bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_0460_orf1
Length=402
Score E
Sequences producing significant alignments: (Bits) Value
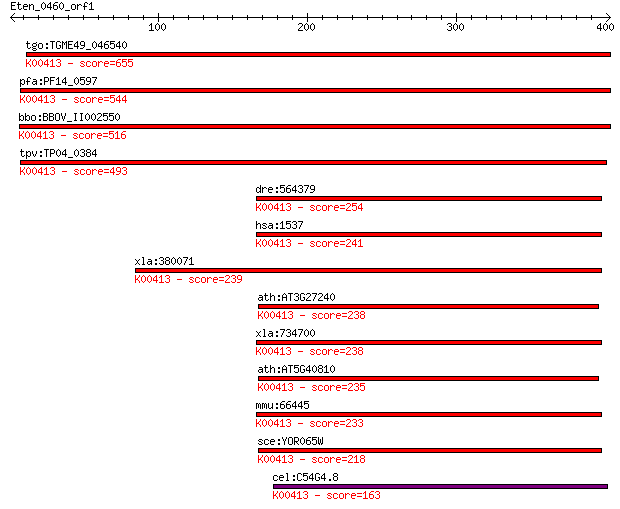
tgo:TGME49_046540 cytochrome C1, putative (EC:1.13.12.7); K004... 655 0.0
pfa:PF14_0597 cytochrome c1 precursor, putative; K00413 ubiqui... 544 2e-154
bbo:BBOV_II002550 18.m06205; cytochrome C1 precursor protein; ... 516 6e-146
tpv:TP04_0384 cytochrome c1 precursor; K00413 ubiquinol-cytoch... 493 4e-139
dre:564379 cyc1, MGC123089, fb80h03, im:6911272, sr:nyz104, wu... 254 6e-67
hsa:1537 CYC1, UQCR4; cytochrome c-1; K00413 ubiquinol-cytochr... 241 4e-63
xla:380071 cyc1, MGC53391; cytochrome c-1; K00413 ubiquinol-cy... 239 1e-62
ath:AT3G27240 cytochrome c1, putative; K00413 ubiquinol-cytoch... 238 2e-62
xla:734700 hypothetical protein MGC116545; K00413 ubiquinol-cy... 238 3e-62
ath:AT5G40810 cytochrome c1, putative; K00413 ubiquinol-cytoch... 235 3e-61
mmu:66445 Cyc1, 2610002H19Rik, AA408921; cytochrome c-1; K0041... 233 1e-60
sce:YOR065W CYT1, CTC1, YOR29-16; Cyt1p; K00413 ubiquinol-cyto... 218 4e-56
cel:C54G4.8 cyc-1; CYtochrome C family member (cyc-1); K00413 ... 163 1e-39
> tgo:TGME49_046540 cytochrome C1, putative (EC:1.13.12.7); K00413
ubiquinol-cytochrome c reductase cytochrome c1 subunit
[EC:1.10.2.2]
Length=396
Score = 655 bits (1690), Expect = 0.0, Method: Compositional matrix adjust.
Identities = 304/391 (77%), Positives = 340/391 (86%), Gaps = 2/391 (0%)
Query 12 ALNKLFPGYKDKIWMKVPVQWRERLVRHWNTTYERQVYAQCVVFNQRLQTYSKTVLDPLR 71
ALNKLFPGYKDKIWMKVP WR+++++HWN +YE+QVY++ V N+ Q ++ VLD L+
Sbjct 8 ALNKLFPGYKDKIWMKVP--WRQQMIQHWNKSYEKQVYSESVALNRTFQARNQLVLDRLK 65
Query 72 PDEKYSLPAVDYKRQKARGTLVEGADFYIPDKHAQSRLARPFEPYTEEEQETRQRYKYQS 131
P Y LPAVDYKRQ +RGTLVEGADFY+P Q RLAR FEPY+E+EQE R+++++QS
Sbjct 66 PSGAYRLPAVDYKRQLSRGTLVEGADFYLPTAQEQQRLARHFEPYSEQEQEERRKFRFQS 125
Query 132 LGFYLAVALGASFVHDCFYQRRPVCWCLEKEPPRPPHYPFWFKSIFHSHDIPSVRRGYEV 191
+ YLAVALGASFVHD FYQRRPV WCLEKEPP PP YPFWFKS+FHSHDIPSVRRGYEV
Sbjct 126 ISVYLAVALGASFVHDYFYQRRPVAWCLEKEPPHPPSYPFWFKSLFHSHDIPSVRRGYEV 185
Query 192 YRKVCSTCHSMEQLHFRHLVGEVLPELRVKQIAAEYDVEDGPNDSGEMYTRPGVAGDAFP 251
YRKVC+TCHSMEQLHFRHLVGEVLPE RVKQIAAEYDV DGPND GEMYTRPG+ GDAFP
Sbjct 186 YRKVCATCHSMEQLHFRHLVGEVLPEKRVKQIAAEYDVTDGPNDQGEMYTRPGILGDAFP 245
Query 252 KPYPNEEAARFANNGAYPPDLSLITGARMHGPDYIMALLNGYHEPPMGVELRPGLYWNVW 311
PYPNEEAAR+AN GAYPPDLSLIT AR GPDY+MALL GY +PP GVELRPGLYWNVW
Sbjct 246 SPYPNEEAARYANGGAYPPDLSLITAARHFGPDYLMALLGGYRDPPEGVELRPGLYWNVW 305
Query 312 FPGNAIAMAPPLQDEMIEYEDGTPCNVSQMSKDVVNFLTWATEPTCDERKLFGLKAVSAA 371
FPGNAIAM PPL DEMI+YEDGTPCN+SQMSKDVVNFLTWATEPT DERKL+GLK VSA
Sbjct 306 FPGNAIAMPPPLMDEMIDYEDGTPCNISQMSKDVVNFLTWATEPTADERKLYGLKCVSAI 365
Query 372 LLGCALMTVWWRFFWVIYATRRIDFGKLKYL 402
+G LMT+WWRF+W +YATRRIDFGKLKYL
Sbjct 366 AIGTVLMTLWWRFYWAMYATRRIDFGKLKYL 396
> pfa:PF14_0597 cytochrome c1 precursor, putative; K00413 ubiquinol-cytochrome
c reductase cytochrome c1 subunit [EC:1.10.2.2]
Length=394
Score = 544 bits (1402), Expect = 2e-154, Method: Compositional matrix adjust.
Identities = 250/395 (63%), Positives = 306/395 (77%), Gaps = 2/395 (0%)
Query 8 GGGGALNKLFPGYKDKIWMKVPVQWRERLVRHWNTTYERQVYAQCVVFNQRLQTYSKTVL 67
GGGA+N LFPGYKDKIW+K+P +R L++ WN +E+ ++ + + N R++ + +L
Sbjct 2 AGGGAMNNLFPGYKDKIWLKLPYHFRLYLIKSWNKNFEKNMF-KAKIKNNRIKNLNYYIL 60
Query 68 DPLRPDEKYSLPAVDYKRQKARGTLVEGADFYIPDKHAQSRLARPFEPYTEEEQETRQRY 127
D +P+E + DYKRQ RGTL EG DFY+PDK +Q RL FEPYTE+E E R++Y
Sbjct 61 DKFKPNENFKNTHTDYKRQICRGTLEEGCDFYLPDKKSQDRLKNHFEPYTEDENEERKKY 120
Query 128 KYQSLGFYLAVALGASFVHDCFYQRRPVCWCLEKEPPRPPHYPFWFKSIFHSHDIPSVRR 187
+Y +L +Y+ ALG + VH+ Q RPV WC++ EPP PHYPFWFKS+FHSHDIPSVRR
Sbjct 121 RYLNLKYYILFALGFTIVHNTI-QSRPVAWCMDSEPPHTPHYPFWFKSMFHSHDIPSVRR 179
Query 188 GYEVYRKVCSTCHSMEQLHFRHLVGEVLPELRVKQIAAEYDVEDGPNDSGEMYTRPGVAG 247
GYEVYR++C+TCHSMEQL FR LV EV PE RVKQIAA YD+ DGP+++GEM+TRPG+
Sbjct 180 GYEVYRQICATCHSMEQLQFRSLVNEVYPENRVKQIAASYDILDGPDETGEMFTRPGILT 239
Query 248 DAFPKPYPNEEAARFANNGAYPPDLSLITGARMHGPDYIMALLNGYHEPPMGVELRPGLY 307
D+FPKPYPNEEAAR+AN GA PPDLS IT AR +GPDYI +LL Y +PP GVELR GLY
Sbjct 240 DSFPKPYPNEEAARYANGGASPPDLSSITTARHNGPDYIFSLLTCYRDPPEGVELRNGLY 299
Query 308 WNVWFPGNAIAMAPPLQDEMIEYEDGTPCNVSQMSKDVVNFLTWATEPTCDERKLFGLKA 367
+N +F G +I+M PPLQD+MIEYEDGTPCNVSQM+KDVVNFL WA EP DERKL GLK
Sbjct 300 YNTYFEGGSISMPPPLQDDMIEYEDGTPCNVSQMAKDVVNFLCWAAEPAHDERKLTGLKL 359
Query 368 VSAALLGCALMTVWWRFFWVIYATRRIDFGKLKYL 402
+S A + LMTVW RFFW IYATRRIDFGK+KYL
Sbjct 360 ISGAFVAMVLMTVWQRFFWTIYATRRIDFGKIKYL 394
> bbo:BBOV_II002550 18.m06205; cytochrome C1 precursor protein;
K00413 ubiquinol-cytochrome c reductase cytochrome c1 subunit
[EC:1.10.2.2]
Length=397
Score = 516 bits (1329), Expect = 6e-146, Method: Compositional matrix adjust.
Identities = 232/396 (58%), Positives = 300/396 (75%), Gaps = 1/396 (0%)
Query 7 GGGGGALNKLFPGYKDKIWMKVPVQWRERLVRHWNTTYERQVYAQCVVFNQRLQTYSKTV 66
GGG ALNK+FPGYKDK+W K+P+ WR +R WN R ++ CV N ++++++K +
Sbjct 2 AGGGSALNKIFPGYKDKLWNKMPLDWRLNKIRRWNAATVRSMFDICVTTNAKIKSFNKFI 61
Query 67 LDPLRPDEKYSLPAVDYKRQKARGTLVEGADFYIPDKHAQSRLARPFEPYTEEEQETRQR 126
LDPL+P +Y AVDYK+Q+ARGTL+EG D+Y+P AQ+RLA FEPYTE+E E R +
Sbjct 62 LDPLKPSYEYRQTAVDYKKQRARGTLIEGVDYYLPTARAQNRLANFFEPYTEDENEDRAK 121
Query 127 YKYQSLGFYLAVALGASFVHDCFYQRRPVCWCLEKEPPRPPHYPFWFKSIFHSHDIPSVR 186
Y+YQSL Y+ VA GA+ +H+ +YQ RP+ WC + PPR P+YPFWFK FH HDI SVR
Sbjct 122 YRYQSLKTYIFVAFGATMLHN-WYQTRPIAWCSDINPPRAPYYPFWFKGPFHGHDIGSVR 180
Query 187 RGYEVYRKVCSTCHSMEQLHFRHLVGEVLPELRVKQIAAEYDVEDGPNDSGEMYTRPGVA 246
RGYEVYR+VC+TCHSM+ L FRHL EV PE R K+IA EY+V+DGPND GEM+ RPG+
Sbjct 181 RGYEVYRQVCATCHSMQYLRFRHLANEVYPEERAKEIAEEYEVQDGPNDEGEMFMRPGIL 240
Query 247 GDAFPKPYPNEEAARFANNGAYPPDLSLITGARMHGPDYIMALLNGYHEPPMGVELRPGL 306
D FP PYPN EAAR+AN GA PPDLSL+ AR GPDY+ ALL GY +PP G+ELR GL
Sbjct 241 TDPFPSPYPNAEAARYANGGAIPPDLSLMASARKTGPDYLFALLTGYCDPPEGIELRQGL 300
Query 307 YWNVWFPGNAIAMAPPLQDEMIEYEDGTPCNVSQMSKDVVNFLTWATEPTCDERKLFGLK 366
Y+N +F G +IAM PPL+D M+EYEDGTP VSQM+KDVV+F+TWA++P DERK G+K
Sbjct 301 YFNTYFTGGSIAMPPPLEDGMVEYEDGTPATVSQMAKDVVSFITWASDPMHDERKNMGMK 360
Query 367 AVSAALLGCALMTVWWRFFWVIYATRRIDFGKLKYL 402
+ ++LG M++W+RFFW AT R+DFG++K++
Sbjct 361 LILGSVLGTVAMSLWYRFFWAYLATSRMDFGRVKHM 396
> tpv:TP04_0384 cytochrome c1 precursor; K00413 ubiquinol-cytochrome
c reductase cytochrome c1 subunit [EC:1.10.2.2]
Length=396
Score = 493 bits (1270), Expect = 4e-139, Method: Compositional matrix adjust.
Identities = 227/392 (57%), Positives = 291/392 (74%), Gaps = 1/392 (0%)
Query 8 GGGGALNKLFPGYKDKIWMKVPVQWRERLVRHWNTTYERQVYAQCVVFNQRLQTYSKTVL 67
GGGALNKLFPGYKDKIW K+P+ R L+ WN V+ + VV N R+++++K VL
Sbjct 2 AGGGALNKLFPGYKDKIWNKLPLTVRLGLINSWNRKLISSVHRESVVTNGRVKSFNKYVL 61
Query 68 DPLRPDEKYSLPAVDYKRQKARGTLVEGADFYIPDKHAQSRLARPFEPYTEEEQETRQRY 127
DPL+P Y PA+DYK+Q+ARGTL+EG D+Y+P AQ RL F PYTEEE R RY
Sbjct 62 DPLKPGYAYRSPAIDYKKQRARGTLIEGVDYYLPTLGAQERLMNFFSPYTEEETAKRSRY 121
Query 128 KYQSLGFYLAVALGASFVHDCFYQRRPVCWCLEKEPPRPPHYPFWFKSIFHSHDIPSVRR 187
+YQSL Y+ ALG + V++ + QRRP+ WC +PP PP YPFWFK++ H HDIPSVRR
Sbjct 122 RYQSLKVYILTALGVTIVYN-YLQRRPIAWCSNLDPPTPPVYPFWFKNVLHGHDIPSVRR 180
Query 188 GYEVYRKVCSTCHSMEQLHFRHLVGEVLPELRVKQIAAEYDVEDGPNDSGEMYTRPGVAG 247
GYEVYR+VC+TCHS+ L FRHL+ EV P +VK+IAAEY++EDGPN+ GEMY+RP +
Sbjct 181 GYEVYRQVCATCHSLNYLKFRHLIDEVYPLEKVKEIAAEYEIEDGPNEQGEMYSRPRIPT 240
Query 248 DAFPKPYPNEEAARFANNGAYPPDLSLITGARMHGPDYIMALLNGYHEPPMGVELRPGLY 307
D FP PYPN EAAR+ANNGA PPDL+L++ AR +GPDYI +LL GY EPP G ELRPGL+
Sbjct 241 DPFPAPYPNSEAARYANNGAIPPDLTLMSSARRNGPDYIFSLLTGYSEPPEGFELRPGLH 300
Query 308 WNVWFPGNAIAMAPPLQDEMIEYEDGTPCNVSQMSKDVVNFLTWATEPTCDERKLFGLKA 367
+N +F G +I+MAPPL+D M+E+EDGTP VSQM+KDV NF+TW ++P DERK K
Sbjct 301 FNNYFNGGSISMAPPLEDGMLEFEDGTPATVSQMAKDVANFITWTSDPMHDERKNLFFKF 360
Query 368 VSAALLGCALMTVWWRFFWVIYATRRIDFGKL 399
+ + L ++++W+RFFW +AT R DF KL
Sbjct 361 FAGSTLCAIVVSMWYRFFWAHFATMRWDFKKL 392
> dre:564379 cyc1, MGC123089, fb80h03, im:6911272, sr:nyz104,
wu:fb80h03, zgc:123089; cytochrome c-1; K00413 ubiquinol-cytochrome
c reductase cytochrome c1 subunit [EC:1.10.2.2]
Length=307
Score = 254 bits (648), Expect = 6e-67, Method: Compositional matrix adjust.
Identities = 125/231 (54%), Positives = 155/231 (67%), Gaps = 0/231 (0%)
Query 166 PPHYPFWFKSIFHSHDIPSVRRGYEVYRKVCSTCHSMEQLHFRHLVGEVLPELRVKQIAA 225
PP YP+ S D SVRRGY+VY++VCS CHSME L FR+LVG E VK +A
Sbjct 73 PPTYPWSHGGFLSSLDHASVRRGYQVYKQVCSACHSMEYLAFRNLVGVSHTEDEVKTLAE 132
Query 226 EYDVEDGPNDSGEMYTRPGVAGDAFPKPYPNEEAARFANNGAYPPDLSLITGARMHGPDY 285
E +V DGP+D+GEM+TRPG D FPKPY N EAAR ANNGA PPDLS I AR G DY
Sbjct 133 EIEVVDGPDDNGEMFTRPGKLSDYFPKPYANPEAARAANNGALPPDLSYIVNARHGGEDY 192
Query 286 IMALLNGYHEPPMGVELRPGLYWNVWFPGNAIAMAPPLQDEMIEYEDGTPCNVSQMSKDV 345
+ +LL GY +PP GV LR GLY+N +FPG AI MAPP+ +E++EY+DGTP +SQ++KDV
Sbjct 193 VFSLLTGYCDPPAGVSLREGLYYNPYFPGQAIGMAPPIYNEVLEYDDGTPATMSQVAKDV 252
Query 346 VNFLTWATEPTCDERKLFGLKAVSAALLGCALMTVWWRFFWVIYATRRIDF 396
FL WA EP D+RK GLK + L L+ R W + +R+I +
Sbjct 253 CTFLRWAAEPEHDDRKRMGLKVLLGGALLIPLIYYIKRHKWSVLKSRKIAY 303
> hsa:1537 CYC1, UQCR4; cytochrome c-1; K00413 ubiquinol-cytochrome
c reductase cytochrome c1 subunit [EC:1.10.2.2]
Length=325
Score = 241 bits (614), Expect = 4e-63, Method: Compositional matrix adjust.
Identities = 119/231 (51%), Positives = 159/231 (68%), Gaps = 0/231 (0%)
Query 166 PPHYPFWFKSIFHSHDIPSVRRGYEVYRKVCSTCHSMEQLHFRHLVGEVLPELRVKQIAA 225
PP YP+ + + S D S+RRG++VY++VC++CHSM+ + +RHLVG E K++AA
Sbjct 91 PPSYPWSHRGLLSSLDHTSIRRGFQVYKQVCASCHSMDFVAYRHLVGVCYTEDEAKELAA 150
Query 226 EYDVEDGPNDSGEMYTRPGVAGDAFPKPYPNEEAARFANNGAYPPDLSLITGARMHGPDY 285
E +V+DGPN+ GEM+ RPG D FPKPYPN EAAR ANNGA PPDLS I AR G DY
Sbjct 151 EVEVQDGPNEDGEMFMRPGKLFDYFPKPYPNSEAARAANNGALPPDLSYIVRARHGGEDY 210
Query 286 IMALLNGYHEPPMGVELRPGLYWNVWFPGNAIAMAPPLQDEMIEYEDGTPCNVSQMSKDV 345
+ +LL GY EPP GV LR GLY+N +FPG AIAMAPP+ +++E++DGTP +SQ++KDV
Sbjct 211 VFSLLTGYCEPPTGVSLREGLYFNPYFPGQAIAMAPPIYTDVLEFDDGTPATMSQIAKDV 270
Query 346 VNFLTWATEPTCDERKLFGLKAVSAALLGCALMTVWWRFFWVIYATRRIDF 396
FL WA+EP D RK GLK + L L+ R W + +R++ +
Sbjct 271 CTFLRWASEPEHDHRKRMGLKMLMMMALLVPLVYTIKRHKWSVLKSRKLAY 321
> xla:380071 cyc1, MGC53391; cytochrome c-1; K00413 ubiquinol-cytochrome
c reductase cytochrome c1 subunit [EC:1.10.2.2]
Length=309
Score = 239 bits (610), Expect = 1e-62, Method: Compositional matrix adjust.
Identities = 133/312 (42%), Positives = 182/312 (58%), Gaps = 15/312 (4%)
Query 85 RQKARGTLVEGADFYIPDKHAQSRLARPFEPYTEEEQETRQRYKYQSLGFYLAVALGASF 144
R+ RG +++G R+A P P R + +L + G S
Sbjct 9 RRSLRGAVLQGG-----------RVAWPVAPQANMSFSALSRGRKVALSTLGILVAGGSG 57
Query 145 VHDCFYQRRPVCWCLEKEPPRPPHYPFWFKSIFHSHDIPSVRRGYEVYRKVCSTCHSMEQ 204
+ +Q LE PP YP+ S D S+RRGY+VY++VC+ CHSME
Sbjct 58 LAFALHQSVKASE-LELHPPS---YPWSHSGFLSSLDHGSIRRGYQVYKQVCAACHSMEY 113
Query 205 LHFRHLVGEVLPELRVKQIAAEYDVEDGPNDSGEMYTRPGVAGDAFPKPYPNEEAARFAN 264
L FR+L+G E K +A E++++DGP+++GEM+ RPG D FPKPY NEEAAR +N
Sbjct 114 LAFRNLIGVSHTEAEAKALAEEFEIQDGPDENGEMFLRPGKLSDYFPKPYANEEAARASN 173
Query 265 NGAYPPDLSLITGARMHGPDYIMALLNGYHEPPMGVELRPGLYWNVWFPGNAIAMAPPLQ 324
NGA PPDLS I AR G DYI +LL GY +PP GV LR GLY+N +FPG A+ MAPP+
Sbjct 174 NGALPPDLSYIANARHGGEDYIFSLLTGYCDPPAGVTLREGLYYNPYFPGQAVGMAPPIY 233
Query 325 DEMIEYEDGTPCNVSQMSKDVVNFLTWATEPTCDERKLFGLKAVSAALLGCALMTVWWRF 384
+E++EYEDGTP +SQ++KDV FL WA+EP D RK GLK + + + L+ R
Sbjct 234 NEVLEYEDGTPATMSQVAKDVSTFLRWASEPEHDHRKRMGLKVLMISSILIPLIYYMKRH 293
Query 385 FWVIYATRRIDF 396
W + +R+I +
Sbjct 294 RWSVLKSRKIAY 305
> ath:AT3G27240 cytochrome c1, putative; K00413 ubiquinol-cytochrome
c reductase cytochrome c1 subunit [EC:1.10.2.2]
Length=307
Score = 238 bits (608), Expect = 2e-62, Method: Compositional matrix adjust.
Identities = 118/228 (51%), Positives = 153/228 (67%), Gaps = 0/228 (0%)
Query 167 PHYPFWFKSIFHSHDIPSVRRGYEVYRKVCSTCHSMEQLHFRHLVGEVLPELRVKQIAAE 226
P YP+ I S+D S+RRG++VY++VC++CHSM + +R LVG E K +AAE
Sbjct 74 PEYPWPHDGILSSYDHASIRRGHQVYQQVCASCHSMSLISYRDLVGVAYTEEEAKAMAAE 133
Query 227 YDVEDGPNDSGEMYTRPGVAGDAFPKPYPNEEAARFANNGAYPPDLSLITGARMHGPDYI 286
+V DGPND GEM+TRPG D FP+PY NE AARFAN GAYPPDLSLIT AR +GP+Y+
Sbjct 134 IEVVDGPNDEGEMFTRPGKLSDRFPQPYANESAARFANGGAYPPDLSLITKARHNGPNYV 193
Query 287 MALLNGYHEPPMGVELRPGLYWNVWFPGNAIAMAPPLQDEMIEYEDGTPCNVSQMSKDVV 346
ALL GY +PP G+ +R GL++N +FPG AIAM L DE +EYEDG P +QM KD+V
Sbjct 194 FALLTGYRDPPAGISIREGLHYNPYFPGGAIAMPKMLNDEAVEYEDGVPATEAQMGKDIV 253
Query 347 NFLTWATEPTCDERKLFGLKAVSAALLGCALMTVWWRFFWVIYATRRI 394
+FL WA EP +ERKL G K + L + R W + +R++
Sbjct 254 SFLAWAAEPEMEERKLMGFKWIFLLSLALLQAAYYRRLKWSVLKSRKL 301
> xla:734700 hypothetical protein MGC116545; K00413 ubiquinol-cytochrome
c reductase cytochrome c1 subunit [EC:1.10.2.2]
Length=309
Score = 238 bits (608), Expect = 3e-62, Method: Compositional matrix adjust.
Identities = 117/231 (50%), Positives = 157/231 (67%), Gaps = 0/231 (0%)
Query 166 PPHYPFWFKSIFHSHDIPSVRRGYEVYRKVCSTCHSMEQLHFRHLVGEVLPELRVKQIAA 225
PP YP+ S D S+RRGY+VY++VC+ CHSME L FR+L+G E K +A
Sbjct 75 PPSYPWSHSGFLSSLDHASIRRGYQVYKQVCAACHSMEYLAFRNLIGVSHTEAEAKALAE 134
Query 226 EYDVEDGPNDSGEMYTRPGVAGDAFPKPYPNEEAARFANNGAYPPDLSLITGARMHGPDY 285
E++++DGP+++GEM+ RPG D FPKPY NEEAAR +NNGA PPDLS I AR G DY
Sbjct 135 EFEIQDGPDENGEMFLRPGKLSDYFPKPYANEEAARASNNGALPPDLSYIANARHGGEDY 194
Query 286 IMALLNGYHEPPMGVELRPGLYWNVWFPGNAIAMAPPLQDEMIEYEDGTPCNVSQMSKDV 345
I +LL GY +PP GV +R GLY+N +FPG A+ MAPP+ +E++EYEDGTP +SQ++KDV
Sbjct 195 IFSLLTGYCDPPAGVTIREGLYYNPYFPGQAVGMAPPIYNEVLEYEDGTPATMSQVAKDV 254
Query 346 VNFLTWATEPTCDERKLFGLKAVSAALLGCALMTVWWRFFWVIYATRRIDF 396
FL WA+EP D RK GLK + + + L+ R W + +R+I +
Sbjct 255 STFLRWASEPEHDHRKRMGLKVLMISSILIPLIYYMKRHRWSVLKSRKIAY 305
> ath:AT5G40810 cytochrome c1, putative; K00413 ubiquinol-cytochrome
c reductase cytochrome c1 subunit [EC:1.10.2.2]
Length=260
Score = 235 bits (599), Expect = 3e-61, Method: Compositional matrix adjust.
Identities = 117/228 (51%), Positives = 155/228 (67%), Gaps = 0/228 (0%)
Query 167 PHYPFWFKSIFHSHDIPSVRRGYEVYRKVCSTCHSMEQLHFRHLVGEVLPELRVKQIAAE 226
P+YP+ + I S+D S+RRG++VY++VC++CHSM + +R LVG E K +AAE
Sbjct 27 PNYPWPHEGILSSYDHASIRRGHQVYQQVCASCHSMSLISYRDLVGVAYTEEEAKAMAAE 86
Query 227 YDVEDGPNDSGEMYTRPGVAGDAFPKPYPNEEAARFANNGAYPPDLSLITGARMHGPDYI 286
+V DGPND GEM+TRPG D P+PY NE AARFAN GAYPPDLSL+T AR +G +Y+
Sbjct 87 IEVVDGPNDEGEMFTRPGKLSDRLPEPYSNESAARFANGGAYPPDLSLVTKARHNGQNYV 146
Query 287 MALLNGYHEPPMGVELRPGLYWNVWFPGNAIAMAPPLQDEMIEYEDGTPCNVSQMSKDVV 346
ALL GY +PP G+ +R GL++N +FPG AIAM L DE +EYEDGTP +QM KDVV
Sbjct 147 FALLTGYRDPPAGISIREGLHYNPYFPGGAIAMPKMLNDEAVEYEDGTPATEAQMGKDVV 206
Query 347 NFLTWATEPTCDERKLFGLKAVSAALLGCALMTVWWRFFWVIYATRRI 394
+FL+WA EP +ERKL G K + L + R W + +R++
Sbjct 207 SFLSWAAEPEMEERKLMGFKWIFLLSLALLQAAYYRRLKWSVLKSRKL 254
> mmu:66445 Cyc1, 2610002H19Rik, AA408921; cytochrome c-1; K00413
ubiquinol-cytochrome c reductase cytochrome c1 subunit [EC:1.10.2.2]
Length=325
Score = 233 bits (593), Expect = 1e-60, Method: Compositional matrix adjust.
Identities = 121/231 (52%), Positives = 155/231 (67%), Gaps = 0/231 (0%)
Query 166 PPHYPFWFKSIFHSHDIPSVRRGYEVYRKVCSTCHSMEQLHFRHLVGEVLPELRVKQIAA 225
PP YP+ + + S D S+RRG++VY++VCS+CHSM+ + +RHLVG E K +A
Sbjct 91 PPSYPWSHRGLLSSLDHTSIRRGFQVYKQVCSSCHSMDYVAYRHLVGVCYTEEEAKALAE 150
Query 226 EYDVEDGPNDSGEMYTRPGVAGDAFPKPYPNEEAARFANNGAYPPDLSLITGARMHGPDY 285
E +V+DGPND GEM+ RPG D FPKPYPN EAAR ANNGA PPDLS I AR G DY
Sbjct 151 EVEVQDGPNDDGEMFMRPGKLSDYFPKPYPNPEAARAANNGALPPDLSYIVRARHGGEDY 210
Query 286 IMALLNGYHEPPMGVELRPGLYWNVWFPGNAIAMAPPLQDEMIEYEDGTPCNVSQMSKDV 345
+ +LL GY EPP GV LR GLY+N +FPG AI MAPP+ E++EY+DGTP +SQ++KDV
Sbjct 211 VFSLLTGYCEPPTGVSLREGLYFNPYFPGQAIGMAPPIYTEVLEYDDGTPATMSQVAKDV 270
Query 346 VNFLTWATEPTCDERKLFGLKAVSAALLGCALMTVWWRFFWVIYATRRIDF 396
FL WA+EP D RK GLK + L L R W + +R++ +
Sbjct 271 ATFLRWASEPEHDHRKRMGLKMLLMMGLLLPLTYAMKRHKWSVLKSRKLAY 321
> sce:YOR065W CYT1, CTC1, YOR29-16; Cyt1p; K00413 ubiquinol-cytochrome
c reductase cytochrome c1 subunit [EC:1.10.2.2]
Length=309
Score = 218 bits (554), Expect = 4e-56, Method: Compositional matrix adjust.
Identities = 114/232 (49%), Positives = 145/232 (62%), Gaps = 3/232 (1%)
Query 167 PHYPFWFKSIFHSHDIPSVRRGYEVYRKVCSTCHSMEQLHFRHLVGEVLPELRVKQIAAE 226
P Y + F + D S+RRGY+VYR+VC+ CHS++++ +R LVG V+ +A E
Sbjct 72 PAYAWSHNGPFETFDHASIRRGYQVYREVCAACHSLDRVAWRTLVGVSHTNEEVRNMAEE 131
Query 227 YDVEDGPNDSGEMYTRPGVAGDAFPKPYPNEEAARFANNGAYPPDLSLITGARMHGPDYI 286
++ +D P++ G RPG D P PYPNE+AAR AN GA PPDLSLI AR G DYI
Sbjct 132 FEYDDEPDEQGNPKKRPGKLSDYIPGPYPNEQAARAANQGALPPDLSLIVKARHGGCDYI 191
Query 287 MALLNGY-HEPPMGVELRPGLYWNVWFPGNAIAMAPPLQDEMIEYEDGTPCNVSQMSKDV 345
+LL GY EPP GV L PG +N +FPG +IAMA L D+M+EYEDGTP SQM+KDV
Sbjct 192 FSLLTGYPDEPPAGVALPPGSNYNPYFPGGSIAMARVLFDDMVEYEDGTPATTSQMAKDV 251
Query 346 VNFLTWATEPTCDERKLFGLKAVSAALLGCALMTVWW-RFFWVIYATRRIDF 396
FL W EP DERK GLK V L L+++W +F W TR+ F
Sbjct 252 TTFLNWCAEPEHDERKRLGLKTV-IILSSLYLLSIWVKKFKWAGIKTRKFVF 302
> cel:C54G4.8 cyc-1; CYtochrome C family member (cyc-1); K00413
ubiquinol-cytochrome c reductase cytochrome c1 subunit [EC:1.10.2.2]
Length=285
Score = 163 bits (412), Expect = 1e-39, Method: Compositional matrix adjust.
Identities = 102/224 (45%), Positives = 135/224 (60%), Gaps = 1/224 (0%)
Query 177 FHSHDIPSVRRGYEVYRKVCSTCHSMEQLHFRHLVGEVLPELRVKQIAAEYDVEDGPNDS 236
F S DI SVRRGYEVY++VC+ CHSM+ LH+RH V ++ E K AA+ + D +D
Sbjct 55 FSSFDIASVRRGYEVYKQVCAACHSMKFLHYRHFVDTIMTEEEAKAEAADALIND-VDDK 113
Query 237 GEMYTRPGVAGDAFPKPYPNEEAARFANNGAYPPDLSLITGARMHGPDYIMALLNGYHEP 296
G RPG+ D P PYPN++AA ANNGA PPDLSL+ AR G DY+ +LL GY E
Sbjct 114 GASIQRPGMLTDKLPNPYPNKKAAAAANNGAAPPDLSLMALARHGGDDYVFSLLTGYLEA 173
Query 297 PMGVELRPGLYWNVWFPGNAIAMAPPLQDEMIEYEDGTPCNVSQMSKDVVNFLTWATEPT 356
P GV++ G +N +FPG I+M L DE IEY+DGTP +SQ +KDV F+ WA EP
Sbjct 174 PAGVKVDDGKAYNPYFPGGIISMPQQLFDEGIEYKDGTPATMSQQAKDVSAFMHWAAEPF 233
Query 357 CDERKLFGLKAVSAALLGCALMTVWWRFFWVIYATRRIDFGKLK 400
D RK + LK + ++ R W +++ F +K
Sbjct 234 HDTRKKWALKIAALIPFVAVVLIYGKRHIWSFTKSQKFLFKTVK 277
Lambda K H
0.322 0.139 0.458
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 18125297876
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40