bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_0478_orf3
Length=118
Score E
Sequences producing significant alignments: (Bits) Value
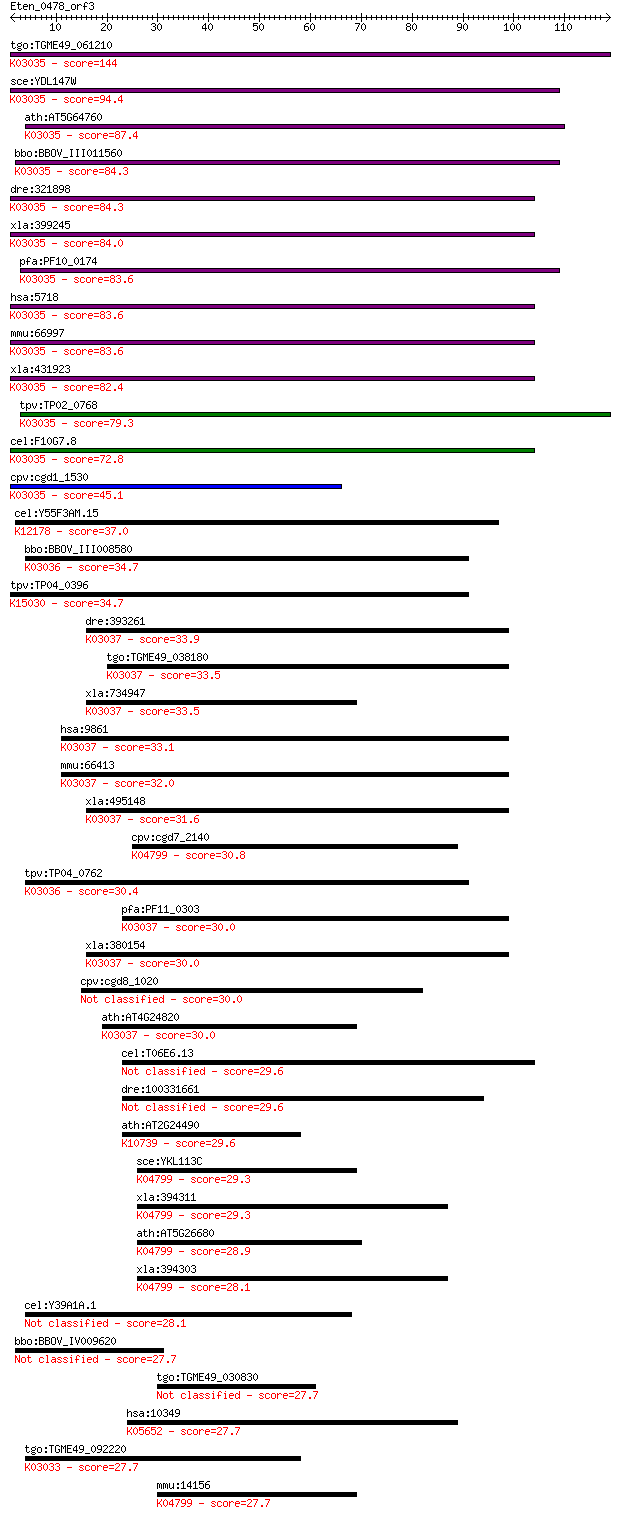
tgo:TGME49_061210 26S proteasome non-ATPase regulatory subunit... 144 7e-35
sce:YDL147W RPN5, NAS5; Essential, non-ATPase regulatory subun... 94.4 8e-20
ath:AT5G64760 RPN5B; RPN5B (REGULATORY PARTICLE NON-ATPASE SUB... 87.4 1e-17
bbo:BBOV_III011560 17.m07987; PCI domain containing protein; K... 84.3 8e-17
dre:321898 psmd12, wu:fb38a06, wu:fc34f08, wu:fc49f02; proteas... 84.3 9e-17
xla:399245 psmd12, MGC80339; proteasome (prosome, macropain) 2... 84.0 1e-16
pfa:PF10_0174 26S proteasome subunit p55, putative; K03035 26S... 83.6 1e-16
hsa:5718 PSMD12, MGC75406, Rpn5, p55; proteasome (prosome, mac... 83.6 1e-16
mmu:66997 Psmd12, 1500002F15Rik, AI480719, P55; proteasome (pr... 83.6 2e-16
xla:431923 hypothetical protein MGC81129; K03035 26S proteasom... 82.4 3e-16
tpv:TP02_0768 26S proteasome subunit p55; K03035 26S proteasom... 79.3 3e-15
cel:F10G7.8 rpn-5; proteasome Regulatory Particle, Non-ATPase-... 72.8 3e-13
cpv:cgd1_1530 Rpn5 like 26S proteasomal regulatory subunit 12,... 45.1 6e-05
cel:Y55F3AM.15 csn-4; COP-9 SigNalosome subunit family member ... 37.0 0.014
bbo:BBOV_III008580 17.m07750; PCI domain containing protein; K... 34.7 0.067
tpv:TP04_0396 hypothetical protein; K15030 translation initiat... 34.7 0.077
dre:393261 psmd6, MGC56471, zgc:56471; proteasome (prosome, ma... 33.9 0.14
tgo:TGME49_038180 26S proteasome non-ATPase regulatory subunit... 33.5 0.17
xla:734947 hypothetical protein MGC131117; K03037 26S proteaso... 33.5 0.18
hsa:9861 PSMD6, KIAA0107, Rpn7, S10, SGA-113M, p44S10; proteas... 33.1 0.21
mmu:66413 Psmd6, 2400006A19Rik; proteasome (prosome, macropain... 32.0 0.51
xla:495148 psmd6, p44s10, rpn7, s10; proteasome (prosome, macr... 31.6 0.58
cpv:cgd7_2140 flap endonuclease 1 ; K04799 flap endonuclease-1... 30.8 1.1
tpv:TP04_0762 proteosome subunit; K03036 26S proteasome regula... 30.4 1.4
pfa:PF11_0303 26S proteasome regulatory complex subunit, putat... 30.0 1.7
xla:380154 p44s10; hypothetical protein MGC53209; K03037 26S p... 30.0 1.8
cpv:cgd8_1020 hypothetical protein 30.0 1.8
ath:AT4G24820 26S proteasome regulatory subunit, putative (RPN... 30.0 2.0
cel:T06E6.13 fbxa-104; F-box A protein family member (fbxa-104) 29.6 2.1
dre:100331661 keratin 7-like 29.6 2.4
ath:AT2G24490 RPA2; RPA2 (REPLICON PROTEIN A2); protein bindin... 29.6 2.4
sce:YKL113C RAD27, ERC11, FEN1, RTH1; 5' to 3' exonuclease, 5'... 29.3 2.9
xla:394311 fen1-b, fen-1, mf1, rad2; flap structure-specific e... 29.3 3.4
ath:AT5G26680 endonuclease, putative; K04799 flap endonuclease... 28.9 4.5
xla:394303 fen1-a, MGC196488, MGC196492, fen-1, fen1, mf1, rad... 28.1 6.2
cel:Y39A1A.1 hypothetical protein 28.1 7.6
bbo:BBOV_IV009620 23.m05751; hypothetical protein 27.7 8.1
tgo:TGME49_030830 dynein heavy chain, putative (EC:3.1.3.48) 27.7 8.8
hsa:10349 ABCA10, EST698739; ATP-binding cassette, sub-family ... 27.7 8.9
tgo:TGME49_092220 26S proteasome non-ATPase regulatory subunit... 27.7 9.2
mmu:14156 Fen1, AW538437; flap structure specific endonuclease... 27.7 9.2
> tgo:TGME49_061210 26S proteasome non-ATPase regulatory subunit,
putative ; K03035 26S proteasome regulatory subunit N5
Length=485
Score = 144 bits (363), Expect = 7e-35, Method: Compositional matrix adjust.
Identities = 72/118 (61%), Positives = 87/118 (73%), Gaps = 0/118 (0%)
Query 1 WTLLRKRVIQHNLRVLCANYSVIHLQRVASLLGIPEEEAESELSELASAGVLYAKIDRPA 60
W LLR+RVIQHN+RV+ YS I + RVASLL I ++EAESE+SEL + + AKIDRPA
Sbjct 368 WALLRRRVIQHNIRVIATYYSCIEMDRVASLLDITKDEAESEISELVCSNFIEAKIDRPA 427
Query 61 RTVAFGKPKTDLVVLEEWSSSLSKLLDKVDLCCHMIQKERMVHAARAKAAAANARMAS 118
TV FGK K L W++ ++ LLD+VDLC H+IQKERMVHAARAK AA AR AS
Sbjct 428 GTVEFGKRKGTFDRLNSWAADVTSLLDRVDLCSHLIQKERMVHAARAKNAALLARNAS 485
> sce:YDL147W RPN5, NAS5; Essential, non-ATPase regulatory subunit
of the 26S proteasome lid, similar to mammalian p55 subunit
and to another S. cerevisiae regulatory subunit, Rpn7p;
K03035 26S proteasome regulatory subunit N5
Length=445
Score = 94.4 bits (233), Expect = 8e-20, Method: Compositional matrix adjust.
Identities = 47/108 (43%), Positives = 73/108 (67%), Gaps = 0/108 (0%)
Query 1 WTLLRKRVIQHNLRVLCANYSVIHLQRVASLLGIPEEEAESELSELASAGVLYAKIDRPA 60
W L+KRVI+HNLRV+ YS I L R+ LL + E + E+ +S+L + G++YAK++RPA
Sbjct 338 WEDLQKRVIEHNLRVISEYYSRITLLRLNELLDLTESQTETYISDLVNQGIIYAKVNRPA 397
Query 61 RTVAFGKPKTDLVVLEEWSSSLSKLLDKVDLCCHMIQKERMVHAARAK 108
+ V F KPK +L EWS ++ +LL+ ++ H+I KE ++H +AK
Sbjct 398 KIVNFEKPKNSSQLLNEWSHNVDELLEHIETIGHLITKEEIMHGLQAK 445
> ath:AT5G64760 RPN5B; RPN5B (REGULATORY PARTICLE NON-ATPASE SUBUNIT
5B); K03035 26S proteasome regulatory subunit N5
Length=442
Score = 87.4 bits (215), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 46/106 (43%), Positives = 64/106 (60%), Gaps = 0/106 (0%)
Query 4 LRKRVIQHNLRVLCANYSVIHLQRVASLLGIPEEEAESELSELASAGVLYAKIDRPARTV 63
L+ R+I+HN+ V+ YS I +R+A LL + EEAE LSE+ + L AKIDRP+ +
Sbjct 337 LKLRIIEHNILVVSKYYSRITFKRLAELLCLTTEEAEKHLSEMVVSKALIAKIDRPSGII 396
Query 64 AFGKPKTDLVVLEEWSSSLSKLLDKVDLCCHMIQKERMVHAARAKA 109
F K +L W+++L KLLD V+ CH I KE MVH +A
Sbjct 397 CFQIVKDSNEILNSWATNLEKLLDLVEKSCHQIHKETMVHKVALRA 442
> bbo:BBOV_III011560 17.m07987; PCI domain containing protein;
K03035 26S proteasome regulatory subunit N5
Length=474
Score = 84.3 bits (207), Expect = 8e-17, Method: Composition-based stats.
Identities = 41/107 (38%), Positives = 64/107 (59%), Gaps = 0/107 (0%)
Query 2 TLLRKRVIQHNLRVLCANYSVIHLQRVASLLGIPEEEAESELSELASAGVLYAKIDRPAR 61
+ L RVIQHN+ V Y+ + + R++ L ++ E E+S + A +YAKIDRPA
Sbjct 357 STLADRVIQHNIMVASKFYTTLQVTRLSELTNTTCDKLEEEISAMVHAKTIYAKIDRPAG 416
Query 62 TVAFGKPKTDLVVLEEWSSSLSKLLDKVDLCCHMIQKERMVHAARAK 108
+ FG+ K +L WS+ ++ L+ VD C ++QKE+M+H AR K
Sbjct 417 LIRFGERKDSDTLLLSWSTDIANLMGLVDQCSRLVQKEKMIHEARLK 463
> dre:321898 psmd12, wu:fb38a06, wu:fc34f08, wu:fc49f02; proteasome
(prosome, macropain) 26S subunit, non-ATPase, 12; K03035
26S proteasome regulatory subunit N5
Length=456
Score = 84.3 bits (207), Expect = 9e-17, Method: Compositional matrix adjust.
Identities = 39/103 (37%), Positives = 63/103 (61%), Gaps = 0/103 (0%)
Query 1 WTLLRKRVIQHNLRVLCANYSVIHLQRVASLLGIPEEEAESELSELASAGVLYAKIDRPA 60
W L+ RV++HN+R++ Y+ I + R+A+LL + +E+E LS L +YAK+DR A
Sbjct 351 WKDLKNRVVEHNIRIMAKYYTSITMGRMAALLDLSVDESEEFLSNLVVNKTIYAKVDRLA 410
Query 61 RTVAFGKPKTDLVVLEEWSSSLSKLLDKVDLCCHMIQKERMVH 103
+ F +PK +L +WS L+ L+ V+ H+I KE M+H
Sbjct 411 GIINFQRPKDPNDLLNDWSQKLNSLMSLVNKTTHLIAKEEMIH 453
> xla:399245 psmd12, MGC80339; proteasome (prosome, macropain)
26S subunit, non-ATPase, 12; K03035 26S proteasome regulatory
subunit N5
Length=441
Score = 84.0 bits (206), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 39/103 (37%), Positives = 63/103 (61%), Gaps = 0/103 (0%)
Query 1 WTLLRKRVIQHNLRVLCANYSVIHLQRVASLLGIPEEEAESELSELASAGVLYAKIDRPA 60
W L+ RV++HN+R++ Y+ I ++R+A LL + +E+E LS L +YAK+DR A
Sbjct 336 WKDLKNRVVEHNIRIMAKYYTRITMKRMAQLLDLSIDESEEFLSSLVVNKTIYAKVDRLA 395
Query 61 RTVAFGKPKTDLVVLEEWSSSLSKLLDKVDLCCHMIQKERMVH 103
+ F +PK +L +WS L+ L+ V+ H+I KE M+H
Sbjct 396 GIINFQRPKDPNDLLNDWSQKLNSLMSLVNKTTHLIAKEEMIH 438
> pfa:PF10_0174 26S proteasome subunit p55, putative; K03035 26S
proteasome regulatory subunit N5
Length=467
Score = 83.6 bits (205), Expect = 1e-16, Method: Composition-based stats.
Identities = 43/106 (40%), Positives = 65/106 (61%), Gaps = 0/106 (0%)
Query 3 LLRKRVIQHNLRVLCANYSVIHLQRVASLLGIPEEEAESELSELASAGVLYAKIDRPART 62
L +K+V+ HN+ V+ Y I L R+A LL E++E+ LSEL SA + +KIDR
Sbjct 351 LFKKKVMHHNIHVISNCYDQISLNRLAQLLNASIEDSENLLSELVSAKFINSKIDRLNGI 410
Query 63 VAFGKPKTDLVVLEEWSSSLSKLLDKVDLCCHMIQKERMVHAARAK 108
+ FG+ K +L WS ++ +LD ++ H+IQKERM+H A+ K
Sbjct 411 IKFGQKKNPENLLNSWSLQINDILDLLEESSHLIQKERMLHEAKLK 456
> hsa:5718 PSMD12, MGC75406, Rpn5, p55; proteasome (prosome, macropain)
26S subunit, non-ATPase, 12; K03035 26S proteasome
regulatory subunit N5
Length=456
Score = 83.6 bits (205), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 38/103 (36%), Positives = 64/103 (62%), Gaps = 0/103 (0%)
Query 1 WTLLRKRVIQHNLRVLCANYSVIHLQRVASLLGIPEEEAESELSELASAGVLYAKIDRPA 60
W L+ RV++HN+R++ Y+ I ++R+A LL + +E+E+ LS L ++AK+DR A
Sbjct 351 WKDLKNRVVEHNIRIMAKYYTRITMKRMAQLLDLSVDESEAFLSNLVVNKTIFAKVDRLA 410
Query 61 RTVAFGKPKTDLVVLEEWSSSLSKLLDKVDLCCHMIQKERMVH 103
+ F +PK +L +WS L+ L+ V+ H+I KE M+H
Sbjct 411 GIINFQRPKDPNNLLNDWSQKLNSLMSLVNKTTHLIAKEEMIH 453
> mmu:66997 Psmd12, 1500002F15Rik, AI480719, P55; proteasome (prosome,
macropain) 26S subunit, non-ATPase, 12; K03035 26S
proteasome regulatory subunit N5
Length=456
Score = 83.6 bits (205), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 38/103 (36%), Positives = 64/103 (62%), Gaps = 0/103 (0%)
Query 1 WTLLRKRVIQHNLRVLCANYSVIHLQRVASLLGIPEEEAESELSELASAGVLYAKIDRPA 60
W L+ RV++HN+R++ Y+ I ++R+A LL + +E+E+ LS L ++AK+DR A
Sbjct 351 WKDLKSRVVEHNIRIMAKYYTRITMKRMAQLLDLSVDESEAFLSNLVVNKTIFAKVDRLA 410
Query 61 RTVAFGKPKTDLVVLEEWSSSLSKLLDKVDLCCHMIQKERMVH 103
+ F +PK +L +WS L+ L+ V+ H+I KE M+H
Sbjct 411 GVINFQRPKDPNNLLNDWSQKLNSLMSLVNKTTHLIAKEEMIH 453
> xla:431923 hypothetical protein MGC81129; K03035 26S proteasome
regulatory subunit N5
Length=441
Score = 82.4 bits (202), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 39/103 (37%), Positives = 62/103 (60%), Gaps = 0/103 (0%)
Query 1 WTLLRKRVIQHNLRVLCANYSVIHLQRVASLLGIPEEEAESELSELASAGVLYAKIDRPA 60
W L+ RV++HN+R++ Y+ I ++R+A LL + E+E LS L +YAK+DR A
Sbjct 336 WKDLKNRVVEHNIRIMAKYYTRITMKRMAQLLDLSVNESEEFLSSLVVNKTIYAKVDRLA 395
Query 61 RTVAFGKPKTDLVVLEEWSSSLSKLLDKVDLCCHMIQKERMVH 103
+ F +PK +L +WS L+ L+ V+ H+I KE M+H
Sbjct 396 GIINFQRPKDPNDLLNDWSQKLNSLMALVNKTTHLIAKEEMIH 438
> tpv:TP02_0768 26S proteasome subunit p55; K03035 26S proteasome
regulatory subunit N5
Length=500
Score = 79.3 bits (194), Expect = 3e-15, Method: Composition-based stats.
Identities = 40/137 (29%), Positives = 75/137 (54%), Gaps = 21/137 (15%)
Query 3 LLRKRVIQHNLRVLCANYSVIHLQRVASLLGI---------------------PEEEAES 41
LL R+IQHN++++ Y+ I L+R+++LL I + + E+
Sbjct 364 LLYDRIIQHNVKIVSKYYNKITLERLSTLLNINSDVNPYYSITLVTHSIHLLPSKSKLEN 423
Query 42 ELSELASAGVLYAKIDRPARTVAFGKPKTDLVVLEEWSSSLSKLLDKVDLCCHMIQKERM 101
E+S + G++ +KI+R V F K ++L W ++++KL++ VD C ++QKE+M
Sbjct 424 EISNMVEMGIIDSKINRITGIVKFQKKLQTEIILNNWVNNITKLMELVDQCSRLVQKEKM 483
Query 102 VHAARAKAAAANARMAS 118
+H AR+K + + +S
Sbjct 484 IHEARSKQIQLDTQFSS 500
> cel:F10G7.8 rpn-5; proteasome Regulatory Particle, Non-ATPase-like
family member (rpn-5); K03035 26S proteasome regulatory
subunit N5
Length=490
Score = 72.8 bits (177), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 39/104 (37%), Positives = 62/104 (59%), Gaps = 1/104 (0%)
Query 1 WTLLRKRVIQHNLRVLCANYSVIHLQRVASLLGIPEEEAESELSELASAG-VLYAKIDRP 59
W+ L RV +HN+R++ Y+ I +R+A LL P +E ES + L G + AK+ RP
Sbjct 377 WSDLHLRVGEHNMRMIAKYYTQITFERLAELLDFPVDEMESFVCNLIVTGQITGAKLHRP 436
Query 60 ARTVAFGKPKTDLVVLEEWSSSLSKLLDKVDLCCHMIQKERMVH 103
+R V K ++ L+ W+S++ KL D ++ H+I KE+MVH
Sbjct 437 SRIVNLRLKKANVEQLDVWASNVHKLTDTLNKVSHLILKEQMVH 480
> cpv:cgd1_1530 Rpn5 like 26S proteasomal regulatory subunit 12,
PINT domain containing protein ; K03035 26S proteasome regulatory
subunit N5
Length=559
Score = 45.1 bits (105), Expect = 6e-05, Method: Composition-based stats.
Identities = 20/65 (30%), Positives = 36/65 (55%), Gaps = 0/65 (0%)
Query 1 WTLLRKRVIQHNLRVLCANYSVIHLQRVASLLGIPEEEAESELSELASAGVLYAKIDRPA 60
+TL +R+ + N+ ++ Y I QRV LL + +E + ++ L G+ AKI++PA
Sbjct 449 FTLFMQRIQERNISIISCYYKTISFQRVQDLLNLDGQELQLVVNHLVERGIFSAKINQPA 508
Query 61 RTVAF 65
+ F
Sbjct 509 GIITF 513
> cel:Y55F3AM.15 csn-4; COP-9 SigNalosome subunit family member
(csn-4); K12178 COP9 signalosome complex subunit 4
Length=412
Score = 37.0 bits (84), Expect = 0.014, Method: Compositional matrix adjust.
Identities = 24/95 (25%), Positives = 45/95 (47%), Gaps = 4/95 (4%)
Query 2 TLLRKRVIQHNLRVLCANYSVIHLQRVASLLGIPEEEAESELSELASAGVLYAKIDRPAR 61
++L+ + +HN+ + Y I + + LLG+ E AES E+ S+ L+ ID+
Sbjct 310 SILKGVIQEHNITAISQLYINISFKTLGQLLGVDTEAAESMAGEMISSERLHGYIDQTNG 369
Query 62 TVAFGKPKTDLVVLEEWSSSLSKLLDKVDLCCHMI 96
+ F D + W S + L++++ MI
Sbjct 370 ILHF----EDSNPMRVWDSQILSTLEQINKVSDMI 400
> bbo:BBOV_III008580 17.m07750; PCI domain containing protein;
K03036 26S proteasome regulatory subunit N6
Length=598
Score = 34.7 bits (78), Expect = 0.067, Method: Compositional matrix adjust.
Identities = 20/88 (22%), Positives = 46/88 (52%), Gaps = 1/88 (1%)
Query 4 LRKRVIQHNLRVLCANYSVIHLQRVASLLGIPEEEAESELSELASAGVLYAKIDRP-ART 62
L +++ N+ + YS++ + ++ L +P+++ E +L+E+ G L ID+ A
Sbjct 501 LYDALLERNITRILKPYSIVQISFLSHKLSLPQDKVEKKLAEMILDGKLRGTIDQGNANL 560
Query 63 VAFGKPKTDLVVLEEWSSSLSKLLDKVD 90
+ F + E+ + ++SKL+ +D
Sbjct 561 LLFDEETVRETFYEDVNHTISKLMSVID 588
> tpv:TP04_0396 hypothetical protein; K15030 translation initiation
factor 3 subunit M
Length=450
Score = 34.7 bits (78), Expect = 0.077, Method: Composition-based stats.
Identities = 24/100 (24%), Positives = 48/100 (48%), Gaps = 10/100 (10%)
Query 1 WTLLRKRVIQHNLRVLCANYSVIHLQRVASLLGIPEEEAESELSELASAGVLYAKIDRPA 60
W L +++ + +C S I ++ + L +P EEAE + + GV+ A ID+ +
Sbjct 344 WKPLVEKLTLLTISTMCQQQSEIPIEMIEKNLQLPPEEAEQMIVNAINKGVMEALIDQNS 403
Query 61 RTV--------AFGKPKTDLVV--LEEWSSSLSKLLDKVD 90
+ V FG + + L++W + ++ L++ VD
Sbjct 404 KKVIINHVVHREFGNEELKQLYNNLKQWRNCINTLINIVD 443
> dre:393261 psmd6, MGC56471, zgc:56471; proteasome (prosome,
macropain) 26S subunit, non-ATPase, 6; K03037 26S proteasome
regulatory subunit N7
Length=386
Score = 33.9 bits (76), Expect = 0.14, Method: Compositional matrix adjust.
Identities = 23/83 (27%), Positives = 38/83 (45%), Gaps = 7/83 (8%)
Query 16 LCANYSVIHLQRVASLLGIPEEEAESELSELASAGVLYAKIDRPARTVAFGKPKTDLVVL 75
L +Y + L +A G+ E + ELS +AG L+ KID+ V +P +
Sbjct 304 LLESYRSLTLGYMAEAFGVSTEFIDQELSRFIAAGRLHCKIDKVNEIVETNRPDS----- 358
Query 76 EEWSSSLSKLLDKVDLCCHMIQK 98
+ W + + K DL + +QK
Sbjct 359 KNW--QYQETIKKGDLLLNRVQK 379
> tgo:TGME49_038180 26S proteasome non-ATPase regulatory subunit,
putative ; K03037 26S proteasome regulatory subunit N7
Length=392
Score = 33.5 bits (75), Expect = 0.17, Method: Compositional matrix adjust.
Identities = 22/79 (27%), Positives = 39/79 (49%), Gaps = 7/79 (8%)
Query 20 YSVIHLQRVASLLGIPEEEAESELSELASAGVLYAKIDRPARTVAFGKPKTDLVVLEEWS 79
Y + ++ +A+ G+ E+E+S ++G L +IDR V +P +E S
Sbjct 314 YKSVTIENMATAFGVSPSFIEAEVSGFIASGKLSCRIDRVNAVVESNRP-------DERS 366
Query 80 SSLSKLLDKVDLCCHMIQK 98
S ++L + DL + IQK
Sbjct 367 RSYLRILKQGDLLLNRIQK 385
> xla:734947 hypothetical protein MGC131117; K03037 26S proteasome
regulatory subunit N7
Length=389
Score = 33.5 bits (75), Expect = 0.18, Method: Composition-based stats.
Identities = 17/53 (32%), Positives = 28/53 (52%), Gaps = 0/53 (0%)
Query 16 LCANYSVIHLQRVASLLGIPEEEAESELSELASAGVLYAKIDRPARTVAFGKP 68
L +Y + LQ +A G+ + + EL+ +AG L+AK+DR V +P
Sbjct 307 LLESYRSLTLQYMAEAFGVTVDYIDQELARFIAAGRLHAKVDRVGGIVETNRP 359
> hsa:9861 PSMD6, KIAA0107, Rpn7, S10, SGA-113M, p44S10; proteasome
(prosome, macropain) 26S subunit, non-ATPase, 6; K03037
26S proteasome regulatory subunit N7
Length=389
Score = 33.1 bits (74), Expect = 0.21, Method: Compositional matrix adjust.
Identities = 24/88 (27%), Positives = 39/88 (44%), Gaps = 7/88 (7%)
Query 11 HNLRVLCANYSVIHLQRVASLLGIPEEEAESELSELASAGVLYAKIDRPARTVAFGKPKT 70
H L +Y + L +A G+ E + ELS +AG L+ KID+ V +P +
Sbjct 302 HAYSQLLESYRSLTLGYMAEAFGVGVEFIDQELSRFIAAGRLHCKIDKVNEIVETNRPDS 361
Query 71 DLVVLEEWSSSLSKLLDKVDLCCHMIQK 98
+ W + + K DL + +QK
Sbjct 362 -----KNW--QYQETIKKGDLLLNRVQK 382
> mmu:66413 Psmd6, 2400006A19Rik; proteasome (prosome, macropain)
26S subunit, non-ATPase, 6; K03037 26S proteasome regulatory
subunit N7
Length=389
Score = 32.0 bits (71), Expect = 0.51, Method: Compositional matrix adjust.
Identities = 23/88 (26%), Positives = 39/88 (44%), Gaps = 7/88 (7%)
Query 11 HNLRVLCANYSVIHLQRVASLLGIPEEEAESELSELASAGVLYAKIDRPARTVAFGKPKT 70
H L +Y + L +A G+ + + ELS +AG L+ KID+ V +P +
Sbjct 302 HAYSQLLESYRSLTLGYMAEAFGVGVDFIDQELSRFIAAGRLHCKIDKVNEIVETNRPDS 361
Query 71 DLVVLEEWSSSLSKLLDKVDLCCHMIQK 98
+ W + + K DL + +QK
Sbjct 362 -----KNW--QYQETIKKGDLLLNRVQK 382
> xla:495148 psmd6, p44s10, rpn7, s10; proteasome (prosome, macropain)
26S subunit, non-ATPase, 6; K03037 26S proteasome regulatory
subunit N7
Length=389
Score = 31.6 bits (70), Expect = 0.58, Method: Composition-based stats.
Identities = 23/83 (27%), Positives = 38/83 (45%), Gaps = 7/83 (8%)
Query 16 LCANYSVIHLQRVASLLGIPEEEAESELSELASAGVLYAKIDRPARTVAFGKPKTDLVVL 75
L +Y + L +A G+ E + ELS +AG L+ KID+ V +P +
Sbjct 307 LLESYRSLTLGHMAEAFGVGVEFIDQELSRFIAAGRLHCKIDKVNEIVETNRPDS----- 361
Query 76 EEWSSSLSKLLDKVDLCCHMIQK 98
+ W + + K DL + +QK
Sbjct 362 KNW--QYQETIKKGDLLLNRVQK 382
> cpv:cgd7_2140 flap endonuclease 1 ; K04799 flap endonuclease-1
[EC:3.-.-.-]
Length=490
Score = 30.8 bits (68), Expect = 1.1, Method: Composition-based stats.
Identities = 19/68 (27%), Positives = 36/68 (52%), Gaps = 6/68 (8%)
Query 25 LQRVASLLGIP----EEEAESELSELASAGVLYAKIDRPARTVAFGKPKTDLVVLEEWSS 80
++++ LG+P EAE++ +EL G++Y + A ++ FG P + +S
Sbjct 143 VKKLLGFLGMPCIDAPSEAEAQCAELCKDGLVYGVVTEDADSLTFGTPIQ--IKQLNFSE 200
Query 81 SLSKLLDK 88
S +K+ DK
Sbjct 201 SSNKITDK 208
> tpv:TP04_0762 proteosome subunit; K03036 26S proteasome regulatory
subunit N6
Length=634
Score = 30.4 bits (67), Expect = 1.4, Method: Composition-based stats.
Identities = 20/87 (22%), Positives = 41/87 (47%), Gaps = 0/87 (0%)
Query 4 LRKRVIQHNLRVLCANYSVIHLQRVASLLGIPEEEAESELSELASAGVLYAKIDRPARTV 63
L +++ N+ + YSV+ +A L + E+ E +L+E+ L ID+ RT+
Sbjct 538 LYDSLLERNILRILKPYSVVQCDFIAQKLQLTSEKVEKKLAEMILDKRLNGTIDQGTRTL 597
Query 64 AFGKPKTDLVVLEEWSSSLSKLLDKVD 90
+ E+ S SL+ + + ++
Sbjct 598 EIYDDVVIHPIYEDVSKSLTHMTEVIE 624
> pfa:PF11_0303 26S proteasome regulatory complex subunit, putative;
K03037 26S proteasome regulatory subunit N7
Length=393
Score = 30.0 bits (66), Expect = 1.7, Method: Composition-based stats.
Identities = 20/76 (26%), Positives = 36/76 (47%), Gaps = 7/76 (9%)
Query 23 IHLQRVASLLGIPEEEAESELSELASAGVLYAKIDRPARTVAFGKPKTDLVVLEEWSSSL 82
+ L+ +A G+ EE E+E+S + G L KID+ ++ +P E ++
Sbjct 318 VTLKNMAFAFGVSEEFIENEISSFIANGKLNCKIDKVNGSIESNQPN-------ERNTMY 370
Query 83 SKLLDKVDLCCHMIQK 98
+ K D+ + IQK
Sbjct 371 QNTIKKGDILLNRIQK 386
> xla:380154 p44s10; hypothetical protein MGC53209; K03037 26S
proteasome regulatory subunit N7
Length=389
Score = 30.0 bits (66), Expect = 1.8, Method: Composition-based stats.
Identities = 22/83 (26%), Positives = 38/83 (45%), Gaps = 7/83 (8%)
Query 16 LCANYSVIHLQRVASLLGIPEEEAESELSELASAGVLYAKIDRPARTVAFGKPKTDLVVL 75
L +Y + L +A G+ E + ELS +AG L+ KID+ V +P +
Sbjct 307 LLESYRSLTLGYMAEAFGVGVEFIDQELSRFIAAGRLHCKIDKVNEIVETNRPDS----- 361
Query 76 EEWSSSLSKLLDKVDLCCHMIQK 98
+ W + + + DL + +QK
Sbjct 362 KNW--QYQETIKRGDLLLNRVQK 382
> cpv:cgd8_1020 hypothetical protein
Length=458
Score = 30.0 bits (66), Expect = 1.8, Method: Composition-based stats.
Identities = 21/72 (29%), Positives = 34/72 (47%), Gaps = 7/72 (9%)
Query 15 VLCANYSVIHLQRVASLLGIPEEEAESELSELASAGVLYAKI-----DRPARTVAFGKPK 69
VL N S ++ +L+GI + + SEL + +G L I D+ RT +
Sbjct 245 VLSYNSSPFQFEK--NLMGIRRDSSNSELQRIVRSGRLSGSIVGNSYDKQYRTNSIATSI 302
Query 70 TDLVVLEEWSSS 81
+D+ +EE SS
Sbjct 303 SDIFCMEEIKSS 314
> ath:AT4G24820 26S proteasome regulatory subunit, putative (RPN7);
K03037 26S proteasome regulatory subunit N7
Length=387
Score = 30.0 bits (66), Expect = 2.0, Method: Composition-based stats.
Identities = 14/50 (28%), Positives = 27/50 (54%), Gaps = 0/50 (0%)
Query 19 NYSVIHLQRVASLLGIPEEEAESELSELASAGVLYAKIDRPARTVAFGKP 68
+Y + ++ +A G+ + + ELS +AG L+ KID+ A + +P
Sbjct 308 SYKSVTVEAMAKAFGVSVDFIDQELSRFIAAGKLHCKIDKVAGVLETNRP 357
> cel:T06E6.13 fbxa-104; F-box A protein family member (fbxa-104)
Length=444
Score = 29.6 bits (65), Expect = 2.1, Method: Composition-based stats.
Identities = 23/97 (23%), Positives = 41/97 (42%), Gaps = 17/97 (17%)
Query 23 IHLQRVASLLGIPEEEAESELSELASAGVLYAKIDRPARTVAFGKPKTDLVVLEEWSS-- 80
+HL+++ + GI EE ++ L L + + D P D+V LE+W +
Sbjct 283 LHLEKI-NFKGINYEEIDTILRHLKTGVLKEISFDYKITNSTIIPPIGDMVYLEQWKNAE 341
Query 81 --------SLSKLLD------KVDLCCHMIQKERMVH 103
SL LLD +D+ CH + +++
Sbjct 342 NLCIHSHISLGSLLDHISHFHTIDILCHTFSRLQILQ 378
> dre:100331661 keratin 7-like
Length=1134
Score = 29.6 bits (65), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 27/78 (34%), Positives = 36/78 (46%), Gaps = 10/78 (12%)
Query 23 IHLQRVASLLGIP-------EEEAESELSELASAGVLYAKIDRPARTVAFGKPKTDLVVL 75
IHLQ A L+ +P E +AES S L G +YA I T+A +P+ L+VL
Sbjct 386 IHLQHYALLIRVPVLELLTLEGKAESLQSML---GRMYAHISAETLTLALQRPQCLLLVL 442
Query 76 EEWSSSLSKLLDKVDLCC 93
+ S L L C
Sbjct 443 DGLEESQKLLCTPSTLIC 460
> ath:AT2G24490 RPA2; RPA2 (REPLICON PROTEIN A2); protein binding;
K10739 replication factor A2
Length=279
Score = 29.6 bits (65), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 12/35 (34%), Positives = 20/35 (57%), Gaps = 0/35 (0%)
Query 23 IHLQRVASLLGIPEEEAESELSELASAGVLYAKID 57
IH+ +A L IP+ + E + L G++Y+ ID
Sbjct 236 IHIDEIAQQLKIPKNKLEGVVQSLEGDGLIYSTID 270
> sce:YKL113C RAD27, ERC11, FEN1, RTH1; 5' to 3' exonuclease,
5' flap endonuclease, required for Okazaki fragment processing
and maturation as well as for long-patch base-excision repair;
member of the S. pombe RAD2/FEN1 family; K04799 flap endonuclease-1
[EC:3.-.-.-]
Length=382
Score = 29.3 bits (64), Expect = 2.9, Method: Composition-based stats.
Identities = 16/47 (34%), Positives = 25/47 (53%), Gaps = 4/47 (8%)
Query 26 QRVASLLGIP----EEEAESELSELASAGVLYAKIDRPARTVAFGKP 68
Q++ L+GIP EAE++ +ELA G +YA T+ + P
Sbjct 140 QKLLGLMGIPYIIAPTEAEAQCAELAKKGKVYAAASEDMDTLCYRTP 186
> xla:394311 fen1-b, fen-1, mf1, rad2; flap structure-specific
endonuclease 1; K04799 flap endonuclease-1 [EC:3.-.-.-]
Length=382
Score = 29.3 bits (64), Expect = 3.4, Method: Composition-based stats.
Identities = 21/65 (32%), Positives = 33/65 (50%), Gaps = 8/65 (12%)
Query 26 QRVASLLGIPEE----EAESELSELASAGVLYAKIDRPARTVAFGKPKTDLVVLEEWSSS 81
+++ SL+GIP EAE+ + L AG +YA + FG P V+L ++S
Sbjct 142 KKLLSLMGIPYVDAPCEAEATCAALVKAGKVYAAATEDMDALTFGTP----VLLRHLTAS 197
Query 82 LSKLL 86
+K L
Sbjct 198 EAKKL 202
> ath:AT5G26680 endonuclease, putative; K04799 flap endonuclease-1
[EC:3.-.-.-]
Length=383
Score = 28.9 bits (63), Expect = 4.5, Method: Composition-based stats.
Identities = 14/48 (29%), Positives = 25/48 (52%), Gaps = 4/48 (8%)
Query 26 QRVASLLGIP----EEEAESELSELASAGVLYAKIDRPARTVAFGKPK 69
+R+ L+G+P EAE++ + L +G +Y ++ FG PK
Sbjct 143 KRLLRLMGVPVVEATSEAEAQCAALCKSGKVYGVASEDMDSLTFGAPK 190
> xla:394303 fen1-a, MGC196488, MGC196492, fen-1, fen1, mf1, rad2,
xFEN1a; flap structure-specific endonuclease 1; K04799
flap endonuclease-1 [EC:3.-.-.-]
Length=382
Score = 28.1 bits (61), Expect = 6.2, Method: Composition-based stats.
Identities = 20/65 (30%), Positives = 33/65 (50%), Gaps = 8/65 (12%)
Query 26 QRVASLLGIPEE----EAESELSELASAGVLYAKIDRPARTVAFGKPKTDLVVLEEWSSS 81
+++ +L+GIP EAE+ + L AG +YA + FG P V+L ++S
Sbjct 142 KKLLTLMGIPYVDAPCEAEATCAALVKAGKVYAAATEDMDALTFGTP----VLLRHLTAS 197
Query 82 LSKLL 86
+K L
Sbjct 198 EAKKL 202
> cel:Y39A1A.1 hypothetical protein
Length=386
Score = 28.1 bits (61), Expect = 7.6, Method: Composition-based stats.
Identities = 18/73 (24%), Positives = 38/73 (52%), Gaps = 9/73 (12%)
Query 4 LRKRVIQHNLRVL----CANY-SVIHLQRVASLLGIPEEEAESELSEL----ASAGVLYA 54
LR+ +Q +L+ + C++Y +V + + GI + E + + + L S+ ++
Sbjct 258 LRRGTVQAHLQCMAFSPCSSYLAVASDKGTLHMFGIRDAEPQKKKNVLERSRGSSSIVKI 317
Query 55 KIDRPARTVAFGK 67
++DRP + FGK
Sbjct 318 QLDRPVMAIGFGK 330
> bbo:BBOV_IV009620 23.m05751; hypothetical protein
Length=1201
Score = 27.7 bits (60), Expect = 8.1, Method: Composition-based stats.
Identities = 11/29 (37%), Positives = 19/29 (65%), Gaps = 0/29 (0%)
Query 2 TLLRKRVIQHNLRVLCANYSVIHLQRVAS 30
T+L R ++ +L+ C+N VI + R+AS
Sbjct 686 TVLEAREVKESLKYACSNSKVISISRIAS 714
> tgo:TGME49_030830 dynein heavy chain, putative (EC:3.1.3.48)
Length=4713
Score = 27.7 bits (60), Expect = 8.8, Method: Compositional matrix adjust.
Identities = 13/31 (41%), Positives = 21/31 (67%), Gaps = 0/31 (0%)
Query 30 SLLGIPEEEAESELSELASAGVLYAKIDRPA 60
SLL +PE+EAE E ++S G+++ RP+
Sbjct 719 SLLVVPEKEAEPEHFTISSTGIVHICPGRPS 749
> hsa:10349 ABCA10, EST698739; ATP-binding cassette, sub-family
A (ABC1), member 10; K05652 ATP-binding cassette, subfamily
A (ABC1), member 10
Length=1543
Score = 27.7 bits (60), Expect = 8.9, Method: Composition-based stats.
Identities = 18/65 (27%), Positives = 33/65 (50%), Gaps = 0/65 (0%)
Query 24 HLQRVASLLGIPEEEAESELSELASAGVLYAKIDRPARTVAFGKPKTDLVVLEEWSSSLS 83
HL+ A++ G+ +E+A +S L A L ++ P +T++ G + VL +
Sbjct 1301 HLELYAAVKGLGKEDAALSISRLVEALKLQEQLKAPVKTLSEGIKRKLCFVLSILGNPSV 1360
Query 84 KLLDK 88
LLD+
Sbjct 1361 VLLDE 1365
> tgo:TGME49_092220 26S proteasome non-ATPase regulatory subunit,
putative ; K03033 26S proteasome regulatory subunit N3
Length=558
Score = 27.7 bits (60), Expect = 9.2, Method: Compositional matrix adjust.
Identities = 20/55 (36%), Positives = 31/55 (56%), Gaps = 1/55 (1%)
Query 4 LRKRVIQHNLRVLCANYSVIHLQRVASLLGIPE-EEAESELSELASAGVLYAKID 57
L VI+ LR++ +YS I L+ VA LG+ AE+ +++ GV+ A ID
Sbjct 419 LHHNVIRAGLRLISLSYSRISLEDVAEKLGLDSAASAENIVAKAILDGVIEAAID 473
> mmu:14156 Fen1, AW538437; flap structure specific endonuclease
1; K04799 flap endonuclease-1 [EC:3.-.-.-]
Length=380
Score = 27.7 bits (60), Expect = 9.2, Method: Compositional matrix adjust.
Identities = 17/43 (39%), Positives = 22/43 (51%), Gaps = 4/43 (9%)
Query 30 SLLGIP----EEEAESELSELASAGVLYAKIDRPARTVAFGKP 68
SL+GIP EAE+ + LA AG +YA + FG P
Sbjct 146 SLMGIPYLDAPSEAEASCAALAKAGKVYAAATEDMDCLTFGSP 188
Lambda K H
0.320 0.129 0.369
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2027872200
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40