bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_0499_orf2
Length=101
Score E
Sequences producing significant alignments: (Bits) Value
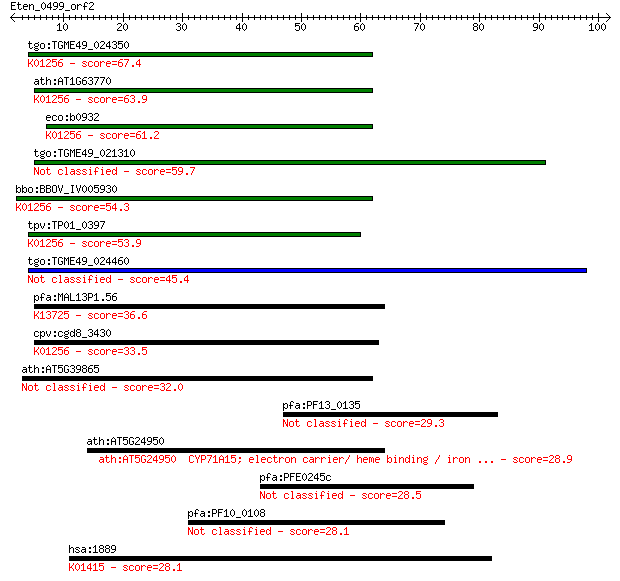
tgo:TGME49_024350 aminopeptidase N, putative (EC:3.4.11.2); K0... 67.4 1e-11
ath:AT1G63770 peptidase M1 family protein; K01256 aminopeptida... 63.9 1e-10
eco:b0932 pepN, ECK0923, JW0915; aminopeptidase N (EC:3.4.11.2... 61.2 8e-10
tgo:TGME49_021310 aminopeptidase N, putative (EC:3.4.11.2) 59.7 2e-09
bbo:BBOV_IV005930 23.m06519; aminopeptidase (EC:3.4.11.2); K01... 54.3 1e-07
tpv:TP01_0397 alpha-aminoacylpeptide hydrolase (EC:3.4.11.2); ... 53.9 1e-07
tgo:TGME49_024460 aminopeptidase N, putative (EC:3.4.11.2) 45.4 4e-05
pfa:MAL13P1.56 M1-family aminopeptidase (EC:3.4.11.-); K13725 ... 36.6 0.022
cpv:cgd8_3430 zincin/aminopeptidase N like metalloprotease ; K... 33.5 0.18
ath:AT5G39865 glutaredoxin family protein 32.0 0.48
pfa:PF13_0135 vacuolar protein sorting 52 homologue 29.3 2.8
ath:AT5G24950 CYP71A15; electron carrier/ heme binding / iron ... 28.9 4.3
pfa:PFE0245c conserved Plasmodium membrane protein, unknown fu... 28.5 5.8
pfa:PF10_0108 conserved Plasmodium protein 28.1 6.7
hsa:1889 ECE1, ECE; endothelin converting enzyme 1 (EC:3.4.24.... 28.1 7.5
> tgo:TGME49_024350 aminopeptidase N, putative (EC:3.4.11.2);
K01256 aminopeptidase N [EC:3.4.11.2]
Length=1419
Score = 67.4 bits (163), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 35/58 (60%), Positives = 41/58 (70%), Gaps = 2/58 (3%)
Query 4 GDPLVLDKWFRAQARSDLPDQVERVAELLNHPAFSLKNPNRLRSLIFTFASLNPLHFH 61
GDPLVLDKWF QA SD+ + E V EL H F+ KNPNRLR+LIF+F + NP FH
Sbjct 1284 GDPLVLDKWFAVQALSDVRNVTETVKELQKHADFTAKNPNRLRALIFSF-TRNP-QFH 1339
> ath:AT1G63770 peptidase M1 family protein; K01256 aminopeptidase
N [EC:3.4.11.2]
Length=987
Score = 63.9 bits (154), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 30/57 (52%), Positives = 42/57 (73%), Gaps = 1/57 (1%)
Query 5 DPLVLDKWFRAQARSDLPDQVERVAELLNHPAFSLKNPNRLRSLIFTFASLNPLHFH 61
D LV++KWF Q+ SD+P VE V +LL+HPAF L+NPN++ SLI F +P++FH
Sbjct 859 DYLVVNKWFLLQSTSDIPGNVENVKKLLDHPAFDLRNPNKVYSLIGGFCG-SPVNFH 914
> eco:b0932 pepN, ECK0923, JW0915; aminopeptidase N (EC:3.4.11.2);
K01256 aminopeptidase N [EC:3.4.11.2]
Length=870
Score = 61.2 bits (147), Expect = 8e-10, Method: Composition-based stats.
Identities = 29/55 (52%), Positives = 36/55 (65%), Gaps = 0/55 (0%)
Query 7 LVLDKWFRAQARSDLPDQVERVAELLNHPAFSLKNPNRLRSLIFTFASLNPLHFH 61
LV+DKWF QA S + +E V LL H +F++ NPNR+RSLI FA NP FH
Sbjct 744 LVMDKWFILQATSPAANVLETVRGLLQHRSFTMSNPNRIRSLIGAFAGSNPAAFH 798
> tgo:TGME49_021310 aminopeptidase N, putative (EC:3.4.11.2)
Length=966
Score = 59.7 bits (143), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 38/93 (40%), Positives = 51/93 (54%), Gaps = 9/93 (9%)
Query 5 DPLVLDKWFRAQARSDLPDQVERVAELLNHPAFSLKNPNRLRSLIFTFASLNPLHFHR-- 62
+P +L KWF QARS LP+ ++R+ EL HP + PN +R+L TF NP FHR
Sbjct 837 NPQLLTKWFALQARSSLPETLDRIRELEKHPEYKPLVPNFVRALYSTFMHGNPSVFHRRD 896
Query 63 -----VFFFFFFFFDKINNAKRKSSRPSTMRLS 90
+ F F D+IN R +SR +T LS
Sbjct 897 GAGYELAFVFLQSMDRIN--PRTASRAATAFLS 927
> bbo:BBOV_IV005930 23.m06519; aminopeptidase (EC:3.4.11.2); K01256
aminopeptidase N [EC:3.4.11.2]
Length=846
Score = 54.3 bits (129), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 28/60 (46%), Positives = 35/60 (58%), Gaps = 2/60 (3%)
Query 2 CPGDPLVLDKWFRAQARSDLPDQVERVAELLNHPAFSLKNPNRLRSLIFTFASLNPLHFH 61
GD LV+DKWF AQA SD D ++RV L H ++ KNPNR SL+ F + FH
Sbjct 709 AAGDALVIDKWFSAQASSDSADCLDRVRMLSTHKDYTNKNPNRANSLVRAFT--RSIRFH 766
> tpv:TP01_0397 alpha-aminoacylpeptide hydrolase (EC:3.4.11.2);
K01256 aminopeptidase N [EC:3.4.11.2]
Length=1020
Score = 53.9 bits (128), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 24/56 (42%), Positives = 34/56 (60%), Gaps = 0/56 (0%)
Query 4 GDPLVLDKWFRAQARSDLPDQVERVAELLNHPAFSLKNPNRLRSLIFTFASLNPLH 59
GD ++DKWF+AQA S+L ++ V +L++H F L NPNR SL+ F H
Sbjct 891 GDAALVDKWFKAQAVSELESSLDTVKDLMSHKDFVLSNPNRFNSLVTVFTYGENFH 946
> tgo:TGME49_024460 aminopeptidase N, putative (EC:3.4.11.2)
Length=970
Score = 45.4 bits (106), Expect = 4e-05, Method: Composition-based stats.
Identities = 28/94 (29%), Positives = 46/94 (48%), Gaps = 1/94 (1%)
Query 4 GDPLVLDKWFRAQARSDLPDQVERVAELLNHPAFSLKNPNRLRSLIFTFASLNPLHFHRV 63
G+ ++++KW QA S D +ERVA+L H AF P SL TF+S +P +
Sbjct 837 GNAVLMEKWMSLQAMSAAADTLERVAQLRQHSAFHPFIPGSAYSLFGTFSSSSPQFHSKT 896
Query 64 FFFFFFFFDKINNAKRKSSRPSTMRLSGSSHSTW 97
+ D + +S S+ R++G + + W
Sbjct 897 GAGYALVLDFLTMIDEHNSLVSS-RIAGVAFANW 929
> pfa:MAL13P1.56 M1-family aminopeptidase (EC:3.4.11.-); K13725
M1-family aminopeptidase [EC:3.4.11.-]
Length=1085
Score = 36.6 bits (83), Expect = 0.022, Method: Composition-based stats.
Identities = 20/59 (33%), Positives = 31/59 (52%), Gaps = 1/59 (1%)
Query 5 DPLVLDKWFRAQARSDLPDQVERVAELLNHPAFSLKNPNRLRSLIFTFASLNPLHFHRV 63
D L+L +W + +RSD D E + +L N KNPN +R++ F + N FH +
Sbjct 956 DELLLQEWLKTVSRSDRKDIYEILKKLENEVLKDSKNPNDIRAVYLPFTN-NLRRFHDI 1013
> cpv:cgd8_3430 zincin/aminopeptidase N like metalloprotease ;
K01256 aminopeptidase N [EC:3.4.11.2]
Length=936
Score = 33.5 bits (75), Expect = 0.18, Method: Compositional matrix adjust.
Identities = 24/61 (39%), Positives = 35/61 (57%), Gaps = 4/61 (6%)
Query 5 DPLVLDKWFRAQARSDLPDQVERVAELL-NHPAF--SLKNPNRLRSLIFTFASLNPLHFH 61
D L+++F QA +PD VERV + ++P F +NP SL+ TFAS N + F+
Sbjct 806 DVSTLNQYFSIQASCLIPDNVERVINIYHSNPQFIKYRENPTIFSSLVGTFAS-NFVAFN 864
Query 62 R 62
R
Sbjct 865 R 865
> ath:AT5G39865 glutaredoxin family protein
Length=390
Score = 32.0 bits (71), Expect = 0.48, Method: Composition-based stats.
Identities = 18/59 (30%), Positives = 28/59 (47%), Gaps = 15/59 (25%)
Query 3 PGDPLVLDKWFRAQARSDLPDQVERVAELLNHPAFSLKNPNRLRSLIFTFASLNPLHFH 61
PG+P ++ W +L + +E V+ L++PN LRS F F + P H H
Sbjct 137 PGEPETINTW-------ELMEGLEDVS--------PLRSPNHLRSFSFDFVRIQPSHDH 180
> pfa:PF13_0135 vacuolar protein sorting 52 homologue
Length=1353
Score = 29.3 bits (64), Expect = 2.8, Method: Composition-based stats.
Identities = 13/36 (36%), Positives = 21/36 (58%), Gaps = 0/36 (0%)
Query 47 SLIFTFASLNPLHFHRVFFFFFFFFDKINNAKRKSS 82
S+ +++ L L V +FFF +KINN K K++
Sbjct 262 SIKYSYIELEKLKKKSVDRIYFFFLEKINNIKNKNT 297
> ath:AT5G24950 CYP71A15; electron carrier/ heme binding / iron
ion binding / monooxygenase/ oxygen binding; K00517 [EC:1.14.-.-]
Length=496
Score = 28.9 bits (63), Expect = 4.3, Method: Composition-based stats.
Identities = 19/50 (38%), Positives = 23/50 (46%), Gaps = 0/50 (0%)
Query 14 RAQARSDLPDQVERVAELLNHPAFSLKNPNRLRSLIFTFASLNPLHFHRV 63
R A+ +LP RV + N SL LRSL + L LHF RV
Sbjct 25 RTVAKDNLPPSPWRVPVIGNLHQLSLHPHRSLRSLSHRYGPLMLLHFGRV 74
> pfa:PFE0245c conserved Plasmodium membrane protein, unknown
function
Length=2975
Score = 28.5 bits (62), Expect = 5.8, Method: Composition-based stats.
Identities = 12/36 (33%), Positives = 18/36 (50%), Gaps = 0/36 (0%)
Query 43 NRLRSLIFTFASLNPLHFHRVFFFFFFFFDKINNAK 78
NR L+F+F + + + F +FDKI N K
Sbjct 1782 NRQEKLLFSFCKYIYYKWKGIMNYLFMYFDKIKNRK 1817
> pfa:PF10_0108 conserved Plasmodium protein
Length=983
Score = 28.1 bits (61), Expect = 6.7, Method: Composition-based stats.
Identities = 15/43 (34%), Positives = 23/43 (53%), Gaps = 0/43 (0%)
Query 31 LLNHPAFSLKNPNRLRSLIFTFASLNPLHFHRVFFFFFFFFDK 73
LL++ S KN +L +I + S + L+F R+ FFF K
Sbjct 55 LLDNNIESFKNAGKLLFIILSGISPSTLYFKRILNLIHFFFHK 97
> hsa:1889 ECE1, ECE; endothelin converting enzyme 1 (EC:3.4.24.71);
K01415 endothelin-converting enzyme [EC:3.4.24.71]
Length=758
Score = 28.1 bits (61), Expect = 7.5, Method: Composition-based stats.
Identities = 14/71 (19%), Positives = 32/71 (45%), Gaps = 1/71 (1%)
Query 11 KWFRAQARSDLPDQVERVAELLNHPAFSLKNPNRLRSLIFTFASLNPLHFHRVFFFFFFF 70
KW + R ++ + + ++ +P F + +P L + + ++ L+F FF F
Sbjct 471 KWMDEETRKSAKEKADAIYNMIGYPNF-IMDPKELDKVFNDYTAVPDLYFENAMRFFNFS 529
Query 71 FDKINNAKRKS 81
+ + RK+
Sbjct 530 WRVTADQLRKA 540
Lambda K H
0.328 0.138 0.457
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2036602604
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40