D/E [EC:6.3.2.19]
Length=147
Score = 67.8 bits (164), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 41/118 (34%), Positives = 51/118 (43%), Gaps = 2/118 (1%)
Query 2 SKAARLLSRFPPSLLQYRAAAVDGVRWLVAIEGAPDTAYANDPFLLSFKFFPKYPIDSPE 61
K L R PP+ D W I G D+ Y F L+ F YP P+
Sbjct 7 QKELNDLQRDPPAQCSAGPVGDDLFHWQATIMGPTDSPYQGGVFFLTIHFPTDYPFKPPK 66
Query 62 LVFVAPFVPVHPHVYSNGHICLSILYDQWSPALGVKACCISLLSMLSSNREKQPPVDD 119
+ F HP++ SNG ICL IL QWSPAL V +S+ S+L P V D
Sbjct 67 VAFTTKIY--HPNINSNGSICLDILRSQWSPALTVSKVLLSICSLLCDPNPDDPLVPD 122
> ath:AT5G41700 UBC8; UBC8 (UBIQUITIN CONJUGATING ENZYME 8); protein
binding / ubiquitin-protein ligase
Length=149
Score = 67.4 bits (163), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 39/116 (33%), Positives = 58/116 (50%), Gaps = 3/116 (2%)
Query 3 KAARLLSRFPPSLLQYRA-AAVDGVRWLVAIEGAPDTAYANDPFLLSFKFFPKYPIDSPE 61
K + L + PP+ + A D W I G ++ Y+ FL++ F P YP P+
Sbjct 8 KELKDLQKDPPTSCIFAGPVAEDMFHWQATIMGPAESPYSGGVFLVTIHFPPDYPFKPPK 67
Query 62 LVFVAPFVPVHPHVYSNGHICLSILYDQWSPALGVKACCISLLSMLSSNREKQPPV 117
+ F HP++ SNG ICL IL +QWSPAL + +S+ S+L+ P V
Sbjct 68 VAFRTKVF--HPNINSNGSICLDILKEQWSPALTISKVLLSICSLLTDPNPDDPLV 121
> xla:403384 ube2d3, ube2d2, ube2d3.1, xubc4; ubiquitin-conjugating
enzyme E2D 3 (UBC4/5 homolog) (EC:6.3.2.19); K06689 ubiquitin-conjugating
enzyme E2 D/E [EC:6.3.2.19]
Length=147
Score = 67.4 bits (163), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 42/131 (32%), Positives = 57/131 (43%), Gaps = 4/131 (3%)
Query 8 LSRFPPSLLQYRAAAVDGVRWLVAIEGAPDTAYANDPFLLSFKFFPKYPIDSPELVFVAP 67
L+R PP+ D W I G D+ Y F L+ F YP P++ F
Sbjct 13 LARDPPAQCSAGPVGDDMFHWQATIMGPNDSPYQGGVFFLTIHFPTDYPFKPPKVAFTTR 72
Query 68 FVPVHPHVYSNGHICLSILYDQWSPALGVKACCISLLSMLSSNREKQPPVDD--RVYCDS 125
HP++ SNG ICL IL QWSPAL + +S+ S+L P V + R+Y
Sbjct 73 IY--HPNINSNGSICLDILRSQWSPALTISKVLLSICSLLCDPNPDDPLVPEIARIYKTD 130
Query 126 RGLRGPKGVKW 136
R +W
Sbjct 131 REKYNRIAREW 141
> mmu:56550 Ube2d2, 1500034D03Rik, Ubc2e, ubc4; ubiquitin-conjugating
enzyme E2D 2 (EC:6.3.2.19); K06689 ubiquitin-conjugating
enzyme E2 D/E [EC:6.3.2.19]
Length=147
Score = 67.4 bits (163), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 42/131 (32%), Positives = 57/131 (43%), Gaps = 4/131 (3%)
Query 8 LSRFPPSLLQYRAAAVDGVRWLVAIEGAPDTAYANDPFLLSFKFFPKYPIDSPELVFVAP 67
L+R PP+ D W I G D+ Y F L+ F YP P++ F
Sbjct 13 LARDPPAQCSAGPVGDDMFHWQATIMGPNDSPYQGGVFFLTIHFPTDYPFKPPKVAFTTR 72
Query 68 FVPVHPHVYSNGHICLSILYDQWSPALGVKACCISLLSMLSSNREKQPPVDD--RVYCDS 125
HP++ SNG ICL IL QWSPAL + +S+ S+L P V + R+Y
Sbjct 73 IY--HPNINSNGSICLDILRSQWSPALTISKVLLSICSLLCDPNPDDPLVPEIARIYKTD 130
Query 126 RGLRGPKGVKW 136
R +W
Sbjct 131 REKYNRIAREW 141
> hsa:7322 UBE2D2, E2(17)KB2, PUBC1, UBC4, UBC4/5, UBCH5B; ubiquitin-conjugating
enzyme E2D 2 (UBC4/5 homolog, yeast) (EC:6.3.2.19);
K06689 ubiquitin-conjugating enzyme E2 D/E [EC:6.3.2.19]
Length=147
Score = 67.4 bits (163), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 42/131 (32%), Positives = 57/131 (43%), Gaps = 4/131 (3%)
Query 8 LSRFPPSLLQYRAAAVDGVRWLVAIEGAPDTAYANDPFLLSFKFFPKYPIDSPELVFVAP 67
L+R PP+ D W I G D+ Y F L+ F YP P++ F
Sbjct 13 LARDPPAQCSAGPVGDDMFHWQATIMGPNDSPYQGGVFFLTIHFPTDYPFKPPKVAFTTR 72
Query 68 FVPVHPHVYSNGHICLSILYDQWSPALGVKACCISLLSMLSSNREKQPPVDD--RVYCDS 125
HP++ SNG ICL IL QWSPAL + +S+ S+L P V + R+Y
Sbjct 73 IY--HPNINSNGSICLDILRSQWSPALTISKVLLSICSLLCDPNPDDPLVPEIARIYKTD 130
Query 126 RGLRGPKGVKW 136
R +W
Sbjct 131 REKYNRIAREW 141
> mmu:66105 Ube2d3, 1100001F19Rik, 9430029A22Rik, AA414951; ubiquitin-conjugating
enzyme E2D 3 (UBC4/5 homolog, yeast) (EC:6.3.2.19);
K06689 ubiquitin-conjugating enzyme E2 D/E [EC:6.3.2.19]
Length=147
Score = 67.4 bits (163), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 43/137 (31%), Positives = 59/137 (43%), Gaps = 4/137 (2%)
Query 2 SKAARLLSRFPPSLLQYRAAAVDGVRWLVAIEGAPDTAYANDPFLLSFKFFPKYPIDSPE 61
+K L+R PP+ D W I G D+ Y F L+ F YP P+
Sbjct 7 NKELSDLARDPPAQCSAGPVGDDMFHWQATIMGPNDSPYQGGVFFLTIHFPTDYPFKPPK 66
Query 62 LVFVAPFVPVHPHVYSNGHICLSILYDQWSPALGVKACCISLLSMLSSNREKQPPVDD-- 119
+ F HP++ SNG ICL IL QWSPAL + +S+ S+L P V +
Sbjct 67 VAFTTRIY--HPNINSNGSICLDILRSQWSPALTISKVLLSICSLLCDPNPDDPLVPEIA 124
Query 120 RVYCDSRGLRGPKGVKW 136
R+Y R +W
Sbjct 125 RIYKTDRDKYNRISREW 141
> hsa:7323 UBE2D3, E2(17)KB3, MGC43926, MGC5416, UBC4/5, UBCH5C;
ubiquitin-conjugating enzyme E2D 3 (UBC4/5 homolog, yeast)
(EC:6.3.2.19); K06689 ubiquitin-conjugating enzyme E2 D/E
[EC:6.3.2.19]
Length=147
Score = 67.4 bits (163), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 43/137 (31%), Positives = 59/137 (43%), Gaps = 4/137 (2%)
Query 2 SKAARLLSRFPPSLLQYRAAAVDGVRWLVAIEGAPDTAYANDPFLLSFKFFPKYPIDSPE 61
+K L+R PP+ D W I G D+ Y F L+ F YP P+
Sbjct 7 NKELSDLARDPPAQCSAGPVGDDMFHWQATIMGPNDSPYQGGVFFLTIHFPTDYPFKPPK 66
Query 62 LVFVAPFVPVHPHVYSNGHICLSILYDQWSPALGVKACCISLLSMLSSNREKQPPVDD-- 119
+ F HP++ SNG ICL IL QWSPAL + +S+ S+L P V +
Sbjct 67 VAFTTRIY--HPNINSNGSICLDILRSQWSPALTISKVLLSICSLLCDPNPDDPLVPEIA 124
Query 120 RVYCDSRGLRGPKGVKW 136
R+Y R +W
Sbjct 125 RIYKTDRDKYNRISREW 141
> xla:496302 ube2c; ubiquitin-conjugating enzyme E2C (EC:6.3.2.19);
K06688 ubiquitin-conjugating enzyme E2 C [EC:6.3.2.19]
Length=179
Score = 67.4 bits (163), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 35/89 (39%), Positives = 51/89 (57%), Gaps = 2/89 (2%)
Query 27 RWLVAIEGAPDTAYANDPFLLSFKFFPKYPIDSPELVFVAPFVPVHPHVYSNGHICLSIL 86
+W+ I+GA T Y N + LS +F YP ++P + FV P HP+V S+G+ICL IL
Sbjct 61 KWIGTIDGAVGTVYENLRYKLSLEFPSGYPYNAPTVKFVTPCF--HPNVDSHGNICLDIL 118
Query 87 YDQWSPALGVKACCISLLSMLSSNREKQP 115
D+WS V+ +SL S+L + P
Sbjct 119 KDKWSALYDVRTILLSLQSLLGEPNNESP 147
> dre:393934 MGC55886, Ube2d2, zgc:77149; zgc:55886 (EC:6.3.2.19);
K06689 ubiquitin-conjugating enzyme E2 D/E [EC:6.3.2.19]
Length=147
Score = 67.0 bits (162), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 41/121 (33%), Positives = 54/121 (44%), Gaps = 4/121 (3%)
Query 8 LSRFPPSLLQYRAAAVDGVRWLVAIEGAPDTAYANDPFLLSFKFFPKYPIDSPELVFVAP 67
L R PP+ D W I G D+ Y F L+ F YP P++ F
Sbjct 13 LGRDPPAQCSAGPVGDDMFHWQATIMGPNDSPYQGGVFFLTIHFPTDYPFKPPKVAFTTR 72
Query 68 FVPVHPHVYSNGHICLSILYDQWSPALGVKACCISLLSMLSSNREKQPPVDD--RVYCDS 125
HP++ SNG ICL IL QWSPAL + +S+ S+L P V + R+Y
Sbjct 73 IY--HPNINSNGSICLDILRSQWSPALTISKVLLSICSLLCDPNPDDPLVPEIARIYKTD 130
Query 126 R 126
R
Sbjct 131 R 131
> bbo:BBOV_IV002040 21.m02810; ubiquitin-conjugating enzyme E2
(EC:6.3.2.19); K06689 ubiquitin-conjugating enzyme E2 D/E [EC:6.3.2.19]
Length=147
Score = 67.0 bits (162), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 40/131 (30%), Positives = 61/131 (46%), Gaps = 4/131 (3%)
Query 8 LSRFPPSLLQYRAAAVDGVRWLVAIEGAPDTAYANDPFLLSFKFFPKYPIDSPELVFVAP 67
L++ PP+ D W I G ++ Y N + L+ F YP P++ F
Sbjct 13 LTKDPPTNCSAGPVGDDMFHWQATIMGPHNSLYQNGVYFLNIHFPSDYPFKPPKVAFTTK 72
Query 68 FVPVHPHVYSNGHICLSILYDQWSPALGVKACCISLLSMLSSNREKQPPVDD--RVYCDS 125
HP++ +NG ICL IL DQWSPAL + +S+ S+L+ P V + ++Y +
Sbjct 73 VY--HPNINNNGAICLDILKDQWSPALTISKVLLSISSLLTDPNPDDPLVPEIAQLYKQN 130
Query 126 RGLRGPKGVKW 136
R L W
Sbjct 131 RKLYESTVRDW 141
> dre:100001914 ube2d4, MGC162263, zgc:162263; ubiquitin-conjugating
enzyme E2D 4 (putative) (EC:6.3.2.19); K06689 ubiquitin-conjugating
enzyme E2 D/E [EC:6.3.2.19]
Length=147
Score = 66.6 bits (161), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 37/100 (37%), Positives = 47/100 (47%), Gaps = 2/100 (2%)
Query 8 LSRFPPSLLQYRAAAVDGVRWLVAIEGAPDTAYANDPFLLSFKFFPKYPIDSPELVFVAP 67
L R PP+ D W I G D+ Y F L+ F YP P++ F
Sbjct 13 LQRDPPAQCSAGPVGEDLFHWQATIMGPNDSPYQGGVFFLTIHFPTDYPFKPPKVAFTTK 72
Query 68 FVPVHPHVYSNGHICLSILYDQWSPALGVKACCISLLSML 107
HP++ SNG ICL IL QWSPAL V +S+ S+L
Sbjct 73 IY--HPNINSNGSICLDILRSQWSPALTVSKVLLSICSLL 110
> hsa:51619 UBE2D4, FLJ32004, HBUCE1; ubiquitin-conjugating enzyme
E2D 4 (putative) (EC:6.3.2.19); K06689 ubiquitin-conjugating
enzyme E2 D/E [EC:6.3.2.19]
Length=147
Score = 66.2 bits (160), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 37/100 (37%), Positives = 47/100 (47%), Gaps = 2/100 (2%)
Query 8 LSRFPPSLLQYRAAAVDGVRWLVAIEGAPDTAYANDPFLLSFKFFPKYPIDSPELVFVAP 67
L R PP+ D W I G D+ Y F L+ F YP P++ F
Sbjct 13 LQRDPPAQCSAGPVGDDLFHWQATIMGPNDSPYQGGVFFLTIHFPTDYPFKPPKVAFTTK 72
Query 68 FVPVHPHVYSNGHICLSILYDQWSPALGVKACCISLLSML 107
HP++ SNG ICL IL QWSPAL V +S+ S+L
Sbjct 73 IY--HPNINSNGSICLDILRSQWSPALTVSKVLLSICSLL 110
> tgo:TGME49_119870 ubiquitin-conjugating enzyme domain-containing
protein (EC:6.3.2.19); K06689 ubiquitin-conjugating enzyme
E2 D/E [EC:6.3.2.19]
Length=345
Score = 65.9 bits (159), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 36/101 (35%), Positives = 50/101 (49%), Gaps = 2/101 (1%)
Query 8 LSRFPPSLLQYRAAAVDGVRWLVAIEGAPDTAYANDPFLLSFKFFPKYPIDSPELVFVAP 67
LS+ PP+ D W I G D+ Y+ F L+ F YP P++ F
Sbjct 211 LSKDPPTNCSAGPVGDDMFHWQATIMGPEDSPYSGGVFFLNIHFPSDYPFKPPKVNFTTK 270
Query 68 FVPVHPHVYSNGHICLSILYDQWSPALGVKACCISLLSMLS 108
HP++ S G ICL IL DQWSPAL + +S+ S+L+
Sbjct 271 IY--HPNINSQGAICLDILKDQWSPALTISKVLLSISSLLT 309
> dre:335444 ube2d2, wu:fj13d01, zgc:73200; ubiquitin-conjugating
enzyme E2D 2 (UBC4/5 homolog, yeast) (EC:6.3.2.-); K06689
ubiquitin-conjugating enzyme E2 D/E [EC:6.3.2.19]
Length=147
Score = 65.9 bits (159), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 40/117 (34%), Positives = 53/117 (45%), Gaps = 4/117 (3%)
Query 8 LSRFPPSLLQYRAAAVDGVRWLVAIEGAPDTAYANDPFLLSFKFFPKYPIDSPELVFVAP 67
L R PP+ D W I G D+ Y F L+ F YP P++ F
Sbjct 13 LGRDPPAQCSAGPVGDDLFHWQATIMGPNDSPYQGGVFFLTIHFPTDYPFKPPKVAFTTR 72
Query 68 FVPVHPHVYSNGHICLSILYDQWSPALGVKACCISLLSMLSSNREKQPPVDD--RVY 122
HP++ SNG ICL IL QWSPAL + +S+ S+L P V + R+Y
Sbjct 73 IY--HPNINSNGSICLDILRSQWSPALTISKVLLSICSLLCDPNPDDPLVPEIARIY 127
> cpv:cgd4_570 ubiquitin-conjugating enzyme ; K06689 ubiquitin-conjugating
enzyme E2 D/E [EC:6.3.2.19]
Length=161
Score = 65.9 bits (159), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 41/131 (31%), Positives = 60/131 (45%), Gaps = 4/131 (3%)
Query 8 LSRFPPSLLQYRAAAVDGVRWLVAIEGAPDTAYANDPFLLSFKFFPKYPIDSPELVFVAP 67
L+R PP+ D W I G D++YA F L+ +F YP P++ F
Sbjct 27 LNRDPPTNCSAGPIGDDMFHWQATIMGPDDSSYAGGVFFLNIQFPSDYPFKPPKVNFTTK 86
Query 68 FVPVHPHVYSNGHICLSILYDQWSPALGVKACCISLLSMLSSNREKQPPVDD--RVYCDS 125
H ++ SNG ICL IL +QWSPAL + +S+ S+L+ P V + +Y
Sbjct 87 IY--HCNINSNGAICLDILKEQWSPALTISKVLLSISSLLTDANPDDPLVPEIAHLYKTD 144
Query 126 RGLRGPKGVKW 136
R +W
Sbjct 145 RSKYEQTAREW 155
> tgo:TGME49_016130 ubiquitin-conjugating enzyme E2, putative
(EC:6.3.2.19); K10573 ubiquitin-conjugating enzyme E2 A [EC:6.3.2.19]
Length=227
Score = 65.9 bits (159), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 40/123 (32%), Positives = 59/123 (47%), Gaps = 4/123 (3%)
Query 6 RLLSRFPPSLLQYRAAAVDGVRWLVAIEGAPDTAYANDPFLLSFKFFPKYPIDSPELVFV 65
R L PP + D ++W I G DT + F L F +YP P + F+
Sbjct 14 RKLQTDPPHGVNGAPVGNDIMKWNAVIFGPEDTPWEGGTFQLEMIFSNEYPNRPPLVKFL 73
Query 66 APFVPVHPHVYSNGHICLSILYDQWSPALGVKACCISLLSMLSSNREKQPPVDD--RVYC 123
+ HP+VY++G+ICL IL QWSP + A S+ S+LS P + R+Y
Sbjct 74 SKLF--HPNVYNDGNICLDILQTQWSPIYDISAILTSIQSLLSDPNPSSPANQEAARLYA 131
Query 124 DSR 126
++R
Sbjct 132 ENR 134
> xla:444775 ube2c, MGC81948; ubiquitin-conjugating enzyme E2
C (EC:6.3.2.19); K06688 ubiquitin-conjugating enzyme E2 C [EC:6.3.2.19]
Length=179
Score = 65.1 bits (157), Expect = 6e-11, Method: Compositional matrix adjust.
Identities = 34/89 (38%), Positives = 51/89 (57%), Gaps = 2/89 (2%)
Query 27 RWLVAIEGAPDTAYANDPFLLSFKFFPKYPIDSPELVFVAPFVPVHPHVYSNGHICLSIL 86
+W+ I+GA T Y + + LS +F YP ++P + FV P HP+V S+G+ICL IL
Sbjct 61 KWIGTIDGAVGTVYEDLRYKLSLEFPSGYPYNAPTVKFVTPCF--HPNVDSHGNICLDIL 118
Query 87 YDQWSPALGVKACCISLLSMLSSNREKQP 115
D+WS V+ +SL S+L + P
Sbjct 119 KDKWSALYDVRTILLSLQSLLGEPNNESP 147
> xla:100101299 ube2d4, HBUCE1, ube2d3.2; ubiquitin-conjugating
enzyme E2D 4 (putative) (EC:6.3.2.19); K06689 ubiquitin-conjugating
enzyme E2 D/E [EC:6.3.2.19]
Length=147
Score = 65.1 bits (157), Expect = 7e-11, Method: Compositional matrix adjust.
Identities = 36/100 (36%), Positives = 47/100 (47%), Gaps = 2/100 (2%)
Query 8 LSRFPPSLLQYRAAAVDGVRWLVAIEGAPDTAYANDPFLLSFKFFPKYPIDSPELVFVAP 67
L R PP+ D W I G D+ + F L+ F YP P++ F
Sbjct 13 LQRDPPAQCSAGPVGEDLFHWQATIMGPNDSPFQGGVFFLTIHFPTDYPFKPPKVAFTTK 72
Query 68 FVPVHPHVYSNGHICLSILYDQWSPALGVKACCISLLSML 107
HP++ SNG ICL IL QWSPAL V +S+ S+L
Sbjct 73 IY--HPNINSNGSICLDILRSQWSPALTVSKVLLSICSLL 110
> dre:100001969 ubiquitin-conjugating enzyme E2D 2-like
Length=147
Score = 64.7 bits (156), Expect = 9e-11, Method: Compositional matrix adjust.
Identities = 39/117 (33%), Positives = 54/117 (46%), Gaps = 4/117 (3%)
Query 8 LSRFPPSLLQYRAAAVDGVRWLVAIEGAPDTAYANDPFLLSFKFFPKYPIDSPELVFVAP 67
L+R PP+ D W I G ++ Y F L+ F YP P++ F
Sbjct 13 LARDPPAQCSAGPVGDDVFHWQATIMGPNESPYQGGVFFLTIHFPTDYPFKPPKVAFTTR 72
Query 68 FVPVHPHVYSNGHICLSILYDQWSPALGVKACCISLLSMLSSNREKQPPVDD--RVY 122
HP++ SNG ICL IL QWSPAL + +S+ S+L P V + R+Y
Sbjct 73 IY--HPNINSNGSICLDILRSQWSPALTISKVLLSICSLLCDPNPDDPLVPEIARIY 127
> dre:321832 eff, fb36f02, wu:fb36f02; zgc:56340 (EC:6.3.2.-);
K01932 [EC:6.3.2.-]
Length=147
Score = 64.7 bits (156), Expect = 9e-11, Method: Compositional matrix adjust.
Identities = 39/117 (33%), Positives = 54/117 (46%), Gaps = 4/117 (3%)
Query 8 LSRFPPSLLQYRAAAVDGVRWLVAIEGAPDTAYANDPFLLSFKFFPKYPIDSPELVFVAP 67
L+R PP+ D W I G ++ Y F L+ F YP P++ F
Sbjct 13 LARDPPAQCSAGPVGDDVFHWQATIMGPNESPYQGGVFFLTIHFPTDYPFKPPKVAFTTR 72
Query 68 FVPVHPHVYSNGHICLSILYDQWSPALGVKACCISLLSMLSSNREKQPPVDD--RVY 122
HP++ SNG ICL IL QWSPAL + +S+ S+L P V + R+Y
Sbjct 73 IY--HPNINSNGSICLDILRSQWSPALTISKVLLSICSLLCDPNPDDPLVPEIARIY 127
> tgo:TGME49_002820 ubiquitin-conjugating enzyme E2, putative
(EC:6.3.2.19); K10575 ubiquitin-conjugating enzyme E2 G1 [EC:6.3.2.19]
Length=168
Score = 64.7 bits (156), Expect = 9e-11, Method: Compositional matrix adjust.
Identities = 37/109 (33%), Positives = 54/109 (49%), Gaps = 16/109 (14%)
Query 21 AAVDGVRWLVAIEGAPDTAYANDPFLLSFKFFPKYPIDSPELVFVAPFVPVHPHVYSNGH 80
A D W V EG PDT Y F + KF P +P PE+ F+ HP++Y +G
Sbjct 32 AGGDFFVWRVCFEGPPDTLYEGGIFTAALKFPPDFPNHPPEMKFLQDM--WHPNIYPDGR 89
Query 81 ICLSILY--------------DQWSPALGVKACCISLLSMLSSNREKQP 115
+C+SIL+ ++W P LGV+A +S++SML + P
Sbjct 90 VCISILHPPGDDVFNEQEKAEERWRPILGVEAILLSVISMLGEPNLESP 138
Lambda K H
0.324 0.140 0.458
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2749206264
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40