bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_0437_orf1
Length=176
Score E
Sequences producing significant alignments: (Bits) Value
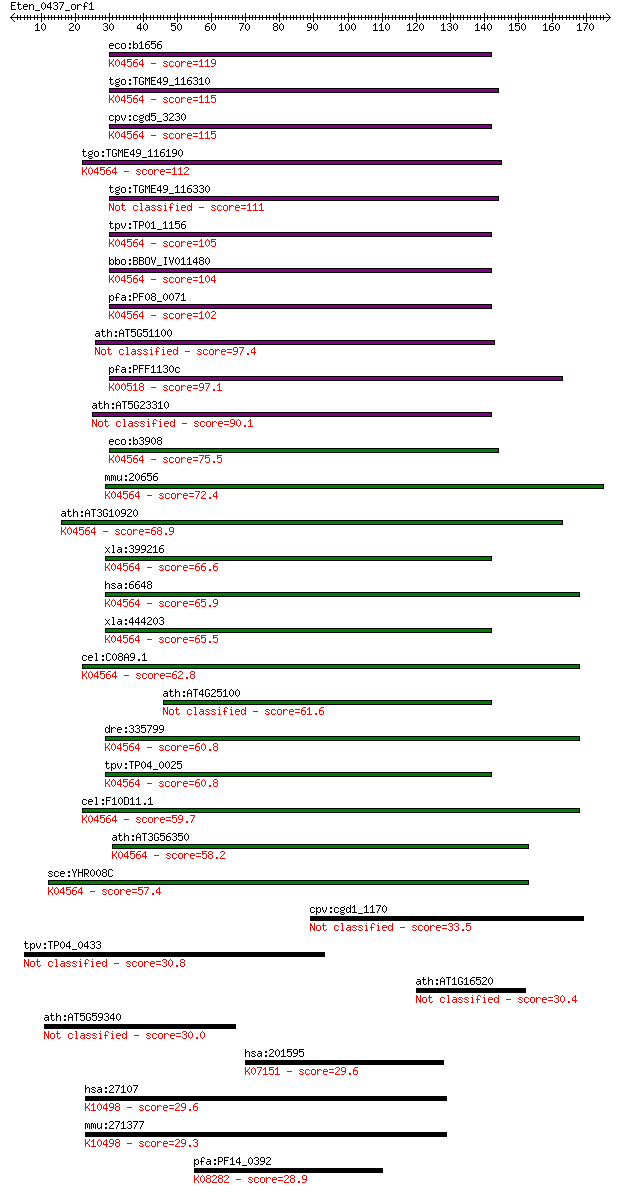
eco:b1656 sodB, ECK1652, JW1648; superoxide dismutase, Fe (EC:... 119 5e-27
tgo:TGME49_116310 superoxide dismutase (EC:1.15.1.1); K04564 s... 115 9e-26
cpv:cgd5_3230 superoxide dismutase ; K04564 superoxide dismuta... 115 1e-25
tgo:TGME49_116190 superoxide dismutase, putative (EC:1.15.1.1)... 112 6e-25
tgo:TGME49_116330 iron-containing superoxide dismutase (EC:1.1... 111 2e-24
tpv:TP01_1156 iron-dependent superoxide dismutase (EC:1.15.1.1... 105 1e-22
bbo:BBOV_IV011480 23.m06274; superoxide dismutase (EC:1.15.1.1... 104 2e-22
pfa:PF08_0071 FeSOD, PfSOD1; Fe-superoxide dismutase (EC:1.15.... 102 5e-22
ath:AT5G51100 FSD2; FSD2 (FE SUPEROXIDE DISMUTASE 2); superoxi... 97.4 2e-20
pfa:PFF1130c PfSOD2; superoxide dismutase (EC:1.15.1.1); K0051... 97.1 3e-20
ath:AT5G23310 FSD3; FSD3 (FE SUPEROXIDE DISMUTASE 3); superoxi... 90.1 3e-18
eco:b3908 sodA, ECK3901, JW3879; superoxide dismutase, Mn (EC:... 75.5 8e-14
mmu:20656 Sod2, MGC6144, MnSOD, Sod-2; superoxide dismutase 2,... 72.4 8e-13
ath:AT3G10920 MSD1; MSD1 (MANGANESE SUPEROXIDE DISMUTASE 1); m... 68.9 8e-12
xla:399216 sod2; superoxide dismutase 2, mitochondrial (EC:1.1... 66.6 4e-11
hsa:6648 SOD2, IPOB, MNSOD, MVCD6; superoxide dismutase 2, mit... 65.9 7e-11
xla:444203 MGC80739 protein; K04564 superoxide dismutase, Fe-M... 65.5 8e-11
cel:C08A9.1 sod-3; SOD (superoxide dismutase) family member (s... 62.8 6e-10
ath:AT4G25100 FSD1; FSD1 (FE SUPEROXIDE DISMUTASE 1); copper i... 61.6 1e-09
dre:335799 sod2, cb463, wu:fj33b01, zgc:73051; superoxide dism... 60.8 2e-09
tpv:TP04_0025 Fe-superoxide dismutase (EC:1.15.1.1); K04564 su... 60.8 2e-09
cel:F10D11.1 sod-2; SOD (superoxide dismutase) family member (... 59.7 6e-09
ath:AT3G56350 superoxide dismutase (Mn), putative / manganese ... 58.2 2e-08
sce:YHR008C SOD2; Mitochondrial superoxide dismutase, protects... 57.4 2e-08
cpv:cgd1_1170 ubiquitin C-terminal hydrolase 33.5 0.35
tpv:TP04_0433 hypothetical protein 30.8 2.5
ath:AT1G16520 hypothetical protein 30.4 2.9
ath:AT5G59340 WOX2; WOX2 (WUSCHEL RELATED HOMEOBOX 2); transcr... 30.0 4.7
hsa:201595 STT3B, FLJ90106, SIMP, STT3-B; STT3, subunit of the... 29.6 5.4
hsa:27107 ZBTB11, FLJ13426, MGC133303, ZNF-U69274, ZNF913; zin... 29.6 6.1
mmu:271377 Zbtb11, 9230110G02Rik, ZNF-U69274; zinc finger and ... 29.3 7.3
pfa:PF14_0392 serine/threonine protein kinase, putative; K0828... 28.9 9.7
> eco:b1656 sodB, ECK1652, JW1648; superoxide dismutase, Fe (EC:1.15.1.1);
K04564 superoxide dismutase, Fe-Mn family [EC:1.15.1.1]
Length=193
Score = 119 bits (298), Expect = 5e-27, Method: Compositional matrix adjust.
Identities = 57/112 (50%), Positives = 75/112 (66%), Gaps = 3/112 (2%)
Query 30 FSLPPLPYKPDALEPYISAKTIEHHYGKHHKAYVDNLNALVKDSNNPIHQKLETIIKNEK 89
F LP LPY DAL P+ISA+TIE+HYGKHH+ YV NLN L+K + + LE II++ +
Sbjct 3 FELPALPYAKDALAPHISAETIEYHYGKHHQTYVTNLNNLIKGTAFE-GKSLEEIIRSSE 61
Query 90 GNIYNQAAQVWNHTFYFNCLGPPSEETAAPLGPTLKRLERDFGSLENFKNEF 141
G ++N AAQVWNHTFY+NCL P + P G + + FGS +FK +F
Sbjct 62 GGVFNNAAQVWNHTFYWNCLAPNA--GGEPTGKVAEAIAASFGSFADFKAQF 111
> tgo:TGME49_116310 superoxide dismutase (EC:1.15.1.1); K04564
superoxide dismutase, Fe-Mn family [EC:1.15.1.1]
Length=201
Score = 115 bits (287), Expect = 9e-26, Method: Compositional matrix adjust.
Identities = 55/115 (47%), Positives = 77/115 (66%), Gaps = 2/115 (1%)
Query 30 FSLPPLPYKPDALEPYISAKTIEHHYGKHHKAYVDNLNALVKDSNNPIHQKLETIIKNEK 89
F+LPPLPY DAL P+IS++T++ H+GKHH YV LN ++ + + LE +I+
Sbjct 3 FTLPPLPYAHDALAPHISSETLQFHHGKHHAGYVAKLNGFIEGTAFA-GKTLEEVIRTST 61
Query 90 GNIYNQAAQVWNHTFYFNCLGPP-SEETAAPLGPTLKRLERDFGSLENFKNEFKK 143
G I+N AAQVWNHTFYF+ + PP S P G L ++++F S+ENFK+EF K
Sbjct 62 GAIFNNAAQVWNHTFYFSSMKPPMSGGGGEPTGRLLDEIKKEFTSVENFKDEFSK 116
> cpv:cgd5_3230 superoxide dismutase ; K04564 superoxide dismutase,
Fe-Mn family [EC:1.15.1.1]
Length=221
Score = 115 bits (287), Expect = 1e-25, Method: Compositional matrix adjust.
Identities = 56/112 (50%), Positives = 70/112 (62%), Gaps = 1/112 (0%)
Query 30 FSLPPLPYKPDALEPYISAKTIEHHYGKHHKAYVDNLNALVKDSNNPIHQKLETIIKNEK 89
F LPPLPY ALEP IS +T+++HYGKHH YV LN+L+K + L +I
Sbjct 27 FELPPLPYDKKALEPVISPETLDYHYGKHHAGYVTKLNSLIKSTEFENETDLMKVIMKSS 86
Query 90 GNIYNQAAQVWNHTFYFNCLGPPSEETAAPLGPTLKRLERDFGSLENFKNEF 141
G IYN A+Q+WNHTFY++CL PS ET P K +E FGS E+FK F
Sbjct 87 GPIYNNASQIWNHTFYWSCLRSPS-ETNKPTSKVSKLIEESFGSFESFKESF 137
> tgo:TGME49_116190 superoxide dismutase, putative (EC:1.15.1.1);
K04564 superoxide dismutase, Fe-Mn family [EC:1.15.1.1]
Length=258
Score = 112 bits (280), Expect = 6e-25, Method: Compositional matrix adjust.
Identities = 57/123 (46%), Positives = 79/123 (64%), Gaps = 3/123 (2%)
Query 22 AALRLSARFSLPPLPYKPDALEPYISAKTIEHHYGKHHKAYVDNLNALVKDSNNPIHQKL 81
A+L + +F LP LPY +ALEPYISAKT++ H+ KHH YV NLN L+K ++ +QKL
Sbjct 44 ASLFSTGKFELPALPYDYNALEPYISAKTLKFHHDKHHATYVKNLNELIKGTDYE-NQKL 102
Query 82 ETIIKNEKGNIYNQAAQVWNHTFYFNCLGPPSEETAAPLGPTLKRLERDFGSLENFKNEF 141
E II+N G +N AAQ WNHTF++N + P P G ++++R F S +K EF
Sbjct 103 EDIIRNADGPTFNNAAQAWNHTFFWNSMKPNG--GGEPTGLVKEQIDRCFSSFSKYKEEF 160
Query 142 KKK 144
+K
Sbjct 161 SEK 163
> tgo:TGME49_116330 iron-containing superoxide dismutase (EC:1.15.1.1)
Length=287
Score = 111 bits (277), Expect = 2e-24, Method: Compositional matrix adjust.
Identities = 57/116 (49%), Positives = 71/116 (61%), Gaps = 4/116 (3%)
Query 30 FSLPPLPYKPDALEPYISAKTIEHHYGKHHKAYVDNLNALVKDSNNPIHQK-LETIIKNE 88
F+LPPLPY DAL P+ISA+T+ H+GKHH AYV+ LN ++ + P K LE I+KN
Sbjct 87 FTLPPLPYAEDALAPHISAETLRFHHGKHHAAYVNKLNGFIQGT--PFADKTLEDIVKNS 144
Query 89 KGNIYNQAAQVWNHTFYFNCLGPPSEETAA-PLGPTLKRLERDFGSLENFKNEFKK 143
G I+N AAQ WNH FYF + P S P G K +E F S FK+EF K
Sbjct 145 SGAIFNNAAQAWNHEFYFKSMKPASAGGGGEPDGLLKKEIEAAFSSFAQFKDEFSK 200
> tpv:TP01_1156 iron-dependent superoxide dismutase (EC:1.15.1.1);
K04564 superoxide dismutase, Fe-Mn family [EC:1.15.1.1]
Length=199
Score = 105 bits (261), Expect = 1e-22, Method: Compositional matrix adjust.
Identities = 52/112 (46%), Positives = 71/112 (63%), Gaps = 3/112 (2%)
Query 30 FSLPPLPYKPDALEPYISAKTIEHHYGKHHKAYVDNLNALVKDSNNPIHQKLETIIKNEK 89
F LPPLPY L P++S +T+ +HY KHH YV+ LN LVK ++ + LE ++K E
Sbjct 3 FKLPPLPYGLKDLVPHLSEETLTYHYTKHHAGYVNKLNGLVKGTSLET-KSLEDLLKTES 61
Query 90 GNIYNQAAQVWNHTFYFNCLGPPSEETAAPLGPTLKRLERDFGSLENFKNEF 141
G ++N AAQVWNHTFY++ + P P G K L++ FGS +NFK EF
Sbjct 62 GAVFNNAAQVWNHTFYWHSMSPSG--GGEPRGTVRKLLDQKFGSFDNFKKEF 111
> bbo:BBOV_IV011480 23.m06274; superoxide dismutase (EC:1.15.1.1);
K04564 superoxide dismutase, Fe-Mn family [EC:1.15.1.1]
Length=199
Score = 104 bits (259), Expect = 2e-22, Method: Compositional matrix adjust.
Identities = 51/113 (45%), Positives = 70/113 (61%), Gaps = 5/113 (4%)
Query 30 FSLPPLPYKPDALEPYISAKTIEHHYGKHHKAYVDNLNALVKDSNNPIHQ-KLETIIKNE 88
F LP LPY L P+IS +T+ HYGKHH YV+ LN+L+K + P+ +E +I +
Sbjct 3 FKLPALPYGMRELIPHISEETLSFHYGKHHAGYVNKLNSLIKGT--PMESCTIEELILGQ 60
Query 89 KGNIYNQAAQVWNHTFYFNCLGPPSEETAAPLGPTLKRLERDFGSLENFKNEF 141
G ++N AAQ+WNHTFY+N +GP P GP K++E FGS FK +F
Sbjct 61 TGAVFNNAAQIWNHTFYWNSMGPNC--GGEPTGPIRKKIEEKFGSFSAFKTDF 111
> pfa:PF08_0071 FeSOD, PfSOD1; Fe-superoxide dismutase (EC:1.15.1.1);
K04564 superoxide dismutase, Fe-Mn family [EC:1.15.1.1]
Length=198
Score = 102 bits (255), Expect = 5e-22, Method: Compositional matrix adjust.
Identities = 51/113 (45%), Positives = 71/113 (62%), Gaps = 5/113 (4%)
Query 30 FSLPPLPYKPDALEPYISAKTIEHHYGKHHKAYVDNLNALVKDSNNPIHQK-LETIIKNE 88
+LP L Y +AL P+IS +T+ HY KHH YV+ LN L+KD+ P +K L I+K
Sbjct 3 ITLPKLKYALNALSPHISEETLNFHYNKHHAGYVNKLNTLIKDT--PFAEKSLLDIVKES 60
Query 89 KGNIYNQAAQVWNHTFYFNCLGPPSEETAAPLGPTLKRLERDFGSLENFKNEF 141
G I+N AAQ+WNHTFY++ +GP P G ++++ DFGS NFK +F
Sbjct 61 SGAIFNNAAQIWNHTFYWDSMGPDC--GGEPHGEIKEKIQEDFGSFNNFKEQF 111
> ath:AT5G51100 FSD2; FSD2 (FE SUPEROXIDE DISMUTASE 2); superoxide
dismutase (EC:1.15.1.1)
Length=305
Score = 97.4 bits (241), Expect = 2e-20, Method: Compositional matrix adjust.
Identities = 49/121 (40%), Positives = 74/121 (61%), Gaps = 6/121 (4%)
Query 26 LSARFSLPPLPYKPDALEPYISAKTIEHHYGKHHKAYVDNLNALVKDSN-NPIHQKLETI 84
++A F L P PY DALEP++S +T+++H+GKHHK YV+NLN + ++ + + + +
Sbjct 49 ITAGFELKPPPYPLDALEPHMSRETLDYHWGKHHKTYVENLNKQILGTDLDALSLEEVVL 108
Query 85 IKNEKGNI---YNQAAQVWNHTFYFNCLGPPSEETAAPLGPTLKRLERDFGSLENFKNEF 141
+ KGN+ +N AAQ WNH F++ + P P G L+ +ERDFGS E F F
Sbjct 109 LSYNKGNMLPAFNNAAQAWNHEFFWESIQPGG--GGKPTGELLRLIERDFGSFEEFLERF 166
Query 142 K 142
K
Sbjct 167 K 167
> pfa:PFF1130c PfSOD2; superoxide dismutase (EC:1.15.1.1); K00518
superoxide dismutase [EC:1.15.1.1]
Length=266
Score = 97.1 bits (240), Expect = 3e-20, Method: Compositional matrix adjust.
Identities = 52/133 (39%), Positives = 73/133 (54%), Gaps = 2/133 (1%)
Query 30 FSLPPLPYKPDALEPYISAKTIEHHYGKHHKAYVDNLNALVKDSNNPIHQKLETIIKNEK 89
FSL LP+ P + P++S + I++HY KHH YV NLN L + + LE IIK
Sbjct 73 FSLMKLPFNPQDMSPFLSEEAIKYHYSKHHATYVKNLNNLSEQHKDLKSLTLEDIIKKYD 132
Query 90 GNIYNQAAQVWNHTFYFNCLGPPSEETAAPLGPTLKRLERDFGSLENFKNEFKKKFSDTS 149
G+I+N AAQ++NH F++ LG + P G ++L+ F S ENFKNEF K+ S
Sbjct 133 GSIHNNAAQIFNHNFFW--LGLKEQGGGMPYGEIKEKLDESFNSFENFKNEFIKQASGHF 190
Query 150 DPGGCGCCTTLRR 162
G R+
Sbjct 191 GSGWIWLIIKDRK 203
> ath:AT5G23310 FSD3; FSD3 (FE SUPEROXIDE DISMUTASE 3); superoxide
dismutase (EC:1.15.1.1)
Length=263
Score = 90.1 bits (222), Expect = 3e-18, Method: Compositional matrix adjust.
Identities = 47/122 (38%), Positives = 72/122 (59%), Gaps = 7/122 (5%)
Query 25 RLSARFSLPPLPYKPDALEPYISAKTIEHHYGKHHKAYVDNLNALVKDSNNPIHQKLETI 84
++ A + L PY DALEPY+S +T+E H+GKHH+ YVDNLN + + +E +
Sbjct 45 KVEAYYGLKTPPYPLDALEPYMSRRTLEVHWGKHHRGYVDNLNKQLGKDDRLYGYTMEEL 104
Query 85 IKNEKGN-----IYNQAAQVWNHTFYFNCLGPPSEETAAPLGPTLKRLERDFGSLENFKN 139
IK N +N AAQV+NH F++ + P +T P L+++++DFGS NF+
Sbjct 105 IKATYNNGNPLPEFNNAAQVYNHDFFWESMQPGGGDT--PQKGVLEQIDKDFGSFTNFRE 162
Query 140 EF 141
+F
Sbjct 163 KF 164
> eco:b3908 sodA, ECK3901, JW3879; superoxide dismutase, Mn (EC:1.15.1.1);
K04564 superoxide dismutase, Fe-Mn family [EC:1.15.1.1]
Length=206
Score = 75.5 bits (184), Expect = 8e-14, Method: Compositional matrix adjust.
Identities = 44/121 (36%), Positives = 71/121 (58%), Gaps = 11/121 (9%)
Query 30 FSLPPLPYKPDALEPYISAKTIEHHYGKHHKAYVDNLNALVKD----SNNPIHQ---KLE 82
++LP LPY DALEP+ +T+E H+ KHH+ YV+N NA ++ +N P+ + KL+
Sbjct 3 YTLPSLPYAYDALEPHFDKQTMEIHHTKHHQTYVNNANAALESLPEFANLPVEELITKLD 62
Query 83 TIIKNEKGNIYNQAAQVWNHTFYFNCLGPPSEETAAPLGPTLKRLERDFGSLENFKNEFK 142
+ ++K + N A NH+ ++ L ++ G +ERDFGS++NFK EF+
Sbjct 63 QLPADKKTVLRNNAGGHANHSLFWKGL----KKGTTLQGDLKAAIERDFGSVDNFKAEFE 118
Query 143 K 143
K
Sbjct 119 K 119
> mmu:20656 Sod2, MGC6144, MnSOD, Sod-2; superoxide dismutase
2, mitochondrial (EC:1.15.1.1); K04564 superoxide dismutase,
Fe-Mn family [EC:1.15.1.1]
Length=222
Score = 72.4 bits (176), Expect = 8e-13, Method: Compositional matrix adjust.
Identities = 46/147 (31%), Positives = 75/147 (51%), Gaps = 16/147 (10%)
Query 29 RFSLPPLPYKPDALEPYISAKTIEHHYGKHHKAYVDNLNALVKDSNNPIHQ-KLETIIKN 87
+ SLP LPY ALEP+I+A+ ++ H+ KHH AYV+NLNA + + + + + T +
Sbjct 25 KHSLPDLPYDYGALEPHINAQIMQLHHSKHHAAYVNNLNATEEKYHEALAKGDVTTQVAL 84
Query 88 EKGNIYNQAAQVWNHTFYFNCLGPPSEETAAPLGPTLKRLERDFGSLENFKNEFKKKFSD 147
+ +N + NHT ++ L P P G L+ ++RDFGS E +FK+K +
Sbjct 85 QPALKFNGGGHI-NHTIFWTNLSPKG--GGEPKGELLEAIKRDFGSFE----KFKEKLTA 137
Query 148 TSDPGGCGCCTTLRRTSWSWRRATTQQ 174
S ++ + W W +Q
Sbjct 138 VS--------VGVQGSGWGWLGFNKEQ 156
> ath:AT3G10920 MSD1; MSD1 (MANGANESE SUPEROXIDE DISMUTASE 1);
metal ion binding / superoxide dismutase (EC:1.15.1.1); K04564
superoxide dismutase, Fe-Mn family [EC:1.15.1.1]
Length=230
Score = 68.9 bits (167), Expect = 8e-12, Method: Compositional matrix adjust.
Identities = 46/149 (30%), Positives = 73/149 (48%), Gaps = 6/149 (4%)
Query 16 EMALRRAALRLSARFSLPPLPYKPDALEPYISAKTIEHHYGKHHKAYVDNLNALVKDSNN 75
E + R +R F+LP LPY ALEP IS + ++ H+ KHH+AYV N N ++ +
Sbjct 17 ETSSRLLRIRGIQTFTLPDLPYDYGALEPAISGEIMQIHHQKHHQAYVTNYNNALEQLDQ 76
Query 76 PIHQ-KLETIIKNEKGNIYNQAAQVWNHTFYFNCLGPPSEETAA-PLGPTLKRLERDFGS 133
+++ T++K + +N V NH+ ++ L P SE P G ++ FGS
Sbjct 77 AVNKGDASTVVKLQSAIKFNGGGHV-NHSIFWKNLAPSSEGGGEPPKGSLGSAIDAHFGS 135
Query 134 LENFKNEFKKKFSDTSDPGGCGCCTTLRR 162
LE KK ++ + G G L +
Sbjct 136 LEGL---VKKMSAEGAAVQGSGWWLGLDK 161
> xla:399216 sod2; superoxide dismutase 2, mitochondrial (EC:1.15.1.1);
K04564 superoxide dismutase, Fe-Mn family [EC:1.15.1.1]
Length=224
Score = 66.6 bits (161), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 38/114 (33%), Positives = 60/114 (52%), Gaps = 4/114 (3%)
Query 29 RFSLPPLPYKPDALEPYISAKTIEHHYGKHHKAYVDNLNALVKDSNNPIHQ-KLETIIKN 87
+ +LP LPY AL+P+ISA+ ++ H+ KHH YV+NLN + + + + T +
Sbjct 27 KHTLPDLPYDYGALQPHISAEIMQLHHSKHHATYVNNLNITEEKYAEALAKGDVTTQVSL 86
Query 88 EKGNIYNQAAQVWNHTFYFNCLGPPSEETAAPLGPTLKRLERDFGSLENFKNEF 141
+ +N + NHT ++ L P P G L ++RDFGS E FK +
Sbjct 87 QAALKFNGGGHI-NHTIFWTNLSPNG--GGEPQGELLDAIKRDFGSFEKFKEKL 137
> hsa:6648 SOD2, IPOB, MNSOD, MVCD6; superoxide dismutase 2, mitochondrial
(EC:1.15.1.1); K04564 superoxide dismutase, Fe-Mn
family [EC:1.15.1.1]
Length=222
Score = 65.9 bits (159), Expect = 7e-11, Method: Compositional matrix adjust.
Identities = 42/140 (30%), Positives = 70/140 (50%), Gaps = 16/140 (11%)
Query 29 RFSLPPLPYKPDALEPYISAKTIEHHYGKHHKAYVDNLNALVKDSNNPIHQ-KLETIIKN 87
+ SLP LPY ALEP+I+A+ ++ H+ KHH AYV+NLN + + + + I
Sbjct 25 KHSLPDLPYDYGALEPHINAQIMQLHHSKHHAAYVNNLNVTEEKYQEALAKGDVTAQIAL 84
Query 88 EKGNIYNQAAQVWNHTFYFNCLGPPSEETAAPLGPTLKRLERDFGSLENFKNEFKKKFSD 147
+ +N + NH+ ++ L P P G L+ ++RDFGS + +FK+K +
Sbjct 85 QPALKFNGGGHI-NHSIFWTNLSPNG--GGEPKGELLEAIKRDFGSFD----KFKEKLTA 137
Query 148 TSDPGGCGCCTTLRRTSWSW 167
S ++ + W W
Sbjct 138 AS--------VGVQGSGWGW 149
> xla:444203 MGC80739 protein; K04564 superoxide dismutase, Fe-Mn
family [EC:1.15.1.1]
Length=224
Score = 65.5 bits (158), Expect = 8e-11, Method: Compositional matrix adjust.
Identities = 37/114 (32%), Positives = 60/114 (52%), Gaps = 4/114 (3%)
Query 29 RFSLPPLPYKPDALEPYISAKTIEHHYGKHHKAYVDNLNALVKDSNNPIHQ-KLETIIKN 87
+ +LP LPY AL+P+ISA+ ++ H+ KHH YV+NLN + + + + T +
Sbjct 27 KHTLPDLPYDYGALQPHISAEIMQLHHSKHHATYVNNLNITEEKYAEALAKGDVTTQVSL 86
Query 88 EKGNIYNQAAQVWNHTFYFNCLGPPSEETAAPLGPTLKRLERDFGSLENFKNEF 141
+ +N + NHT ++ L P P G L ++RDFGS + FK +
Sbjct 87 QAALKFNGGGHI-NHTIFWTNLSPNG--GGEPQGELLDTIKRDFGSFQKFKEKL 137
> cel:C08A9.1 sod-3; SOD (superoxide dismutase) family member
(sod-3); K04564 superoxide dismutase, Fe-Mn family [EC:1.15.1.1]
Length=218
Score = 62.8 bits (151), Expect = 6e-10, Method: Compositional matrix adjust.
Identities = 41/147 (27%), Positives = 72/147 (48%), Gaps = 17/147 (11%)
Query 22 AALRLSARFSLPPLPYKPDALEPYISAKTIEHHYGKHHKAYVDNLNALVKDSNNPIHQ-K 80
L + ++ +LP LP+ LEP IS + ++ H+ KHH YV+NLN + + + + +
Sbjct 18 GVLAVRSKHTLPDLPFDYADLEPVISHEIMQLHHQKHHATYVNNLNQIEEKLHEAVSKGN 77
Query 81 LETIIKNEKGNIYNQAAQVWNHTFYFNCLGPPSEETAAPLGPTLKRLERDFGSLENFKNE 140
L+ I + +N + NH+ ++ L E + L T+K RDFGSL+N
Sbjct 78 LKEAIALQPALKFNGGGHI-NHSIFWTNLAKDGGEPSKELMDTIK---RDFGSLDN---- 129
Query 141 FKKKFSDTSDPGGCGCCTTLRRTSWSW 167
+K+ SD + ++ + W W
Sbjct 130 LQKRLSDIT--------IAVQGSGWGW 148
> ath:AT4G25100 FSD1; FSD1 (FE SUPEROXIDE DISMUTASE 1); copper
ion binding / superoxide dismutase
Length=186
Score = 61.6 bits (148), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 40/101 (39%), Positives = 53/101 (52%), Gaps = 8/101 (7%)
Query 46 ISAKTIEHHYGKHHKAYVDNLNALVKDSNNPIHQKLETIIKNEKGN-----IYNQAAQVW 100
+S +T+E H+GKHH+AYVDNL V + + LE II + N +N AAQ W
Sbjct 1 MSKQTLEFHWGKHHRAYVDNLKKQVLGTELE-GKPLEHIIHSTYNNGDLLPAFNNAAQAW 59
Query 101 NHTFYFNCLGPPSEETAAPLGPTLKRLERDFGSLENFKNEF 141
NH F++ + P + L LERDF S E F EF
Sbjct 60 NHEFFWESMKPGGGGKPSG--ELLALLERDFTSYEKFYEEF 98
> dre:335799 sod2, cb463, wu:fj33b01, zgc:73051; superoxide dismutase
2, mitochondrial (EC:1.15.1.1); K04564 superoxide dismutase,
Fe-Mn family [EC:1.15.1.1]
Length=224
Score = 60.8 bits (146), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 39/140 (27%), Positives = 67/140 (47%), Gaps = 16/140 (11%)
Query 29 RFSLPPLPYKPDALEPYISAKTIEHHYGKHHKAYVDNLNALVKDSNNPIHQ-KLETIIKN 87
+ +LP L Y ALEP+I A+ ++ H+ KHH YV+NLN + + + + T +
Sbjct 27 KHALPDLTYDYGALEPHICAEIMQLHHSKHHATYVNNLNVTEEKYQEALAKGDVTTQVSL 86
Query 88 EKGNIYNQAAQVWNHTFYFNCLGPPSEETAAPLGPTLKRLERDFGSLENFKNEFKKKFSD 147
+ +N + NHT ++ L P P G L+ ++RDFGS + + K+K S
Sbjct 87 QPALKFNGGGHI-NHTIFWTNLSPNG--GGEPQGELLEAIKRDFGSFQ----KMKEKISA 139
Query 148 TSDPGGCGCCTTLRRTSWSW 167
+ ++ + W W
Sbjct 140 AT--------VAVQGSGWGW 151
> tpv:TP04_0025 Fe-superoxide dismutase (EC:1.15.1.1); K04564
superoxide dismutase, Fe-Mn family [EC:1.15.1.1]
Length=241
Score = 60.8 bits (146), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 39/123 (31%), Positives = 55/123 (44%), Gaps = 27/123 (21%)
Query 29 RFSLPPLPY----------KPDALEPYISAKTIEHHYGKHHKAYVDNLNALVKDSNNPIH 78
RF L LPY K D L P +S + + HY KHH+ YV+NLN L++ +
Sbjct 47 RFELMELPYVRKQVVHIFQKLDELSPDLSEEAVRFHYLKHHQGYVNNLNKLIRSA----- 101
Query 79 QKLETIIKNEKGNIYNQAAQVWNHTFYFNCLGPPSEETAAPLGPTLKRLERDFGSLENFK 138
+G +Y A Q+WNH Y+ L P S P + F + +NFK
Sbjct 102 ----------EGKVYENACQIWNHNQYWLGLDPHS--AGKPTKFVEDVINLTFQTYDNFK 149
Query 139 NEF 141
+F
Sbjct 150 QQF 152
> cel:F10D11.1 sod-2; SOD (superoxide dismutase) family member
(sod-2); K04564 superoxide dismutase, Fe-Mn family [EC:1.15.1.1]
Length=221
Score = 59.7 bits (143), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 45/157 (28%), Positives = 70/157 (44%), Gaps = 39/157 (24%)
Query 22 AALRLSARFSLPPLPYKPDALEPYISAKTIEHHYGKHHKAYVDNLNALVKDSNNPIHQKL 81
AA+R ++ SLP LPY LEP IS + ++ H+ KHH YV+NLN I +KL
Sbjct 20 AAVR--SKHSLPDLPYDYADLEPVISHEIMQLHHQKHHATYVNNLNQ--------IEEKL 69
Query 82 ETIIKNEKGNI-----------YNQAAQVWNHTFYFNCLGPPSEETAAPLGPTLKRLERD 130
+ KGN+ +N + NH+ ++ L E +A L L ++ D
Sbjct 70 HEAV--SKGNVKEAIALQPALKFNGGGHI-NHSIFWTNLAKDGGEPSAEL---LTAIKSD 123
Query 131 FGSLENFKNEFKKKFSDTSDPGGCGCCTTLRRTSWSW 167
FGSL+N + + ++ + W W
Sbjct 124 FGSLDNLQKQLS------------ASTVAVQGSGWGW 148
> ath:AT3G56350 superoxide dismutase (Mn), putative / manganese
superoxide dismutase, putative; K04564 superoxide dismutase,
Fe-Mn family [EC:1.15.1.1]
Length=241
Score = 58.2 bits (139), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 40/130 (30%), Positives = 61/130 (46%), Gaps = 15/130 (11%)
Query 31 SLPPLPYKPDALEPYISAKTIEHHYGKHHKAYV-------DNLNALVKDSNNPIHQKLET 83
SLP LPY DALEP IS + + H+ KHH+ YV ++L + + D ++ KL++
Sbjct 37 SLPDLPYAYDALEPAISEEIMRLHHQKHHQTYVTQYNKALNSLRSAMADGDHSSVVKLQS 96
Query 84 IIKNEKGNIYNQAAQVWNHTFYFNCLGPPSEETAA-PLGPTLKRLERDFGSLENFKNEFK 142
+IK +N V NH ++ L P E P P ++ FGSLE +
Sbjct 97 LIK------FNGGGHV-NHAIFWKNLAPVHEGGGKPPHDPLASAIDAHFGSLEGLIQKMN 149
Query 143 KKFSDTSDPG 152
+ + G
Sbjct 150 AEGAAVQGSG 159
> sce:YHR008C SOD2; Mitochondrial superoxide dismutase, protects
cells against oxygen toxicity; phosphorylated (EC:1.15.1.1);
K04564 superoxide dismutase, Fe-Mn family [EC:1.15.1.1]
Length=233
Score = 57.4 bits (137), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 43/154 (27%), Positives = 64/154 (41%), Gaps = 19/154 (12%)
Query 12 TQKTEMALRRAALRLSARFSLPPLPYKPDALEPYISAKTIEHHYGKHHKAYVDNLNALVK 71
T+K ++L R + + +LP L + ALEPYIS + E HY KHH+ YV+ N V
Sbjct 11 TKKGGLSLLSTTARRT-KVTLPDLKWDFGALEPYISGQINELHYTKHHQTYVNGFNTAVD 69
Query 72 DSNNPIHQKLETIIKNE------------KGNIYNQAAQVWNHTFYFNCLGPPSEETAA- 118
Q+L ++ E + NI NH ++ L P S+
Sbjct 70 Q-----FQELSDLLAKEPSPANARKMIAIQQNIKFHGGGFTNHCLFWENLAPESQGGGEP 124
Query 119 PLGPTLKRLERDFGSLENFKNEFKKKFSDTSDPG 152
P G K ++ FGSL+ K + G
Sbjct 125 PTGALAKAIDEQFGSLDELIKLTNTKLAGVQGSG 158
> cpv:cgd1_1170 ubiquitin C-terminal hydrolase
Length=398
Score = 33.5 bits (75), Expect = 0.35, Method: Compositional matrix adjust.
Identities = 30/85 (35%), Positives = 38/85 (44%), Gaps = 10/85 (11%)
Query 89 KGNIYNQAAQVWNHTFYFNCLGPPSEETAAPLGPTLKRLERDFGS-LENFKNEFKKKFSD 147
KGN ++Q + FY N + + T A L L RL+ D GS LE F KKFS
Sbjct 88 KGNHFSQPIEAPPGMFYANQVINNACATQAILSIILNRLDIDIGSHLEEF-----KKFSS 142
Query 148 TSDPGGCGCCT----TLRRTSWSWR 168
+ DP G LR S+R
Sbjct 143 SFDPMTKGLVIGNSEVLRTAHNSFR 167
> tpv:TP04_0433 hypothetical protein
Length=465
Score = 30.8 bits (68), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 26/91 (28%), Positives = 47/91 (51%), Gaps = 11/91 (12%)
Query 5 YAFGAPETQKTEMALRRAALRLSARFSLPPLPYKPDALEPYISAKTIEHHYGKHHKAYVD 64
Y ++ K E++LR A ++ S P KP+ + Y+S + I+H G + Y++
Sbjct 316 YFIERVQSSKEEVSLRSLAYLIN---SCHP---KPELFD-YVSRRIIDHK-GPIGEDYLE 367
Query 65 NLNALVKDS---NNPIHQKLETIIKNEKGNI 92
N + S N+P+++ L +II NE N+
Sbjct 368 NFAFVFARSYYTNDPLYEALASIINNESTNL 398
> ath:AT1G16520 hypothetical protein
Length=325
Score = 30.4 bits (67), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 14/32 (43%), Positives = 17/32 (53%), Gaps = 0/32 (0%)
Query 120 LGPTLKRLERDFGSLENFKNEFKKKFSDTSDP 151
L T+ +L RD LE FK + K SD S P
Sbjct 100 LAMTVTKLTRDLAKLETFKRQLIKSLSDESGP 131
> ath:AT5G59340 WOX2; WOX2 (WUSCHEL RELATED HOMEOBOX 2); transcription
factor
Length=260
Score = 30.0 bits (66), Expect = 4.7, Method: Compositional matrix adjust.
Identities = 17/57 (29%), Positives = 26/57 (45%), Gaps = 1/57 (1%)
Query 11 ETQKTE-MALRRAALRLSARFSLPPLPYKPDALEPYISAKTIEHHYGKHHKAYVDNL 66
+ QK E MA L ++RF PP + PY + +HH +H Y ++L
Sbjct 72 QKQKQERMAYFNRLLHKTSRFFYPPPCSNVGCVSPYYLQQASDHHMNQHGSVYTNDL 128
> hsa:201595 STT3B, FLJ90106, SIMP, STT3-B; STT3, subunit of the
oligosaccharyltransferase complex, homolog B (S. cerevisiae)
(EC:2.4.1.119); K07151 dolichyl-diphosphooligosaccharide--protein
glycosyltransferase [EC:2.4.1.119]
Length=826
Score = 29.6 bits (65), Expect = 5.4, Method: Composition-based stats.
Identities = 16/58 (27%), Positives = 27/58 (46%), Gaps = 7/58 (12%)
Query 70 VKDSNNPIHQKLETIIKNEKGNIYNQAAQVWNHTFYFNCLGPPSEETAAPLGPTLKRL 127
+K N P+ + K +GN+Y++A +V H E+T LGP +K +
Sbjct 488 MKRENPPVEDSSDEDDKRNQGNLYDKAGKVRKH-------ATEQEKTEEGLGPNIKSI 538
> hsa:27107 ZBTB11, FLJ13426, MGC133303, ZNF-U69274, ZNF913; zinc
finger and BTB domain containing 11; K10498 zinc finger
and BTB domain-containing protein 11
Length=1053
Score = 29.6 bits (65), Expect = 6.1, Method: Compositional matrix adjust.
Identities = 30/114 (26%), Positives = 40/114 (35%), Gaps = 24/114 (21%)
Query 23 ALRLSARFSLPPLPYK-PDALEPYISAKTIEHHYGKHHKAYVDNLNALVKDSNNPIHQKL 81
+L+ R PY P E YI A+T+ H K H+ YV L KD+
Sbjct 900 SLKRHVRTHTGERPYVCPVCSEAYIDARTLRKHMTKFHRDYVPCKIMLEKDTL------- 952
Query 82 ETIIKNEKGNIYNQAAQVWNHTFYFNC-------LGPPSEETAAPLGPTLKRLE 128
+NQ QV + GP ET G T++ LE
Sbjct 953 ---------QFHNQGTQVAHAVSILTAGMQEQESSGPQELETVVVTGETMEALE 997
> mmu:271377 Zbtb11, 9230110G02Rik, ZNF-U69274; zinc finger and
BTB domain containing 11; K10498 zinc finger and BTB domain-containing
protein 11
Length=1050
Score = 29.3 bits (64), Expect = 7.3, Method: Compositional matrix adjust.
Identities = 30/114 (26%), Positives = 40/114 (35%), Gaps = 24/114 (21%)
Query 23 ALRLSARFSLPPLPYK-PDALEPYISAKTIEHHYGKHHKAYVDNLNALVKDSNNPIHQKL 81
+L+ R PY P E YI A+T+ H K H+ YV L KD+
Sbjct 897 SLKRHVRTHTGERPYVCPVCSEAYIDARTLRKHMTKFHRDYVPCKIMLEKDTL------- 949
Query 82 ETIIKNEKGNIYNQAAQVWNHTFYFNC-------LGPPSEETAAPLGPTLKRLE 128
+NQ QV + GP ET G T++ LE
Sbjct 950 ---------QFHNQGTQVEHAVSILTADMQEQESSGPQELETVVVTGETMEVLE 994
> pfa:PF14_0392 serine/threonine protein kinase, putative; K08282
non-specific serine/threonine protein kinase [EC:2.7.11.1]
Length=2247
Score = 28.9 bits (63), Expect = 9.7, Method: Composition-based stats.
Identities = 18/59 (30%), Positives = 27/59 (45%), Gaps = 5/59 (8%)
Query 55 YGKHHKAY----VDNLNALVKDSNNPIHQKLETIIKNEKGNIYNQAAQVWNHTFYFNCL 109
YGK+ K + N N + D N I + I N NI+NQ +++ H NC+
Sbjct 1543 YGKNEKKNDIINIRNFNDCISDYKNSICDGFQDIKNNSVKNIFNQDKKIY-HDLSNNCV 1600
Lambda K H
0.317 0.132 0.414
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4600750868
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40