bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_0480_orf1
Length=219
Score E
Sequences producing significant alignments: (Bits) Value
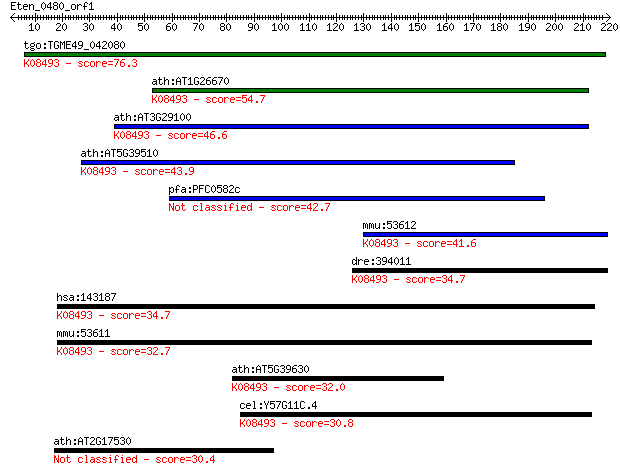
tgo:TGME49_042080 vesicle transport v-SNARE, putative ; K08493... 76.3 8e-14
ath:AT1G26670 VTI1B; VTI1B; SNARE binding / receptor/ soluble ... 54.7 2e-07
ath:AT3G29100 VTI13; VTI13; SNARE binding / receptor; K08493 v... 46.6 6e-05
ath:AT5G39510 SGR4; SGR4 (SHOOT GRAVITROPSIM 4); receptor; K08... 43.9 5e-04
pfa:PFC0582c vesicle transport V-SNARE protein, putative 42.7 0.001
mmu:53612 Vti1b, AU015348, GES30, MVti1b, SNARE, Vti1-rp1; ves... 41.6 0.002
dre:394011 vti1b, MGC63525, zgc:63525; vesicle transport throu... 34.7 0.29
hsa:143187 VTI1A, MVti1, Vti1-rp2; vesicle transport through i... 34.7 0.30
mmu:53611 Vti1a, 1110014F16Rik, 1110018K19Rik, 4921537J05Rik, ... 32.7 0.89
ath:AT5G39630 vesicle transport v-SNARE family protein; K08493... 32.0 1.8
cel:Y57G11C.4 hypothetical protein; K08493 vesicle transport t... 30.8 4.0
ath:AT2G17530 protein kinase family protein 30.4 5.6
> tgo:TGME49_042080 vesicle transport v-SNARE, putative ; K08493
vesicle transport through interaction with t-SNAREs 1
Length=281
Score = 76.3 bits (186), Expect = 8e-14, Method: Compositional matrix adjust.
Identities = 70/255 (27%), Positives = 109/255 (42%), Gaps = 55/255 (21%)
Query 6 AHAPAAATAAYKTTNYRALEQQLKL---RMQNAQSALKRAELTPRSPHVAAALATALSDA 62
AH P++A + N+R L L +MQN +A + E A A A
Sbjct 35 AHLPSSALFQDQERNWRRHYASLTLCLQKMQNGDAAAEDVE----------AGTEACEGA 84
Query 63 DVTLQQLSIEARGSPSGQALAAELAAYGDAL----------------------------- 93
D++LQQL EAR SP+ QAL + Y
Sbjct 85 DISLQQLETEARTSPNSQALLQAVRRYQSDFEQARSVFTRLVQQWQRQQLLQGPGSRRGQ 144
Query 94 ----ARLRQQSEEWTSRAVAGETPRQQLEPEELAE-------VSSRLILESNSSALQTED 142
R+ + +E S A ++ + ++ E L+ ESN A+++E
Sbjct 145 DTQRTRIPRSAESPFSFASTADSGNHSFDADDADEKALAVHHAGGTLLGESNRLAVESEH 204
Query 143 IGLHIMQQLRTQRDAIIKTNRLAEGTGAEGARGRQAIVKMIRDHWLNRLLLTGTILLLAL 202
+GL +M QLRTQR+++I + LA+ T E R + M+R ++L+L I+ L+L
Sbjct 205 VGLAVMGQLRTQRESLISSRSLAQKTYGELQDSRSLLTAMLRQSLFSKLILVAIIVFLSL 264
Query 203 CILLVLLHKARLFRF 217
I+ VL+H RL R
Sbjct 265 AIVCVLIH--RLLRL 277
> ath:AT1G26670 VTI1B; VTI1B; SNARE binding / receptor/ soluble
NSF attachment protein; K08493 vesicle transport through interaction
with t-SNAREs 1
Length=222
Score = 54.7 bits (130), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 45/181 (24%), Positives = 90/181 (49%), Gaps = 23/181 (12%)
Query 53 AALATALSDADVTLQQLSIEARG-SPSGQALA-AELAAYGDALARLRQQSEEWTSRAVAG 110
A + + + +ADV ++++ +EAR PS +A+ ++L Y L +L+++ + +S A A
Sbjct 40 AEIKSGIDEADVLIRKMDLEARSLQPSAKAVCLSKLREYKSDLNQLKKEFKRVSS-ADAK 98
Query 111 ETPRQQLEPEELAEV--------------------SSRLILESNSSALQTEDIGLHIMQQ 150
+ R++L +A++ SS I ES L+TE++G+ I+Q
Sbjct 99 PSSREELMESGMADLHAVSADQRGRLAMSVERLDQSSDRIRESRRLMLETEEVGISIVQD 158
Query 151 LRTQRDAIIKTNRLAEGTGAEGARGRQAIVKMIRDHWLNRLLLTGTILLLALCILLVLLH 210
L QR ++ + G + ++ + M R N+ ++T I+ L L I+L++ +
Sbjct 159 LSQQRQTLLHAHNKLHGVDDAIDKSKKVLTAMSRRMTRNKWIITSVIVALVLAIILIISY 218
Query 211 K 211
K
Sbjct 219 K 219
> ath:AT3G29100 VTI13; VTI13; SNARE binding / receptor; K08493
vesicle transport through interaction with t-SNAREs 1
Length=195
Score = 46.6 bits (109), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 43/180 (23%), Positives = 77/180 (42%), Gaps = 20/180 (11%)
Query 39 LKRAELTPRS--PHVAAALATALSDADVTLQQLSIEARGSPSGQALAA---EL--AAYGD 91
+K+ +L R+ P+V ++L L + L E + SG A EL A D
Sbjct 26 VKKMDLEARNLPPNVKSSLLVKLREYKSDLNNFKTEVKRITSGNLNATARDELLEAGMAD 85
Query 92 ALARLRQQSEEWTSRAVAGETPRQQLEPEELAEVSSRLILESNSSALQTEDIGLHIMQQL 151
L A A + R + + L + R I +S + L+TE++G+ I+Q L
Sbjct 86 TLT------------ASADQRSRLMMSTDHLGRTTDR-IKDSRRTILETEELGVSILQDL 132
Query 152 RTQRDAIIKTNRLAEGTGAEGARGRQAIVKMIRDHWLNRLLLTGTILLLALCILLVLLHK 211
QR ++++ + G + ++ + M R N+ + I +L L I+ +L K
Sbjct 133 HGQRQSLLRAHETLHGVDDNVGKSKKILTTMTRRMNRNKWTIGAIITVLVLAIIFILYFK 192
> ath:AT5G39510 SGR4; SGR4 (SHOOT GRAVITROPSIM 4); receptor; K08493
vesicle transport through interaction with t-SNAREs 1
Length=221
Score = 43.9 bits (102), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 39/163 (23%), Positives = 75/163 (46%), Gaps = 16/163 (9%)
Query 27 QLKLRMQNAQSALKRAELTPRS--PHVAAALATALSDADVTLQQLSIEARGSPSGQALAA 84
++K ++NA+ +++ +L R+ P++ ++L L + L E + SGQ AA
Sbjct 40 EIKSGLENAEVLIRKMDLEARTLPPNLKSSLLVKLREFKSDLNNFKTEVKRITSGQLNAA 99
Query 85 ---ELAAYGDALARLRQQSEEWTSRAVAGETPRQQLEPEELAEVSSRLILESNSSALQTE 141
EL G A T A A + R + E L + R + +S + ++TE
Sbjct 100 ARDELLEAGMA----------DTKTASADQRARLMMSTERLGRTTDR-VKDSRRTMMETE 148
Query 142 DIGLHIMQQLRTQRDAIIKTNRLAEGTGAEGARGRQAIVKMIR 184
+IG+ I+Q L QR ++++ + G + ++ + M R
Sbjct 149 EIGVSILQDLHGQRQSLLRAHETLHGVDDNIGKSKKILTDMTR 191
> pfa:PFC0582c vesicle transport V-SNARE protein, putative
Length=198
Score = 42.7 bits (99), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 33/140 (23%), Positives = 66/140 (47%), Gaps = 8/140 (5%)
Query 59 LSDADVTLQQLSIEARGSPSG-QALAAELAAYGDALARLRQQSEEWTSRAVAGE--TPRQ 115
+ +A+ ++Q++IE P G + ++ Y L + + ++ ++ + E RQ
Sbjct 42 IFEAETCIKQINIEINSLPKGSNKIINQINTYNLDLKKYKNIVQKMSADYYSEEYLNSRQ 101
Query 116 QLEPEELAEVSSRLILESNSSALQTEDIGLHIMQQLRTQRDAIIKTNRLAEGTGAEGARG 175
++ E + ES A EDIG IM +L +QR AI++T + T E R
Sbjct 102 YDMTKKYKE-GVDFLKESERRAQDIEDIGYTIMSELNSQRSAILRTKHHTDETRQEQNRV 160
Query 176 RQAIVKMIRDHWLNRLLLTG 195
++ ++ + + N+L+ G
Sbjct 161 KRMLLSI----YQNKLVFKG 176
> mmu:53612 Vti1b, AU015348, GES30, MVti1b, SNARE, Vti1-rp1; vesicle
transport through interaction with t-SNAREs 1B homolog;
K08493 vesicle transport through interaction with t-SNAREs
1
Length=232
Score = 41.6 bits (96), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 29/89 (32%), Positives = 49/89 (55%), Gaps = 5/89 (5%)
Query 130 ILESNSSALQTEDIGLHIMQQLRTQRDAIIKTNRLAEGTGAEGARGRQAIVKMIRDHWLN 189
I S+ A +T+ IG I+++L QRD + +T T ++ R+ + M R N
Sbjct 146 IERSHRIATETDQIGTEIIEELGEQRDQLERTKSRLVNTNENLSKSRKILRSMSRKVITN 205
Query 190 RLLLTGTILLLALCILLVLLHKARLFRFF 218
+LLL+ I+LL L IL+ L++ ++FF
Sbjct 206 KLLLS-VIILLELAILVGLVY----YKFF 229
> dre:394011 vti1b, MGC63525, zgc:63525; vesicle transport through
interaction with t-SNAREs homolog 1B (yeast); K08493 vesicle
transport through interaction with t-SNAREs 1
Length=225
Score = 34.7 bits (78), Expect = 0.29, Method: Compositional matrix adjust.
Identities = 27/93 (29%), Positives = 50/93 (53%), Gaps = 5/93 (5%)
Query 126 SSRLILESNSSALQTEDIGLHIMQQLRTQRDAIIKTNRLAEGTGAEGARGRQAIVKMIRD 185
+S+ I S A +T+ IG I+++L QR+ + +T TG +R R+ + M R
Sbjct 136 ASKSIERSQRIAAETDQIGTDIIEELGEQREQLDRTRDRLVNTGENLSRSRKILRAMSRR 195
Query 186 HWLNRLLLTGTILLLALCILLVLLHKARLFRFF 218
N+LLL+ I+++ + IL +++ +FF
Sbjct 196 IVTNKLLLS-IIIIMEVAILGGVVY----LKFF 223
> hsa:143187 VTI1A, MVti1, Vti1-rp2; vesicle transport through
interaction with t-SNAREs homolog 1A (yeast); K08493 vesicle
transport through interaction with t-SNAREs 1
Length=217
Score = 34.7 bits (78), Expect = 0.30, Method: Compositional matrix adjust.
Identities = 46/219 (21%), Positives = 88/219 (40%), Gaps = 28/219 (12%)
Query 18 TTNYRALEQQLKLRMQNAQSALKRAELTPRSP-----HVAAALATALSDADVTLQQLSIE 72
++++ EQ + S + R PR P + A + L +A L+Q+ +E
Sbjct 2 SSDFEGYEQDFAVLTAEITSKIARV---PRLPPDEKKQMVANVEKQLEEAKELLEQMDLE 58
Query 73 ARGSP--SGQALAAELAAYGDALARLRQQSEEWTSRAVAGETPRQQL------------- 117
R P S + + +Y + +L +++ SR + R +L
Sbjct 59 VREIPPQSRGMYSNRMRSYKQEMGKL--ETDFKRSRIAYSDEVRNELLGDDGNSSENQRA 116
Query 118 ---EPEELAEVSSRLILESNSSALQTEDIGLHIMQQLRTQRDAIIKTNRLAEGTGAEGAR 174
+ E E SSR + A++TE IG +++ L R+ I + T A +
Sbjct 117 HLLDNTERLERSSRRLEAGYQIAVETEQIGQEMLENLSHDREKIQRARERLRETDANLGK 176
Query 175 GRQAIVKMIRDHWLNRLLLTGTILLLALCILLVLLHKAR 213
+ + M+R NR+LL +++ + IL+ + R
Sbjct 177 SSRILTGMLRRIIQNRILLVILGIIVVITILMAITFSVR 215
> mmu:53611 Vti1a, 1110014F16Rik, 1110018K19Rik, 4921537J05Rik,
MGC102006, MVti1, MVti1a, Vti1, Vti1-rp2; vesicle transport
through interaction with t-SNAREs homolog 1A (yeast); K08493
vesicle transport through interaction with t-SNAREs 1
Length=217
Score = 32.7 bits (73), Expect = 0.89, Method: Compositional matrix adjust.
Identities = 51/219 (23%), Positives = 91/219 (41%), Gaps = 28/219 (12%)
Query 18 TTNYRALEQQLKLRMQNAQSALKRAELTPRSP-----HVAAALATALSDADVTLQQLSIE 72
++++ EQ + S + R PR P + A + L +A L+Q+ +E
Sbjct 2 SSDFEGYEQDFAVLTAEITSKIAR---VPRLPPDEKKQMVANVEKQLEEARELLEQMDLE 58
Query 73 ARGSP--SGQALAAELAAYGDALARLRQQ--------SEEWTSRAV--AGETPRQQ---- 116
R P S + + +Y + +L S+E + + AG + Q
Sbjct 59 VREIPPQSRGMYSNRMRSYKQEMGKLETDFKRSRIAYSDEVRNELLGDAGNSSENQRAHL 118
Query 117 LEPEELAEVSSRLILESNSSALQTEDIGLHIMQQLRTQRDAIIKT-NRLAEGTGAEGARG 175
L+ E E SSR + A++TE IG +++ L R+ I + +RL + G
Sbjct 119 LDNTERLERSSRRLEAGYQIAVETEQIGQEMLENLSHDREKIQRARDRLRDADANLGKSS 178
Query 176 RQAIVKMIRDHWLNRLLLT--GTILLLALCILLVLLHKA 212
R + M+R NR+LL G I+++A+ + K
Sbjct 179 R-ILTGMLRRIIQNRILLVILGIIVVIAILTAIAFFVKG 216
> ath:AT5G39630 vesicle transport v-SNARE family protein; K08493
vesicle transport through interaction with t-SNAREs 1
Length=207
Score = 32.0 bits (71), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 27/92 (29%), Positives = 46/92 (50%), Gaps = 15/92 (16%)
Query 82 LAAELAAYGDALARLRQQSEEWTSRAVAGETPRQQLEPE--ELAEVSSRL---------- 129
L +L +L RLR + + TS + T + LE E +LA+ SRL
Sbjct 70 LLEKLKESKSSLKRLRNEIKRNTSENLKVTTREEVLEAEKADLADQRSRLMKSTEGLVRT 129
Query 130 ---ILESNSSALQTEDIGLHIMQQLRTQRDAI 158
I +S L+TE+IG+ I++ L+ Q++++
Sbjct 130 REMIKDSQRKLLETENIGISILENLQRQKESL 161
> cel:Y57G11C.4 hypothetical protein; K08493 vesicle transport
through interaction with t-SNAREs 1
Length=224
Score = 30.8 bits (68), Expect = 4.0, Method: Compositional matrix adjust.
Identities = 36/128 (28%), Positives = 62/128 (48%), Gaps = 15/128 (11%)
Query 85 ELAAYGDALARLRQQSEEWTSRAVAGETPRQQLEPEELAEVSSRLILESNSSALQTEDIG 144
EL A+ D L RQ+ + + T R E S+R + +++ A++TE IG
Sbjct 111 ELLAFDDQLDEHRQEDQ------LIANTQR--------LERSTRKVQDAHRIAVETEQIG 156
Query 145 LHIMQQLRTQRDAIIKTNRLAEGTGAEGARGRQAIVKMIRDHWLNRLLLTGTILLLALCI 204
++ L +QR+ I ++ + A+ R + + MIR NRLLL LL+ +
Sbjct 157 AEMLSNLASQRETIGRSRDRLRQSNADLGRANKTLSSMIRRAIQNRLLLLIVTFLLSF-M 215
Query 205 LLVLLHKA 212
L +++KA
Sbjct 216 FLYIVYKA 223
> ath:AT2G17530 protein kinase family protein
Length=354
Score = 30.4 bits (67), Expect = 5.6, Method: Compositional matrix adjust.
Identities = 26/80 (32%), Positives = 38/80 (47%), Gaps = 7/80 (8%)
Query 17 KTTNYRALEQQLKLRMQNAQSALKRAELTPRSPHVAAALATALSDADVTLQQLSIEARGS 76
+T+NY AL+ Q K +Q AQ+AL EL + AA + ++ +
Sbjct 59 RTSNYVALKIQ-KSALQFAQAALHEIEL------LQAAADGDPENTKCVIRLIDDFKHAG 111
Query 77 PSGQALAAELAAYGDALARL 96
P+GQ L L GD+L RL
Sbjct 112 PNGQHLCMVLEFLGDSLLRL 131
Lambda K H
0.318 0.128 0.349
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 7137946020
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40