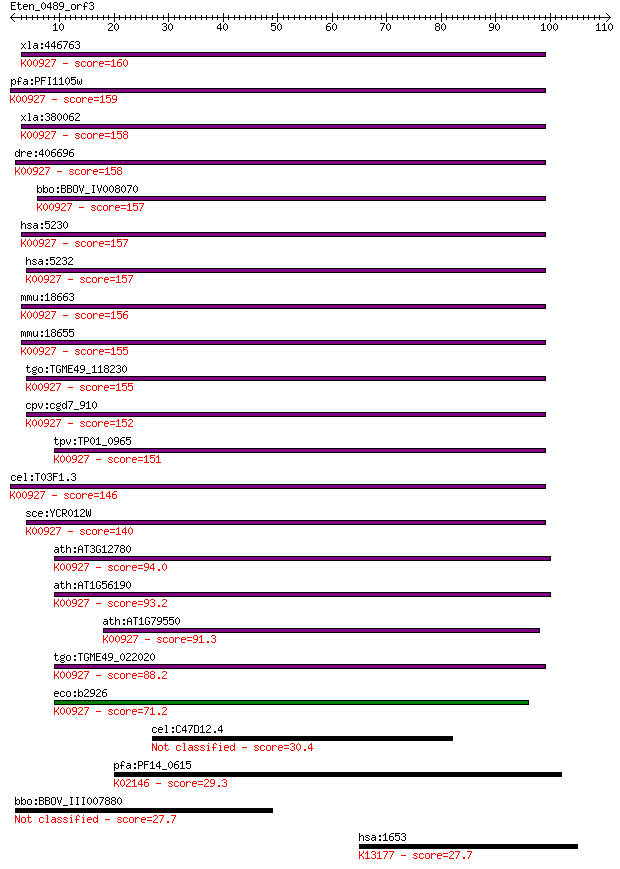
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_0489_orf3
Length=110
Score E
Sequences producing significant alignments: (Bits) Value
xla:446763 pgk1, MGC132300, MGC80128, pgk2, pgka; phosphoglyce... 160 7e-40
pfa:PFI1105w PGK; phosphoglycerate kinase (EC:2.7.2.3); K00927... 159 2e-39
xla:380062 pgk1, MGC53000; phosphoglycerate kinase 1 (EC:2.7.2... 158 4e-39
dre:406696 pgk1, wu:fd59b07, wu:fj36g06, zgc:56252, zgc:77899;... 158 5e-39
bbo:BBOV_IV008070 23.m06017; phosphoglycerate kinase (EC:2.7.2... 157 9e-39
hsa:5230 PGK1, MGC117307, MGC142128, MGC8947, MIG10, PGKA; pho... 157 9e-39
hsa:5232 PGK2, PGKB, PGKPS, dJ417L20.2; phosphoglycerate kinas... 157 1e-38
mmu:18663 Pgk2, Pgk-2, Tcp-2; phosphoglycerate kinase 2 (EC:2.... 156 2e-38
mmu:18655 Pgk1, MGC118097, Pgk-1; phosphoglycerate kinase 1 (E... 155 2e-38
tgo:TGME49_118230 phosphoglycerate kinase, putative (EC:2.7.2.... 155 4e-38
cpv:cgd7_910 phosphoglycerate kinase 1 ; K00927 phosphoglycera... 152 2e-37
tpv:TP01_0965 phosphoglycerate kinase; K00927 phosphoglycerate... 151 6e-37
cel:T03F1.3 pgk-1; PhosphoGlycerate Kinase family member (pgk-... 146 1e-35
sce:YCR012W PGK1; 3-phosphoglycerate kinase, catalyzes transfe... 140 1e-33
ath:AT3G12780 PGK1; PGK1 (PHOSPHOGLYCERATE KINASE 1); phosphog... 94.0 1e-19
ath:AT1G56190 phosphoglycerate kinase, putative; K00927 phosph... 93.2 2e-19
ath:AT1G79550 PGK; PGK (PHOSPHOGLYCERATE KINASE); phosphoglyce... 91.3 8e-19
tgo:TGME49_022020 phosphoglycerate kinase, putative (EC:2.7.2.... 88.2 6e-18
eco:b2926 pgk, ECK2922, JW2893; phosphoglycerate kinase (EC:2.... 71.2 7e-13
cel:C47D12.4 hypothetical protein 30.4 1.3
pfa:PF14_0615 ATP synthase (C/AC39) subunit, putative; K02146 ... 29.3 3.1
bbo:BBOV_III007880 17.m10587; hypothetical protein 27.7 8.3
hsa:1653 DDX1, DBP-RB, UKVH5d; DEAD (Asp-Glu-Ala-Asp) box poly... 27.7 9.4
> xla:446763 pgk1, MGC132300, MGC80128, pgk2, pgka; phosphoglycerate
kinase 1 (EC:2.7.2.3); K00927 phosphoglycerate kinase
[EC:2.7.2.3]
Length=417
Score = 160 bits (406), Expect = 7e-40, Method: Compositional matrix adjust.
Identities = 73/96 (76%), Positives = 88/96 (91%), Gaps = 0/96 (0%)
Query 3 ANGNKVKADSKQVEAFRNSLTKLGDIFVNDAFGTAHRAHASMVGVNLPVKAAGLLMKKEL 62
A+GNK+KAD+ +V+ FR SL+KLGD+++NDAFGTAHRAH+SMVGV LP +AAG LMKKEL
Sbjct 135 ASGNKIKADAAKVDTFRASLSKLGDVYINDAFGTAHRAHSSMVGVKLPQRAAGFLMKKEL 194
Query 63 DYFSKALENPQKPFLAILGGAKVKDKIQLIKNLLSK 98
DYF+KALENP++PFLAILGGAKVKDKIQLI N+L K
Sbjct 195 DYFAKALENPERPFLAILGGAKVKDKIQLISNMLDK 230
> pfa:PFI1105w PGK; phosphoglycerate kinase (EC:2.7.2.3); K00927
phosphoglycerate kinase [EC:2.7.2.3]
Length=416
Score = 159 bits (402), Expect = 2e-39, Method: Compositional matrix adjust.
Identities = 76/98 (77%), Positives = 85/98 (86%), Gaps = 0/98 (0%)
Query 1 TSANGNKVKADSKQVEAFRNSLTKLGDIFVNDAFGTAHRAHASMVGVNLPVKAAGLLMKK 60
ANGNKVKA+ + VE F+N LTKL D+F+NDAFGTAHRAH+SMVGV L VKA+G LMKK
Sbjct 132 VDANGNKVKANKEDVEKFQNDLTKLADVFINDAFGTAHRAHSSMVGVKLNVKASGFLMKK 191
Query 61 ELDYFSKALENPQKPFLAILGGAKVKDKIQLIKNLLSK 98
EL+YFSKALENPQ+P LAILGGAKV DKIQLIKNLL K
Sbjct 192 ELEYFSKALENPQRPLLAILGGAKVSDKIQLIKNLLDK 229
> xla:380062 pgk1, MGC53000; phosphoglycerate kinase 1 (EC:2.7.2.3);
K00927 phosphoglycerate kinase [EC:2.7.2.3]
Length=417
Score = 158 bits (400), Expect = 4e-39, Method: Compositional matrix adjust.
Identities = 72/96 (75%), Positives = 87/96 (90%), Gaps = 0/96 (0%)
Query 3 ANGNKVKADSKQVEAFRNSLTKLGDIFVNDAFGTAHRAHASMVGVNLPVKAAGLLMKKEL 62
A GNK+KAD+ V++FR SL+KLGD+++NDAFGTAHRAH+SMVGV LP +AAG LMKKEL
Sbjct 135 AAGNKIKADAANVDSFRASLSKLGDVYINDAFGTAHRAHSSMVGVKLPQRAAGFLMKKEL 194
Query 63 DYFSKALENPQKPFLAILGGAKVKDKIQLIKNLLSK 98
DYF+KALENP++PFLAILGGAKVKDKIQLI N+L +
Sbjct 195 DYFAKALENPERPFLAILGGAKVKDKIQLISNMLDQ 230
> dre:406696 pgk1, wu:fd59b07, wu:fj36g06, zgc:56252, zgc:77899;
phosphoglycerate kinase 1 (EC:2.7.2.3); K00927 phosphoglycerate
kinase [EC:2.7.2.3]
Length=417
Score = 158 bits (399), Expect = 5e-39, Method: Compositional matrix adjust.
Identities = 74/97 (76%), Positives = 85/97 (87%), Gaps = 0/97 (0%)
Query 2 SANGNKVKADSKQVEAFRNSLTKLGDIFVNDAFGTAHRAHASMVGVNLPVKAAGLLMKKE 61
A+GNK KA +++AFR SL+KLGD++VNDAFGTAHRAH+SMVGVNLP KAAG LMKKE
Sbjct 134 DASGNKTKASQAEIDAFRASLSKLGDVYVNDAFGTAHRAHSSMVGVNLPQKAAGFLMKKE 193
Query 62 LDYFSKALENPQKPFLAILGGAKVKDKIQLIKNLLSK 98
LDYF+ ALE PQ+PFLAILGGAKVKDKIQLI N+L K
Sbjct 194 LDYFAMALEKPQRPFLAILGGAKVKDKIQLINNMLDK 230
> bbo:BBOV_IV008070 23.m06017; phosphoglycerate kinase (EC:2.7.2.3);
K00927 phosphoglycerate kinase [EC:2.7.2.3]
Length=415
Score = 157 bits (397), Expect = 9e-39, Method: Compositional matrix adjust.
Identities = 70/93 (75%), Positives = 85/93 (91%), Gaps = 0/93 (0%)
Query 6 NKVKADSKQVEAFRNSLTKLGDIFVNDAFGTAHRAHASMVGVNLPVKAAGLLMKKELDYF 65
+KVK D ++AFRNSLTKLGDI+VNDAFGTAHRAH+SMVGVN+P+K AGLLMKKELDYF
Sbjct 136 DKVKVDDSIIQAFRNSLTKLGDIYVNDAFGTAHRAHSSMVGVNVPIKVAGLLMKKELDYF 195
Query 66 SKALENPQKPFLAILGGAKVKDKIQLIKNLLSK 98
+K LE P++PF++ILGGAKVKDKIQLIK+++ K
Sbjct 196 AKILETPKRPFMSILGGAKVKDKIQLIKSIMRK 228
> hsa:5230 PGK1, MGC117307, MGC142128, MGC8947, MIG10, PGKA; phosphoglycerate
kinase 1 (EC:2.7.2.3); K00927 phosphoglycerate
kinase [EC:2.7.2.3]
Length=417
Score = 157 bits (396), Expect = 9e-39, Method: Compositional matrix adjust.
Identities = 73/96 (76%), Positives = 87/96 (90%), Gaps = 0/96 (0%)
Query 3 ANGNKVKADSKQVEAFRNSLTKLGDIFVNDAFGTAHRAHASMVGVNLPVKAAGLLMKKEL 62
A+GNKVKA+ ++EAFR SL+KLGD++VNDAFGTAHRAH+SMVGVNLP KA G LMKKEL
Sbjct 135 ASGNKVKAEPAKIEAFRASLSKLGDVYVNDAFGTAHRAHSSMVGVNLPQKAGGFLMKKEL 194
Query 63 DYFSKALENPQKPFLAILGGAKVKDKIQLIKNLLSK 98
+YF+KALE+P++PFLAILGGAKV DKIQLI N+L K
Sbjct 195 NYFAKALESPERPFLAILGGAKVADKIQLINNMLDK 230
> hsa:5232 PGK2, PGKB, PGKPS, dJ417L20.2; phosphoglycerate kinase
2 (EC:2.7.2.3); K00927 phosphoglycerate kinase [EC:2.7.2.3]
Length=417
Score = 157 bits (396), Expect = 1e-38, Method: Compositional matrix adjust.
Identities = 73/95 (76%), Positives = 86/95 (90%), Gaps = 0/95 (0%)
Query 4 NGNKVKADSKQVEAFRNSLTKLGDIFVNDAFGTAHRAHASMVGVNLPVKAAGLLMKKELD 63
+G K+KA+ ++EAFR SL+KLGD++VNDAFGTAHRAH+SMVGVNLP KA+G LMKKELD
Sbjct 136 SGKKIKAEPDKIEAFRASLSKLGDVYVNDAFGTAHRAHSSMVGVNLPHKASGFLMKKELD 195
Query 64 YFSKALENPQKPFLAILGGAKVKDKIQLIKNLLSK 98
YF+KALENP +PFLAILGGAKV DKIQLIKN+L K
Sbjct 196 YFAKALENPVRPFLAILGGAKVADKIQLIKNMLDK 230
> mmu:18663 Pgk2, Pgk-2, Tcp-2; phosphoglycerate kinase 2 (EC:2.7.2.3);
K00927 phosphoglycerate kinase [EC:2.7.2.3]
Length=417
Score = 156 bits (394), Expect = 2e-38, Method: Compositional matrix adjust.
Identities = 73/96 (76%), Positives = 86/96 (89%), Gaps = 0/96 (0%)
Query 3 ANGNKVKADSKQVEAFRNSLTKLGDIFVNDAFGTAHRAHASMVGVNLPVKAAGLLMKKEL 62
++G K+ AD +VEAF+ SL+KLGD++VNDAFGTAHRAH+S VGVNLP KA+G LMKKEL
Sbjct 135 SSGKKISADPAKVEAFQASLSKLGDVYVNDAFGTAHRAHSSTVGVNLPQKASGFLMKKEL 194
Query 63 DYFSKALENPQKPFLAILGGAKVKDKIQLIKNLLSK 98
DYFSKALE P++PFLAILGGAKVKDKIQLIKN+L K
Sbjct 195 DYFSKALEKPERPFLAILGGAKVKDKIQLIKNMLDK 230
> mmu:18655 Pgk1, MGC118097, Pgk-1; phosphoglycerate kinase 1
(EC:2.7.2.3); K00927 phosphoglycerate kinase [EC:2.7.2.3]
Length=417
Score = 155 bits (393), Expect = 2e-38, Method: Compositional matrix adjust.
Identities = 72/96 (75%), Positives = 87/96 (90%), Gaps = 0/96 (0%)
Query 3 ANGNKVKADSKQVEAFRNSLTKLGDIFVNDAFGTAHRAHASMVGVNLPVKAAGLLMKKEL 62
A+GNKVKA+ +++AFR SL+KLGD++VNDAFGTAHRAH+SMVGVNLP KA G LMKKEL
Sbjct 135 ASGNKVKAEPAKIDAFRASLSKLGDVYVNDAFGTAHRAHSSMVGVNLPQKAGGFLMKKEL 194
Query 63 DYFSKALENPQKPFLAILGGAKVKDKIQLIKNLLSK 98
+YF+KALE+P++PFLAILGGAKV DKIQLI N+L K
Sbjct 195 NYFAKALESPERPFLAILGGAKVADKIQLINNMLDK 230
> tgo:TGME49_118230 phosphoglycerate kinase, putative (EC:2.7.2.3);
K00927 phosphoglycerate kinase [EC:2.7.2.3]
Length=417
Score = 155 bits (391), Expect = 4e-38, Method: Compositional matrix adjust.
Identities = 70/95 (73%), Positives = 84/95 (88%), Gaps = 0/95 (0%)
Query 4 NGNKVKADSKQVEAFRNSLTKLGDIFVNDAFGTAHRAHASMVGVNLPVKAAGLLMKKELD 63
GNK+KA + + F+ LTKLGDI++NDAFGTAHRAHASMVG+ +PV+AAGLLMKKELD
Sbjct 136 QGNKIKASPEAIAKFKADLTKLGDIYINDAFGTAHRAHASMVGIEVPVRAAGLLMKKELD 195
Query 64 YFSKALENPQKPFLAILGGAKVKDKIQLIKNLLSK 98
YFSKALE P+KPFLAILGGAKV+DKIQLI+N+L +
Sbjct 196 YFSKALECPEKPFLAILGGAKVRDKIQLIENMLDR 230
> cpv:cgd7_910 phosphoglycerate kinase 1 ; K00927 phosphoglycerate
kinase [EC:2.7.2.3]
Length=404
Score = 152 bits (384), Expect = 2e-37, Method: Compositional matrix adjust.
Identities = 72/95 (75%), Positives = 84/95 (88%), Gaps = 0/95 (0%)
Query 4 NGNKVKADSKQVEAFRNSLTKLGDIFVNDAFGTAHRAHASMVGVNLPVKAAGLLMKKELD 63
NG KVKA +++EAFR SL+ LGDI+VNDAFGTAHRAH+SMVGV+L K AGLL+KKEL+
Sbjct 122 NGEKVKATPEEIEAFRKSLSSLGDIYVNDAFGTAHRAHSSMVGVDLTPKVAGLLLKKELE 181
Query 64 YFSKALENPQKPFLAILGGAKVKDKIQLIKNLLSK 98
YFSKALE+P +PFLAI+GGAKV DKIQLIKNLL K
Sbjct 182 YFSKALESPTRPFLAIMGGAKVADKIQLIKNLLLK 216
> tpv:TP01_0965 phosphoglycerate kinase; K00927 phosphoglycerate
kinase [EC:2.7.2.3]
Length=415
Score = 151 bits (381), Expect = 6e-37, Method: Compositional matrix adjust.
Identities = 70/90 (77%), Positives = 80/90 (88%), Gaps = 0/90 (0%)
Query 9 KADSKQVEAFRNSLTKLGDIFVNDAFGTAHRAHASMVGVNLPVKAAGLLMKKELDYFSKA 68
K D +V+ FR SLTKLGD++VNDAFGTAHRAH+SMVGVNLP++ AG LMKKELDYF+KA
Sbjct 139 KVDDVRVQRFRESLTKLGDVYVNDAFGTAHRAHSSMVGVNLPLRVAGFLMKKELDYFAKA 198
Query 69 LENPQKPFLAILGGAKVKDKIQLIKNLLSK 98
LENPQKPFLAILGGAKVKDK++LI LL K
Sbjct 199 LENPQKPFLAILGGAKVKDKVKLILALLEK 228
> cel:T03F1.3 pgk-1; PhosphoGlycerate Kinase family member (pgk-1);
K00927 phosphoglycerate kinase [EC:2.7.2.3]
Length=417
Score = 146 bits (369), Expect = 1e-35, Method: Compositional matrix adjust.
Identities = 72/98 (73%), Positives = 81/98 (82%), Gaps = 0/98 (0%)
Query 1 TSANGNKVKADSKQVEAFRNSLTKLGDIFVNDAFGTAHRAHASMVGVNLPVKAAGLLMKK 60
A+G KVKADS V+ FR SLTKLGDI+VNDAFGTAHRAH+SMVGV +A+G L+K
Sbjct 132 VDASGAKVKADSAAVKKFRESLTKLGDIYVNDAFGTAHRAHSSMVGVEHSQRASGFLLKN 191
Query 61 ELDYFSKALENPQKPFLAILGGAKVKDKIQLIKNLLSK 98
EL YFSKAL+NP +PFLAILGGAKV DKIQLIKNLL K
Sbjct 192 ELSYFSKALDNPARPFLAILGGAKVADKIQLIKNLLDK 229
> sce:YCR012W PGK1; 3-phosphoglycerate kinase, catalyzes transfer
of high-energy phosphoryl groups from the acyl phosphate
of 1,3-bisphosphoglycerate to ADP to produce ATP; key enzyme
in glycolysis and gluconeogenesis (EC:2.7.2.3); K00927 phosphoglycerate
kinase [EC:2.7.2.3]
Length=416
Score = 140 bits (352), Expect = 1e-33, Method: Compositional matrix adjust.
Identities = 64/95 (67%), Positives = 78/95 (82%), Gaps = 0/95 (0%)
Query 4 NGNKVKADSKQVEAFRNSLTKLGDIFVNDAFGTAHRAHASMVGVNLPVKAAGLLMKKELD 63
+G KVKA + V+ FR+ L+ L D+++NDAFGTAHRAH+SMVG +LP +AAG L++KEL
Sbjct 134 DGQKVKASKEDVQKFRHELSSLADVYINDAFGTAHRAHSSMVGFDLPQRAAGFLLEKELK 193
Query 64 YFSKALENPQKPFLAILGGAKVKDKIQLIKNLLSK 98
YF KALENP +PFLAILGGAKV DKIQLI NLL K
Sbjct 194 YFGKALENPTRPFLAILGGAKVADKIQLIDNLLDK 228
> ath:AT3G12780 PGK1; PGK1 (PHOSPHOGLYCERATE KINASE 1); phosphoglycerate
kinase; K00927 phosphoglycerate kinase [EC:2.7.2.3]
Length=481
Score = 94.0 bits (232), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 48/93 (51%), Positives = 61/93 (65%), Gaps = 2/93 (2%)
Query 9 KADSKQVEAFRNSLTKLGDIFVNDAFGTAHRAHASMVGVN--LPVKAAGLLMKKELDYFS 66
K + K F L L D++VNDAFGTAHRAHAS GV L AG L++KELDY
Sbjct 200 KEEEKNDPEFAKKLASLADLYVNDAFGTAHRAHASTEGVTKFLKPSVAGFLLQKELDYLV 259
Query 67 KALENPQKPFLAILGGAKVKDKIQLIKNLLSKS 99
A+ NP++PF AI+GG+KV KI +I++LL K
Sbjct 260 GAVSNPKRPFAAIVGGSKVSSKIGVIESLLEKC 292
> ath:AT1G56190 phosphoglycerate kinase, putative; K00927 phosphoglycerate
kinase [EC:2.7.2.3]
Length=405
Score = 93.2 bits (230), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 48/93 (51%), Positives = 61/93 (65%), Gaps = 2/93 (2%)
Query 9 KADSKQVEAFRNSLTKLGDIFVNDAFGTAHRAHASMVGVN--LPVKAAGLLMKKELDYFS 66
K + K F L L D++VNDAFGTAHRAHAS GV L AG L++KELDY
Sbjct 124 KEEEKNEPDFAKKLASLADLYVNDAFGTAHRAHASTEGVTKFLKPSVAGFLLQKELDYLV 183
Query 67 KALENPQKPFLAILGGAKVKDKIQLIKNLLSKS 99
A+ NP++PF AI+GG+KV KI +I++LL K
Sbjct 184 GAVSNPKRPFAAIVGGSKVSSKIGVIESLLEKC 216
> ath:AT1G79550 PGK; PGK (PHOSPHOGLYCERATE KINASE); phosphoglycerate
kinase (EC:2.7.7.2 2.7.2.3); K00927 phosphoglycerate
kinase [EC:2.7.2.3]
Length=401
Score = 91.3 bits (225), Expect = 8e-19, Method: Compositional matrix adjust.
Identities = 47/82 (57%), Positives = 58/82 (70%), Gaps = 2/82 (2%)
Query 18 FRNSLTKLGDIFVNDAFGTAHRAHASMVGVN--LPVKAAGLLMKKELDYFSKALENPQKP 75
F L L D++VNDAFGTAHRAHAS GV L AG LM+KELDY A+ NP+KP
Sbjct 134 FAKKLAALADVYVNDAFGTAHRAHASTEGVAKFLKPSVAGFLMQKELDYLVGAVANPKKP 193
Query 76 FLAILGGAKVKDKIQLIKNLLS 97
F AI+GG+KV KI +I++LL+
Sbjct 194 FAAIVGGSKVSTKIGVIESLLN 215
> tgo:TGME49_022020 phosphoglycerate kinase, putative (EC:2.7.2.3);
K00927 phosphoglycerate kinase [EC:2.7.2.3]
Length=551
Score = 88.2 bits (217), Expect = 6e-18, Method: Compositional matrix adjust.
Identities = 42/92 (45%), Positives = 59/92 (64%), Gaps = 2/92 (2%)
Query 9 KADSKQVEAFRNSLTKLGDIFVNDAFGTAHRAHASMVGVNLPVK--AAGLLMKKELDYFS 66
K ++K E L D+FVNDAFGTAHRAH+S GV V AG L+++EL+Y S
Sbjct 271 KGETKNDEELSRKFANLADMFVNDAFGTAHRAHSSTAGVTQFVSRSVAGFLLQQELEYLS 330
Query 67 KALENPQKPFLAILGGAKVKDKIQLIKNLLSK 98
+ NP +PF A++GGAKV K+ +++ L+ K
Sbjct 331 TVVANPVRPFAALVGGAKVSSKLGVLRALIEK 362
> eco:b2926 pgk, ECK2922, JW2893; phosphoglycerate kinase (EC:2.7.2.3);
K00927 phosphoglycerate kinase [EC:2.7.2.3]
Length=387
Score = 71.2 bits (173), Expect = 7e-13, Method: Compositional matrix adjust.
Identities = 39/89 (43%), Positives = 52/89 (58%), Gaps = 2/89 (2%)
Query 9 KADSKQVEAFRNSLTKLGDIFVNDAFGTAHRAHASMVGVN--LPVKAAGLLMKKELDYFS 66
K + K E L D+FV DAFGTAHRA AS G+ V AG L+ ELD
Sbjct 116 KGEKKDDETLSKKYAALCDVFVMDAFGTAHRAQASTHGIGKFADVACAGPLLAAELDALG 175
Query 67 KALENPQKPFLAILGGAKVKDKIQLIKNL 95
KAL+ P +P +AI+GG+KV K+ ++ +L
Sbjct 176 KALKEPARPMVAIVGGSKVSTKLTVLDSL 204
> cel:C47D12.4 hypothetical protein
Length=290
Score = 30.4 bits (67), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 18/61 (29%), Positives = 31/61 (50%), Gaps = 6/61 (9%)
Query 27 DIFVNDAFGTAHRAHASMVGVNLPVKAAGLLMKK-----ELDYFS-KALENPQKPFLAIL 80
DI +ND H H ++ G+NL +L+KK L+YF + +E+P + + +
Sbjct 175 DIQLNDINLMVHCWHVALAGINLNSDHLNVLLKKWKENSSLEYFCAREMESPLEKTIILE 234
Query 81 G 81
G
Sbjct 235 G 235
> pfa:PF14_0615 ATP synthase (C/AC39) subunit, putative; K02146
V-type H+-transporting ATPase subunit AC39 [EC:3.6.3.14]
Length=382
Score = 29.3 bits (64), Expect = 3.1, Method: Composition-based stats.
Identities = 21/83 (25%), Positives = 34/83 (40%), Gaps = 1/83 (1%)
Query 20 NSLTKLGDIFVNDAFGTAHRAHASMVGVNLPVKAAGLLMKKELDYFSKALENPQKPFLAI 79
++L L + + +GT S + V + M E +Y E P + FL
Sbjct 37 DTLEDLKSVLEDTDYGTFMMDEPSPIAVTTIAQKCKEKMAHEFNYIRAQAEEPLRTFLDY 96
Query 80 LGGAKVKDK-IQLIKNLLSKSAA 101
+ K+ D I LI+ L+K A
Sbjct 97 IAKEKMIDNVISLIQGTLNKKPA 119
> bbo:BBOV_III007880 17.m10587; hypothetical protein
Length=1139
Score = 27.7 bits (60), Expect = 8.3, Method: Composition-based stats.
Identities = 14/47 (29%), Positives = 24/47 (51%), Gaps = 0/47 (0%)
Query 2 SANGNKVKADSKQVEAFRNSLTKLGDIFVNDAFGTAHRAHASMVGVN 48
SAN N+ ++SK+VE L ++ F +D H+S G++
Sbjct 163 SANANRKYSNSKRVEEVLQQLDEITSCFCSDRCAMDFMRHSSYWGID 209
> hsa:1653 DDX1, DBP-RB, UKVH5d; DEAD (Asp-Glu-Ala-Asp) box polypeptide
1 (EC:3.6.4.13); K13177 ATP-dependent RNA helicase
DDX1 [EC:3.6.4.13]
Length=740
Score = 27.7 bits (60), Expect = 9.4, Method: Composition-based stats.
Identities = 11/40 (27%), Positives = 22/40 (55%), Gaps = 0/40 (0%)
Query 65 FSKALENPQKPFLAILGGAKVKDKIQLIKNLLSKSAAAPA 104
F K ++NP+ L I+GG +D++ +++N + P
Sbjct 309 FKKYIDNPKLRELLIIGGVAARDQLSVLENGVDIVVGTPG 348
Lambda K H
0.313 0.127 0.339
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2067351240
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40