bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_0589_orf1
Length=121
Score E
Sequences producing significant alignments: (Bits) Value
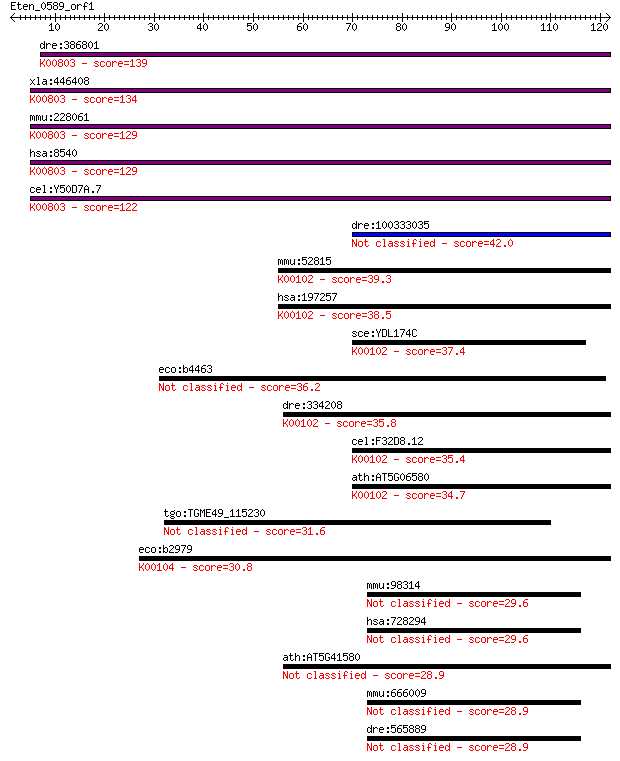
dre:386801 agps, fd16e06, wu:fd16e06, zgc:56718; alkylglyceron... 139 2e-33
xla:446408 agps, MGC83829; alkylglycerone phosphate synthase (... 134 9e-32
mmu:228061 Agps, 5832437L22, 9930035G10Rik, AW123847, Adaps, A... 129 2e-30
hsa:8540 AGPS, ADAP-S, ADAS, ADHAPS, ADPS, ALDHPSY, DKFZp762O2... 129 3e-30
cel:Y50D7A.7 ads-1; Alkyl-Dihydroxyacetonephosphate Synthase f... 122 4e-28
dre:100333035 lactate dehydrogenase D-like 42.0 5e-04
mmu:52815 Ldhd, 4733401P21Rik, D8Bwg1320e; lactate dehydrogena... 39.3 0.003
hsa:197257 LDHD, DLD, MGC57726; lactate dehydrogenase D (EC:1.... 38.5 0.005
sce:YDL174C DLD1; D-lactate dehydrogenase, oxidizes D-lactate ... 37.4 0.011
eco:b4463 ygcU, ECK2767, JW5442, ygcT, ygcV; predicted FAD con... 36.2 0.029
dre:334208 ldhd, wu:fi36b04, zgc:55447; lactate dehydrogenase ... 35.8 0.030
cel:F32D8.12 hypothetical protein; K00102 D-lactate dehydrogen... 35.4 0.040
ath:AT5G06580 FAD linked oxidase family protein (EC:1.1.2.4); ... 34.7 0.071
tgo:TGME49_115230 hypothetical protein 31.6 0.66
eco:b2979 glcD, ECK2974, gox, JW2946, yghM; glycolate oxidase ... 30.8 1.2
mmu:98314 D2hgdh, AA408776, AA408778, AI325464; D-2-hydroxyglu... 29.6 2.1
hsa:728294 D2HGDH, D2HGD, FLJ42195, MGC25181; D-2-hydroxygluta... 29.6 2.4
ath:AT5G41580 zinc ion binding 28.9 3.7
mmu:666009 Gm7889, EG666009; predicted gene 7889 28.9
dre:565889 d2hgdh, zgc:158661; D-2-hydroxyglutarate dehydrogen... 28.9 3.9
> dre:386801 agps, fd16e06, wu:fd16e06, zgc:56718; alkylglycerone
phosphate synthase (EC:2.5.1.26); K00803 alkyldihydroxyacetonephosphate
synthase [EC:2.5.1.26]
Length=629
Score = 139 bits (350), Expect = 2e-33, Method: Compositional matrix adjust.
Identities = 60/115 (52%), Positives = 86/115 (74%), Gaps = 0/115 (0%)
Query 7 VKDRIKSDCSRLGIKYPPLVMVRLTQVYDGGAVLYFYFGFNWAGLREPIKVFSEVEHNAR 66
VK+RI +C G+++PPL R+TQ YD GA +YFYF FN+ GL +P+ V+ +VEH AR
Sbjct 515 VKERIIRECKERGVQFPPLSTCRVTQTYDAGACVYFYFAFNYRGLSDPVHVYEQVEHAAR 574
Query 67 QVILDCKGSISHHHGVGKLRKEFMAKAVGKTGMELLRATKDAVDPQNIFGNGNMI 121
+ IL GS+SHHHGVGKLRKE+M ++V G+ ++++ K+ VDPQNIFG+ N++
Sbjct 575 EEILANGGSLSHHHGVGKLRKEWMKESVSGVGLGMIQSVKEFVDPQNIFGSRNLL 629
> xla:446408 agps, MGC83829; alkylglycerone phosphate synthase
(EC:2.5.1.26); K00803 alkyldihydroxyacetonephosphate synthase
[EC:2.5.1.26]
Length=627
Score = 134 bits (336), Expect = 9e-32, Method: Compositional matrix adjust.
Identities = 57/117 (48%), Positives = 83/117 (70%), Gaps = 0/117 (0%)
Query 5 QRVKDRIKSDCSRLGIKYPPLVMVRLTQVYDGGAVLYFYFGFNWAGLREPIKVFSEVEHN 64
+ VK+RI +C G+++PPL R+TQ YD GA +YFYF FN+ GL +P+ V+ E+E
Sbjct 511 RNVKERIVRECKERGVQFPPLSTCRVTQTYDAGACVYFYFAFNYRGLSDPVHVYEEIEFA 570
Query 65 ARQVILDCKGSISHHHGVGKLRKEFMAKAVGKTGMELLRATKDAVDPQNIFGNGNMI 121
AR+ IL GS+SHHHGVGKLRK+++ ++ G+ +LR+ K+ VDP N FGN N++
Sbjct 571 AREEILANGGSLSHHHGVGKLRKQWLKDSISDVGIGMLRSVKNFVDPNNTFGNRNLL 627
> mmu:228061 Agps, 5832437L22, 9930035G10Rik, AW123847, Adaps,
Adas, Adhaps, Adps, Aldhpsy, bs2; alkylglycerone phosphate
synthase (EC:2.5.1.26); K00803 alkyldihydroxyacetonephosphate
synthase [EC:2.5.1.26]
Length=671
Score = 129 bits (325), Expect = 2e-30, Method: Compositional matrix adjust.
Identities = 54/117 (46%), Positives = 82/117 (70%), Gaps = 0/117 (0%)
Query 5 QRVKDRIKSDCSRLGIKYPPLVMVRLTQVYDGGAVLYFYFGFNWAGLREPIKVFSEVEHN 64
+ VK+RI+ +C G+++ PL R+TQ YD GA +YFYF FN+ G+ +P+ VF E
Sbjct 555 RNVKERIRRECKERGVQFAPLSTCRVTQTYDAGACIYFYFAFNYRGISDPLTVFEHTEAA 614
Query 65 ARQVILDCKGSISHHHGVGKLRKEFMAKAVGKTGMELLRATKDAVDPQNIFGNGNMI 121
AR+ IL GS+SHHHGVGK+RK+++ +++ G +L++ K+ VDP NIFGN N++
Sbjct 615 AREEILANGGSLSHHHGVGKIRKQWLKESISDVGFGMLKSVKEYVDPSNIFGNRNLL 671
> hsa:8540 AGPS, ADAP-S, ADAS, ADHAPS, ADPS, ALDHPSY, DKFZp762O2215,
FLJ99755; alkylglycerone phosphate synthase (EC:2.5.1.26);
K00803 alkyldihydroxyacetonephosphate synthase [EC:2.5.1.26]
Length=658
Score = 129 bits (323), Expect = 3e-30, Method: Compositional matrix adjust.
Identities = 54/117 (46%), Positives = 81/117 (69%), Gaps = 0/117 (0%)
Query 5 QRVKDRIKSDCSRLGIKYPPLVMVRLTQVYDGGAVLYFYFGFNWAGLREPIKVFSEVEHN 64
+ VK+RI +C G+++ P R+TQ YD GA +YFYF FN+ G+ +P+ VF + E
Sbjct 542 RNVKERITRECKEKGVQFAPFSTCRVTQTYDAGACIYFYFAFNYRGISDPLTVFEQTEAA 601
Query 65 ARQVILDCKGSISHHHGVGKLRKEFMAKAVGKTGMELLRATKDAVDPQNIFGNGNMI 121
AR+ IL GS+SHHHGVGKLRK+++ +++ G +L++ K+ VDP NIFGN N++
Sbjct 602 AREEILANGGSLSHHHGVGKLRKQWLKESISDVGFGMLKSVKEYVDPNNIFGNRNLL 658
> cel:Y50D7A.7 ads-1; Alkyl-Dihydroxyacetonephosphate Synthase
family member (ads-1); K00803 alkyldihydroxyacetonephosphate
synthase [EC:2.5.1.26]
Length=597
Score = 122 bits (305), Expect = 4e-28, Method: Compositional matrix adjust.
Identities = 56/117 (47%), Positives = 78/117 (66%), Gaps = 0/117 (0%)
Query 5 QRVKDRIKSDCSRLGIKYPPLVMVRLTQVYDGGAVLYFYFGFNWAGLREPIKVFSEVEHN 64
+ VK+ +K + G+ +P L R+TQVYD GA +YFYFGFN GL+ ++V+ +E
Sbjct 471 RNVKELMKREAKAQGVTHPVLANCRVTQVYDAGACVYFYFGFNARGLKNGLEVYDRIETA 530
Query 65 ARQVILDCKGSISHHHGVGKLRKEFMAKAVGKTGMELLRATKDAVDPQNIFGNGNMI 121
AR I+ C GSISHHHGVGK+RK++M G G+ LL+A K +DP NIF + N+I
Sbjct 531 ARDEIIACGGSISHHHGVGKIRKQWMLTTNGAVGIALLKAIKSELDPANIFASANLI 587
> dre:100333035 lactate dehydrogenase D-like
Length=484
Score = 42.0 bits (97), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 19/52 (36%), Positives = 31/52 (59%), Gaps = 1/52 (1%)
Query 70 LDCKGSISHHHGVGKLRKEFMAKAVGKTGMELLRATKDAVDPQNIFGNGNMI 121
L C G+ + HG+G+ + F+ + G + L+R K AVDP+NIF G ++
Sbjct 430 LACGGTCTGEHGIGQGKMSFLEEEAGP-ALGLMRTIKRAVDPKNIFNPGKIL 480
> mmu:52815 Ldhd, 4733401P21Rik, D8Bwg1320e; lactate dehydrogenase
D (EC:1.1.2.4); K00102 D-lactate dehydrogenase (cytochrome)
[EC:1.1.2.4]
Length=484
Score = 39.3 bits (90), Expect = 0.003, Method: Composition-based stats.
Identities = 18/67 (26%), Positives = 37/67 (55%), Gaps = 3/67 (4%)
Query 55 IKVFSEVEHNARQVILDCKGSISHHHGVGKLRKEFMAKAVGKTGMELLRATKDAVDPQNI 114
+K F+E N + L G+ + HG+G +++ + + VG G+E +R K+ +DP+ +
Sbjct 421 VKAFAE---NLGRRALALGGTCTGEHGIGLGKRQLLQEEVGPVGVETMRQLKNTLDPRGL 477
Query 115 FGNGNMI 121
G ++
Sbjct 478 MNPGKVL 484
> hsa:197257 LDHD, DLD, MGC57726; lactate dehydrogenase D (EC:1.1.2.4);
K00102 D-lactate dehydrogenase (cytochrome) [EC:1.1.2.4]
Length=507
Score = 38.5 bits (88), Expect = 0.005, Method: Composition-based stats.
Identities = 19/67 (28%), Positives = 37/67 (55%), Gaps = 3/67 (4%)
Query 55 IKVFSEVEHNARQVILDCKGSISHHHGVGKLRKEFMAKAVGKTGMELLRATKDAVDPQNI 114
+K F+E + R + L G+ + HG+G +++ + + VG G+E +R K +DPQ +
Sbjct 444 VKAFAE-QLGRRALAL--HGTCTGEHGIGMGKRQLLQEEVGAVGVETMRQLKAVLDPQGL 500
Query 115 FGNGNMI 121
G ++
Sbjct 501 MNPGKVL 507
> sce:YDL174C DLD1; D-lactate dehydrogenase, oxidizes D-lactate
to pyruvate, transcription is heme-dependent, repressed by
glucose, and derepressed in ethanol or lactate; located in
the mitochondrial inner membrane (EC:1.1.2.4); K00102 D-lactate
dehydrogenase (cytochrome) [EC:1.1.2.4]
Length=587
Score = 37.4 bits (85), Expect = 0.011, Method: Compositional matrix adjust.
Identities = 15/47 (31%), Positives = 31/47 (65%), Gaps = 0/47 (0%)
Query 70 LDCKGSISHHHGVGKLRKEFMAKAVGKTGMELLRATKDAVDPQNIFG 116
L+ +G+ + HGVG ++E++ + +G+ ++L+R K A+DP+ I
Sbjct 524 LNAEGTCTGEHGVGIGKREYLLEELGEAPVDLMRKIKLAIDPKRIMN 570
> eco:b4463 ygcU, ECK2767, JW5442, ygcT, ygcV; predicted FAD containing
dehydrogenase
Length=484
Score = 36.2 bits (82), Expect = 0.029, Method: Compositional matrix adjust.
Identities = 24/94 (25%), Positives = 41/94 (43%), Gaps = 6/94 (6%)
Query 31 TQVYDGGAVLYFYFGFNWAGLREPIKVFSEVEHNARQVILD----CKGSISHHHGVGKLR 86
+ Y G +YF + +N + P + + + ++I + GS+ HHHG+GK R
Sbjct 388 SHSYQNGTNMYFVYDYNVVDCK-PEEEIDKYHNPLNKIICEETIRLGGSMVHHHGIGKHR 446
Query 87 KEFMAKAVGKTGMELLRATKDAVDPQNIFGNGNM 120
+ +K + LL K DP I G +
Sbjct 447 VHW-SKLEHGSAWALLEGLKKQFDPNGIMNTGTI 479
> dre:334208 ldhd, wu:fi36b04, zgc:55447; lactate dehydrogenase
D (EC:1.1.2.4); K00102 D-lactate dehydrogenase (cytochrome)
[EC:1.1.2.4]
Length=497
Score = 35.8 bits (81), Expect = 0.030, Method: Compositional matrix adjust.
Identities = 17/66 (25%), Positives = 37/66 (56%), Gaps = 1/66 (1%)
Query 56 KVFSEVEHNARQVILDCKGSISHHHGVGKLRKEFMAKAVGKTGMELLRATKDAVDPQNIF 115
+V S E AR+ L G+ + HG+G ++ + + +G +E+++ K ++DP+N+
Sbjct 424 RVHSFTERLARRA-LAMDGTCTGEHGIGLGKRALLREELGPLAIEVMKGLKASLDPRNLM 482
Query 116 GNGNMI 121
G ++
Sbjct 483 NPGKLL 488
> cel:F32D8.12 hypothetical protein; K00102 D-lactate dehydrogenase
(cytochrome) [EC:1.1.2.4]
Length=460
Score = 35.4 bits (80), Expect = 0.040, Method: Compositional matrix adjust.
Identities = 14/52 (26%), Positives = 30/52 (57%), Gaps = 0/52 (0%)
Query 70 LDCKGSISHHHGVGKLRKEFMAKAVGKTGMELLRATKDAVDPQNIFGNGNMI 121
L G+ + HG+G +++++ + +G+ + L+ K A+DP NI G ++
Sbjct 406 LAADGTCTGEHGIGLGKRKYLREELGENTVRLMHTIKHALDPNNIMNPGKVL 457
> ath:AT5G06580 FAD linked oxidase family protein (EC:1.1.2.4);
K00102 D-lactate dehydrogenase (cytochrome) [EC:1.1.2.4]
Length=567
Score = 34.7 bits (78), Expect = 0.071, Method: Compositional matrix adjust.
Identities = 14/52 (26%), Positives = 28/52 (53%), Gaps = 0/52 (0%)
Query 70 LDCKGSISHHHGVGKLRKEFMAKAVGKTGMELLRATKDAVDPQNIFGNGNMI 121
L G+ + HGVG + +++ K +G ++ ++ K +DP +I G +I
Sbjct 510 LSMDGTCTGEHGVGTGKMKYLEKELGIEALQTMKRIKKTLDPNDIMNPGKLI 561
> tgo:TGME49_115230 hypothetical protein
Length=628
Score = 31.6 bits (70), Expect = 0.66, Method: Composition-based stats.
Identities = 26/82 (31%), Positives = 39/82 (47%), Gaps = 6/82 (7%)
Query 32 QVYDGGAVLYFYFGFNWAGLREPIKVFSE--VEHNARQVILDCKGSISHHHGVGKLRKEF 89
Q YDGG + Y GF LRE ++ E VEH+A+ ++L C + G + + F
Sbjct 153 QAYDGGQMQQSYRGFR--RLRETLEHADEQSVEHSAKALVLGCMYTALFRVGEYREARRF 210
Query 90 MAKA--VGKTGMELLRATKDAV 109
A A + + G + L A V
Sbjct 211 AADARDLSRAGHQSLAADTATV 232
> eco:b2979 glcD, ECK2974, gox, JW2946, yghM; glycolate oxidase
subunit, FAD-linked; K00104 glycolate oxidase [EC:1.1.3.15]
Length=499
Score = 30.8 bits (68), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 28/106 (26%), Positives = 42/106 (39%), Gaps = 14/106 (13%)
Query 27 MVRLTQVYDGGAVLYFYFGFNWAGLREPIKVFSEVEHN--AR---------QVILDCKGS 75
+ RL+Q YD F+ G G P+ +F E AR ++ ++ GS
Sbjct 370 IARLSQQYDLRVANVFHAG---DGNMHPLILFDANEPGEFARAEELGGKILELCVEVGGS 426
Query 76 ISHHHGVGKLRKEFMAKAVGKTGMELLRATKDAVDPQNIFGNGNMI 121
IS HG+G+ + M + A K A DP + G I
Sbjct 427 ISGEHGIGREKINQMCAQFNSDEITTFHAVKAAFDPDGLLNPGKNI 472
> mmu:98314 D2hgdh, AA408776, AA408778, AI325464; D-2-hydroxyglutarate
dehydrogenase (EC:1.1.99.-)
Length=535
Score = 29.6 bits (65), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 12/43 (27%), Positives = 24/43 (55%), Gaps = 0/43 (0%)
Query 73 KGSISHHHGVGKLRKEFMAKAVGKTGMELLRATKDAVDPQNIF 115
+GS+S HG+G +K+ + + + L++ K +DP+ I
Sbjct 483 RGSVSAEHGLGFKKKDVLGYSKPPVAVTLMQQLKAMLDPEGIL 525
> hsa:728294 D2HGDH, D2HGD, FLJ42195, MGC25181; D-2-hydroxyglutarate
dehydrogenase (EC:1.1.99.-)
Length=521
Score = 29.6 bits (65), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 12/43 (27%), Positives = 25/43 (58%), Gaps = 0/43 (0%)
Query 73 KGSISHHHGVGKLRKEFMAKAVGKTGMELLRATKDAVDPQNIF 115
+GS+S HGVG +++ + + ++L++ K +DP+ I
Sbjct 469 QGSVSAEHGVGFRKRDVLGYSKPPGALQLMQQLKALLDPKGIL 511
> ath:AT5G41580 zinc ion binding
Length=760
Score = 28.9 bits (63), Expect = 3.7, Method: Composition-based stats.
Identities = 17/66 (25%), Positives = 29/66 (43%), Gaps = 12/66 (18%)
Query 56 KVFSEVEHNARQVILDCKGSISHHHGVGKLRKEFMAKAVGKTGMELLRATKDAVDPQNIF 115
K+ +VEHNA VI+D G+ + K G+T + D DP ++
Sbjct 372 KILKDVEHNAADVIIDAGGTWK------------VTKNTGETPEPVREIIHDLEDPMSLL 419
Query 116 GNGNMI 121
+G ++
Sbjct 420 NSGPVV 425
> mmu:666009 Gm7889, EG666009; predicted gene 7889
Length=189
Score = 28.9 bits (63), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 12/43 (27%), Positives = 24/43 (55%), Gaps = 0/43 (0%)
Query 73 KGSISHHHGVGKLRKEFMAKAVGKTGMELLRATKDAVDPQNIF 115
+GS+S HG+G +K+ + + + L++ K +DP+ I
Sbjct 137 RGSVSAEHGLGFKKKDVLGYSKPSVAVTLMQQLKAMLDPKGIL 179
> dre:565889 d2hgdh, zgc:158661; D-2-hydroxyglutarate dehydrogenase
(EC:1.1.99.-)
Length=533
Score = 28.9 bits (63), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 13/43 (30%), Positives = 24/43 (55%), Gaps = 0/43 (0%)
Query 73 KGSISHHHGVGKLRKEFMAKAVGKTGMELLRATKDAVDPQNIF 115
KGSIS HG+G ++ ++ + + L+ + K +DP+ I
Sbjct 480 KGSISAEHGLGLKKRNYIYYSKPSEAVALMGSIKAMLDPKGIL 522
Lambda K H
0.324 0.141 0.430
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2013067560
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40