bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_0406_orf1
Length=352
Score E
Sequences producing significant alignments: (Bits) Value
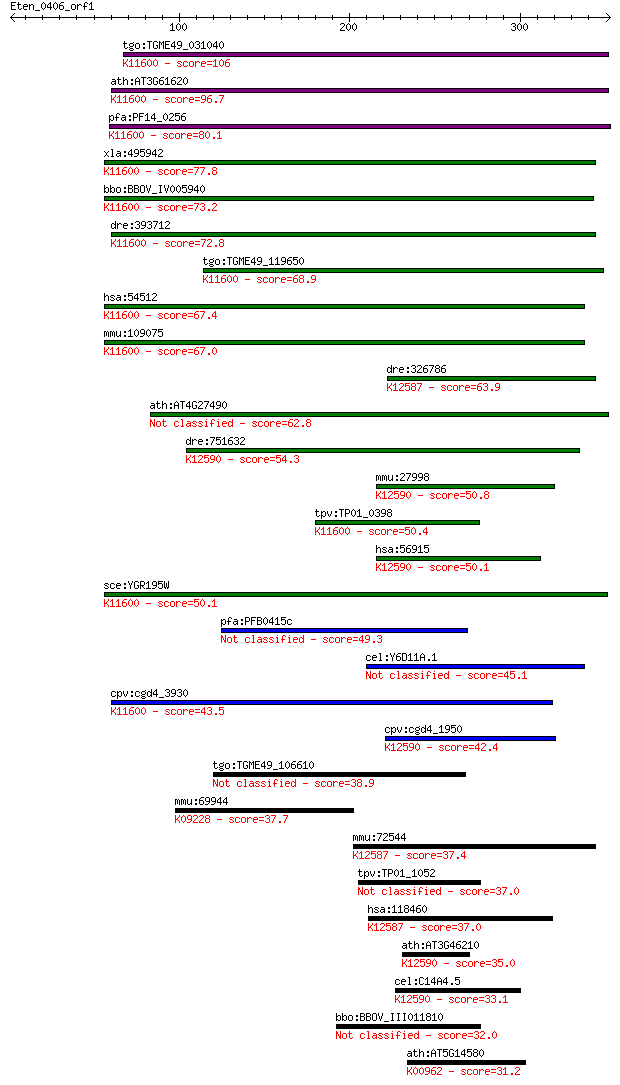
tgo:TGME49_031040 exosome complex exonuclease, putative ; K116... 106 1e-22
ath:AT3G61620 RRP41; RRP41; 3'-5'-exoribonuclease/ RNA binding... 96.7 1e-19
pfa:PF14_0256 exosome complex exonuclease rrp41, putative; K11... 80.1 1e-14
xla:495942 exosc4; exosome component 4; K11600 exosome complex... 77.8 6e-14
bbo:BBOV_IV005940 23.m05757; exosome complex exonuclease rrp41... 73.2 1e-12
dre:393712 exosc4, MGC73175, zgc:73175; exosome component 4; K... 72.8 2e-12
tgo:TGME49_119650 3' exoribonuclease, putative (EC:2.7.7.56); ... 68.9 3e-11
hsa:54512 EXOSC4, FLJ20591, RRP41, RRP41A, Rrp41p, SKI6, Ski6p... 67.4 9e-11
mmu:109075 Exosc4, 1110039I09Rik, 1500001N04Rik, Rrp41; exosom... 67.0 9e-11
dre:326786 exosc6, id:ibd1130, wu:fe17d05, zgc:110071; exosome... 63.9 9e-10
ath:AT4G27490 3' exoribonuclease family domain 1-containing pr... 62.8 2e-09
dre:751632 exosc5, MGC153707, si:ch211-194n6.2, zgc:153707; ex... 54.3 7e-07
mmu:27998 Exosc5, D7Wsu180e; exosome component 5; K12590 exoso... 50.8 7e-06
tpv:TP01_0398 exosome complex exonuclease Rrp41; K11600 exosom... 50.4 1e-05
hsa:56915 EXOSC5, MGC111224, MGC12901, RRP41B, RRP46, Rrp46p, ... 50.1 1e-05
sce:YGR195W SKI6, ECM20, RRP41; Exosome non-catalytic core com... 50.1 1e-05
pfa:PFB0415c 3' exoribonuclease, putative 49.3 2e-05
cel:Y6D11A.1 exos-4.2; EXOSome (multiexonuclease complex) comp... 45.1 4e-04
cpv:cgd4_3930 archeo-eukaryotic exosomal RNAse ; K11600 exosom... 43.5 0.001
cpv:cgd4_1950 RPR46-like RNAse PH domain ; K12590 exosome comp... 42.4 0.003
tgo:TGME49_106610 3' exoribonuclease family, domain 1 containi... 38.9 0.033
mmu:69944 2810021J22Rik, AI449137; RIKEN cDNA 2810021J22 gene;... 37.7 0.065
mmu:72544 Exosc6, 2610510N21Rik, C76919, MGC151362, Mtr3; exos... 37.4 0.078
tpv:TP01_1052 hypothetical protein 37.0 0.11
hsa:118460 EXOSC6, EAP4, MTR3, Mtr3p, hMtr3p, p11; exosome com... 37.0 0.12
ath:AT3G46210 3' exoribonuclease family domain 1-containing pr... 35.0 0.40
cel:C14A4.5 crn-5; Cell-death-Related Nuclease family member (... 33.1 1.6
bbo:BBOV_III011810 17.m10653; hypothetical protein 32.0 3.5
ath:AT5G14580 polyribonucleotide nucleotidyltransferase, putat... 31.2 6.0
> tgo:TGME49_031040 exosome complex exonuclease, putative ; K11600
exosome complex component RRP41
Length=352
Score = 106 bits (265), Expect = 1e-22, Method: Compositional matrix adjust.
Identities = 104/311 (33%), Positives = 155/311 (49%), Gaps = 35/311 (11%)
Query 67 FRADGRLVYECRQLRCRTSVPS---------AAASD---LTTSAEVPDGPAIATAGASGS 114
RADGR +E R+LR T VPS A+ASD +S E + +A + G
Sbjct 21 LRADGRRPHELRELRFSTCVPSLLPSPSARPASASDPRKADSSVESEEN-NLAPFASGGV 79
Query 115 LASCDGRAVVSLGCTXVMALVYGPQP----SSNMS-TLSTSSSNTWS----VSCTDNTSA 165
DGR++VS G T V A V+GP+P + N S L ++S WS + + S
Sbjct 80 GTHADGRSLVSFGSTKVAAFVFGPRPLPAGAGNASRALVSASGGGWSERGVLEEGFDASG 139
Query 166 AMQMMHEGKQGLARGELAVPVSVVCSVGLVDSCVR-----TRSRYTPDAAEIAAAVRTAA 220
A ++ +G + G S++C VG+ TRS + E+A VR
Sbjct 140 ADSVLGQGGAAGSGGAAER-ASLLCRVGIAPFSGDWRADVTRSGGAAEEHEVALGVRKVV 198
Query 221 EGIILRRLYTQTRITISILVLADDGSILSASLIAASLALADAGVAMRDLLPSCTVLLLPQ 280
E +IL ++ I + + V+ +DG IL+AS+ AASLAL DAG+A +D + + + + +P+
Sbjct 199 ESVILADTCPRSLICLFVHVVENDGGILAASISAASLALVDAGIATKDFVAAMSCVYMPR 258
Query 281 QFPQQQQEPLLLVDPTSDETRSGGPCLTLGVTAETNNIVALQLTGRVDRQTYEKMFEVCL 340
Q PLL DP E ++G P LTL + + + LQL G+V +E+M+E C
Sbjct 259 QL-----TPLL--DPPRAELQTGAPSLTLAMLVSSGEVSLLQLDGQVSTDVFEQMYEACA 311
Query 341 RGCLATAECMK 351
GC E MK
Sbjct 312 AGCREVGEAMK 322
> ath:AT3G61620 RRP41; RRP41; 3'-5'-exoribonuclease/ RNA binding;
K11600 exosome complex component RRP41
Length=241
Score = 96.7 bits (239), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 84/294 (28%), Positives = 126/294 (42%), Gaps = 72/294 (24%)
Query 60 EAVSPCGFRADGRLVYECRQLRCRTSVPSAAASDLTTSAEVPDGPAIATAGASGSLASCD 119
E V+P G R DGR E RQ+ AEV G ++ D
Sbjct 2 EYVNPEGLRLDGRRFNEMRQI----------------VAEV------------GVVSKAD 33
Query 120 GRAVVSLGCTXVMALVYGPQPSSNMSTLSTSSSNTWSVSCTDNTSAAMQMMHEGKQGLAR 179
G AV +G T V+A VYGP+ N S N +V
Sbjct 34 GSAVFEMGNTKVIAAVYGPREIQNKSQ---QKKNDHAV---------------------- 68
Query 180 GELAVPVSVVC--SVGLVDSCVRTRSRYTPDAAEIAAAVRTAAEGIILRRLYTQTRITIS 237
V+C S+ + R R ++ + E++ +R E IL L ++I I
Sbjct 69 --------VLCEYSMAQFSTGDRRRQKFDRRSTELSLVIRQTMEACILTELMPHSQIDIF 120
Query 238 ILVLADDGSILSASLIAASLALADAGVAMRDLLPSCTVLLLPQQFPQQQQEPLLLVDPTS 297
+ VL DG SA + AA+LALADAG+ MRDL SC+ L PLL ++
Sbjct 121 LQVLQADGGTRSACINAATLALADAGIPMRDLAVSCSAGYL-------NSTPLLDLNYVE 173
Query 298 DETRSGGPCLTLGVTAETNNIVALQLTGRVDRQTYEKMFEVCLRGCLATAECMK 351
D +GG +T+G+ + + + LQ+ ++ +T+E +F + GC A AE ++
Sbjct 174 DS--AGGADVTVGILPKLDKVSLLQMDAKLPMETFETVFALASEGCKAIAERIR 225
> pfa:PF14_0256 exosome complex exonuclease rrp41, putative; K11600
exosome complex component RRP41
Length=246
Score = 80.1 bits (196), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 67/294 (22%), Positives = 112/294 (38%), Gaps = 69/294 (23%)
Query 59 AEAVSPCGFRADGRLVYECRQLRCRTSVPSAAASDLTTSAEVPDGPAIATAGASGSLASC 118
+ ++ G+R DGR ECR ++ + G L
Sbjct 4 VQYINDEGYRIDGRKNDECRLIKI-------------------------SLGNGNELIDV 38
Query 119 DGRAVVSLGCTXVMALVYGPQPSSNMSTLSTSSSNTWSVSCTDNTSAAMQMMHEGKQGLA 178
DG + +G T + A + GP V C N + + E K+ +
Sbjct 39 DGFSFFEIGNTKLFAYIQGPNEYR-------RPDEKCLVKC--NVFLSPFNILEKKRKKS 89
Query 179 RGELAVPVSVVCSVGLVDSCVRTRSRYTPDAAEIAAAVRTAAEGIILRRLYTQTRITISI 238
+ D+ R EI++ +R IIL LY + I I +
Sbjct 90 K----------------DNVTR----------EISSYIRNICNHIILLDLYKNSEINIFL 123
Query 239 LVLADDGSILSASLIAASLALADAGVAMRDLLPSCTVLLLPQQFPQQQQEPLLLVDPTSD 298
++ DG + +A++ LAL DAG+A++ + + +VL L ++VD
Sbjct 124 YIIERDGGLKAAAVNTCILALIDAGIAIKYFISASSVLYLQNN---------IIVDGNQF 174
Query 299 ETRSGGPCLTLGVTAETNNIVALQLTGRVDRQTYEKMFEVCLRGCLATAECMKV 352
E SG P LTL + +NNIV L+ V ++ M + C + C+ MK+
Sbjct 175 EVNSGSPELTLAIDMSSNNIVLLEFDAEVPIDIFQSMLKTCAQACIHVGNIMKL 228
> xla:495942 exosc4; exosome component 4; K11600 exosome complex
component RRP41
Length=249
Score = 77.8 bits (190), Expect = 6e-14, Method: Compositional matrix adjust.
Identities = 68/288 (23%), Positives = 111/288 (38%), Gaps = 67/288 (23%)
Query 56 MFKAEAVSPCGFRADGRLVYECRQLRCRTSVPSAAASDLTTSAEVPDGPAIATAGASGSL 115
M E +S G+R DGR E R++R R G
Sbjct 1 MAGMELLSDQGYRVDGRKAGELRKIRARM----------------------------GVF 32
Query 116 ASCDGRAVVSLGCTXVMALVYGPQPSSNMSTLSTSSSNTWSVSCTDNTSAAMQMMHEGKQ 175
A DG A + G T +A+VYGP + S S C N +M G++
Sbjct 33 AQADGSAYIEQGNTKALAVVYGPHE------IRGSRSKMLHDRCVINCQYSMATFSTGER 86
Query 176 GLARGELAVPVSVVCSVGLVDSCVRTRSRYTPDAAEIAAAVRTAAEGIILRRLYTQTRIT 235
+ R ++E+ ++ E IL +LY +++I
Sbjct 87 ------------------------KRRPHGDRKSSEMTLHLKQTFEAAILTQLYPRSQID 122
Query 236 ISILVLADDGSILSASLIAASLALADAGVAMRDLLPSCTVLLLPQQFPQQQQEPLLLVDP 295
I + +L DG + AA+LA+ DAG+ MRD + +C+ + E L D
Sbjct 123 IYVQILQADGGNYCTCVNAATLAVIDAGIPMRDYVCACSAGFI---------EDTPLADL 173
Query 296 TSDETRSGGPCLTLGVTAETNNIVALQLTGRVDRQTYEKMFEVCLRGC 343
+ E +GGP L L + +++ I L++ R+ E++ E + C
Sbjct 174 SYVEEATGGPQLALALLPKSDQIALLEMNSRLHEDHLERVMEAVSKAC 221
> bbo:BBOV_IV005940 23.m05757; exosome complex exonuclease rrp41
(EC:3.1.13.-); K11600 exosome complex component RRP41
Length=259
Score = 73.2 bits (178), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 69/287 (24%), Positives = 112/287 (39%), Gaps = 61/287 (21%)
Query 56 MFKAEAVSPCGFRADGRLVYECRQLRCRTSVPSAAASDLTTSAEVPDGPAIATAGASGSL 115
M K E +S G R DGR E R + ++ GP +
Sbjct 1 MSKIEYISLEGLRIDGRRPSEVRHI------------------DILCGPECGV-----DI 37
Query 116 ASCDGRAVVSLGCTXVMALVYGPQPSSNMSTLSTSSSNTWSVSCTDNTSAAMQMMHEGKQ 175
+ DG A V+ G T V A + GP T K+
Sbjct 38 INYDGVAQVTQGLTKVQAFINGPTDIGRSKT---------------------------KE 70
Query 176 GLARGELAVPVSVVCSVGLVDSCVRTRSRYTPDAAEIAAAVRTAAEGIILRRLYTQTRIT 235
G + PV + C V + R EI+ AV E +I+ LY + I
Sbjct 71 GFETAD--SPVEIRCEVCIPSERKSMGHRNNDATVEISRAVVGTFEPVIISHLYKNSTIH 128
Query 236 ISILVLADDGSILSASLIAASLALADAGVAMRDLLPSCTVLLLPQQFPQQQQEPLLLVDP 295
I + VL DG + + + A +AL DAG+AM+D++ +CT ++L + LL+DP
Sbjct 129 IFVNVLEADGGVKATVINAVLIALIDAGIAMKDIIVACTAVMLNET---------LLIDP 179
Query 296 TSDETRSGGPCLTLGVTAETNNIVALQLTGRVDRQTYEKMFEVCLRG 342
E + LTL ++ ++ + L + ++Y ++ E ++G
Sbjct 180 NQLEINASIMELTLALSVSNEELIHIDLRSKYPIKSYAEIIEKAIKG 226
> dre:393712 exosc4, MGC73175, zgc:73175; exosome component 4;
K11600 exosome complex component RRP41
Length=245
Score = 72.8 bits (177), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 67/284 (23%), Positives = 108/284 (38%), Gaps = 67/284 (23%)
Query 60 EAVSPCGFRADGRLVYECRQLRCRTSVPSAAASDLTTSAEVPDGPAIATAGASGSLASCD 119
E +S G+R DGR E R+++ R SV A D
Sbjct 5 ELLSDQGYRLDGRKATELRKVQARMSV----------------------------FAQAD 36
Query 120 GRAVVSLGCTXVMALVYGPQPSSNMSTLSTSSSNTWSVSCTDNTSAAMQMMHEGKQGLAR 179
G A + G T +A+VYGP + S S + N +M ++
Sbjct 37 GSAYLEQGNTKALAVVYGPHE------IRGSRSKSLHDRAIINCQYSMATFSTAER---- 86
Query 180 GELAVPVSVVCSVGLVDSCVRTRSRYTPDAAEIAAAVRTAAEGIILRRLYTQTRITISIL 239
+ R ++E++ ++ E +L LY +++I I +
Sbjct 87 --------------------KRRPHGDRKSSEMSLHLKQTFEAAVLTELYPRSQIDIYVK 126
Query 240 VLADDGSILSASLIAASLALADAGVAMRDLLPSCTVLLLPQQFPQQQQEPLLLVDPTSDE 299
+L DG SA + AA+LAL DAG+ MRD + +C+ + E L D E
Sbjct 127 ILQADGGNYSACVNAATLALVDAGIPMRDYVCACSAGFV---------EDTPLADLCHAE 177
Query 300 TRSGGPCLTLGVTAETNNIVALQLTGRVDRQTYEKMFEVCLRGC 343
GG L L + + NI LQ+ R+ + + + E + C
Sbjct 178 ESGGGTALALALLPRSGNIALLQMDARLHQDHLDTLMEAAMTAC 221
> tgo:TGME49_119650 3' exoribonuclease, putative (EC:2.7.7.56);
K11600 exosome complex component RRP41
Length=303
Score = 68.9 bits (167), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 60/240 (25%), Positives = 102/240 (42%), Gaps = 40/240 (16%)
Query 114 SLASCDGRAVVSLGCTXVMALVYGPQPSSNMSTLSTSSSNTWSVSCTDNTSAAMQMMHEG 173
SLA+ G A VS+G T + VYGP+P+ ++ S N
Sbjct 65 SLAAAAGSAFVSVGKTKLNCAVYGPRPNMKHASQDRGSIN-------------------- 104
Query 174 KQGLARGELAVPVSVVCSVGLVDSCVRTRSRYTPDAAEIAAAVRTAAEGIILRRLYTQTR 233
L + + G D+C D A + + A ++ LY ++
Sbjct 105 --------LEFRFAPFATTG-KDACSER------DTAHLTTLLHQALNAVVRLDLYAKST 149
Query 234 ITISILVLADDGSILSASLIAASLALADAGVAMRDLLPSCTVLLLPQQFPQQQQEPLLLV 293
I +S+LVL DDG ++SA+L LALADAG+ M D++ + + P +L+
Sbjct 150 IAVSVLVLEDDGGLISAALTCIGLALADAGIEMLDVVTGASACVFSVGHPDSPPRTCVLL 209
Query 294 DPTSDETRSGGP----CLTLGVTAETNNIVALQLTGR-VDRQTYEKMFEVCLRGCLATAE 348
DP ++E R+ + LG ++ + +G + ++ E+M +C C A A+
Sbjct 210 DPDAEERRAFADKNCTFVDLGYCPALASVCFIHASGTLLATESGEQMLRLCEAACYAVAD 269
> hsa:54512 EXOSC4, FLJ20591, RRP41, RRP41A, Rrp41p, SKI6, Ski6p,
hRrp41p, p12A; exosome component 4; K11600 exosome complex
component RRP41
Length=245
Score = 67.4 bits (163), Expect = 9e-11, Method: Compositional matrix adjust.
Identities = 68/285 (23%), Positives = 109/285 (38%), Gaps = 73/285 (25%)
Query 56 MFKAEAVSPCGFRADGRLVYECRQLRCRTSVPSAAASDLTTSAEVPDGPAIATAGASGSL 115
M E +S G+R DGR E R+++ R V
Sbjct 1 MAGLELLSDQGYRVDGRRAGELRKIQARMGV----------------------------F 32
Query 116 ASCDGRAVVSLGCTXVMALVYGPQP---SSNMSTLSTSSSNTWSVSCTDNTSAAMQMMHE 172
A DG A + G T +A+VYGP S + + N S T +T + H
Sbjct 33 AQADGSAYIEQGNTKALAVVYGPHEIRGSRARALPDRALVNCQYSSATFSTGERKRRPHG 92
Query 173 GKQGLARGELAVPVSVVCSVGLVDSCVRTRSRYTPDAAEIAAAVRTAAEGIILRRLYTQT 232
++ C +GL +R E IL +L+ ++
Sbjct 93 DRKS-------------CEMGL--------------------QLRQTFEAAILTQLHPRS 119
Query 233 RITISILVLADDGSILSASLIAASLALADAGVAMRDLLPSCTVLLLPQQFPQQQQEPLLL 292
+I I + VL DG +A + AA+LA+ DAG+ MRD + +C+ + + L
Sbjct 120 QIDIYVQVLQADGGTYAACVNAATLAVLDAGIPMRDFVCACSAGFV---------DGTAL 170
Query 293 VDPTSDETRSGGPCLTLGVTAETNNIVALQLTGRVDRQTYEKMFE 337
D + E +GGP L L + + I L++ R+ E++ E
Sbjct 171 ADLSHVEEAAGGPQLALALLPASGQIALLEMDARLHEDHLERVLE 215
> mmu:109075 Exosc4, 1110039I09Rik, 1500001N04Rik, Rrp41; exosome
component 4; K11600 exosome complex component RRP41
Length=245
Score = 67.0 bits (162), Expect = 9e-11, Method: Compositional matrix adjust.
Identities = 68/285 (23%), Positives = 110/285 (38%), Gaps = 73/285 (25%)
Query 56 MFKAEAVSPCGFRADGRLVYECRQLRCRTSVPSAAASDLTTSAEVPDGPAIATAGASGSL 115
M E +S G+R DGR E R+++ R V
Sbjct 1 MAGLELLSDQGYRIDGRRAGELRKIQARMGV----------------------------F 32
Query 116 ASCDGRAVVSLGCTXVMALVYGPQP---SSNMSTLSTSSSNTWSVSCTDNTSAAMQMMHE 172
A DG A + G T +A+VYGP S + + + N S T +T + H
Sbjct 33 AQADGSAYIEQGNTKALAVVYGPHEIRGSRSRALPDRALVNCQYSSATFSTGERKRRPHG 92
Query 173 GKQGLARGELAVPVSVVCSVGLVDSCVRTRSRYTPDAAEIAAAVRTAAEGIILRRLYTQT 232
++ C +GL +R E IL +L+ ++
Sbjct 93 DRKS-------------CEMGL--------------------QLRQTFEAAILTQLHPRS 119
Query 233 RITISILVLADDGSILSASLIAASLALADAGVAMRDLLPSCTVLLLPQQFPQQQQEPLLL 292
+I I + VL DG +A + AA+LA+ DAG+ MRD + +C+ + + L
Sbjct 120 QIDIYVQVLQADGGTYAACVNAATLAVMDAGIPMRDFVCACSAGFV---------DGTAL 170
Query 293 VDPTSDETRSGGPCLTLGVTAETNNIVALQLTGRVDRQTYEKMFE 337
D + E +GGP L L + + I L++ R+ E++ E
Sbjct 171 ADLSHVEEAAGGPQLALALLPASGQIALLEMDSRLHEDHLEQVLE 215
> dre:326786 exosc6, id:ibd1130, wu:fe17d05, zgc:110071; exosome
component 6 (EC:3.1.13.-); K12587 exosome complex component
MTR3, animal type
Length=271
Score = 63.9 bits (154), Expect = 9e-10, Method: Compositional matrix adjust.
Identities = 39/130 (30%), Positives = 65/130 (50%), Gaps = 18/130 (13%)
Query 222 GIILRRLYTQTRITISILVLADDGSILSASLIAASLALADAGVAMRDLLPSCTVLLLPQQ 281
G+ L R Y +++I ++++VL +DGS+L+ ++ AS+ALADAG+ M D++ CT+
Sbjct 128 GVCLHR-YPRSQIDVNVMVLENDGSVLAHAVTCASMALADAGIEMYDIVLGCTL------ 180
Query 282 FPQQQQEPLLLVDPTSDET--------RSGGPCLTLGVTAETNNIVALQLTGRVDRQTYE 333
+Q LVDP+ E C+TL + N + L G + T
Sbjct 181 ---RQSGNACLVDPSYAEECGSWQEGYGDNQGCVTLALLPNLNQVSGLNADGEMREDTLT 237
Query 334 KMFEVCLRGC 343
+ C+ GC
Sbjct 238 EAMRTCMDGC 247
> ath:AT4G27490 3' exoribonuclease family domain 1-containing
protein
Length=256
Score = 62.8 bits (151), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 69/276 (25%), Positives = 106/276 (38%), Gaps = 57/276 (20%)
Query 83 RTSVPSAAASDLTTSAEVPDG-------PAIATAGASGSLASCDGRAVVSLGCTXVMALV 135
R+ +P SDL S PDG PA+ GA ++S G A G T V+ V
Sbjct 19 RSRLPIFKDSDLDWSR--PDGRGFHQCRPALLQTGA---VSSASGSAYAEFGNTKVIVSV 73
Query 136 YGPQPSSNMSTLSTSSSNTWSVSCTDNTSAAMQMMHEGKQGLARGELAVPVSVVCSVGLV 195
+GP+ S S +VS T+ S + QG
Sbjct 74 FGPRESKKAMVYSDVGRLNCNVSYTNFASPTL------GQGT------------------ 109
Query 196 DSCVRTRSRYTPDAAEIAAAVRTAAEGIILRRLYTQTRITISILVLADDGSILSASLIAA 255
D E ++ + A EG+I+ + +T + + LVL GS LS + A
Sbjct 110 ------------DHKEYSSMLHKALEGVIMMETFPKTTVDVFALVLESGGSDLSVLISCA 157
Query 256 SLALADAGVAMRDLLPSCTVLLLPQQFPQQQQEPLLLVDPTSDETRSGGPCLTLGVTAET 315
SLALADAG+ M DL+ + +V + + L++DP ++E +
Sbjct 158 SLALADAGIMMYDLITAVSVSCIGKS---------LMIDPVTEEEGCEDGSFMMTCMPSR 208
Query 316 NNIVALQLTGRVDRQTYEKMFEVCLRGCLATAECMK 351
I L +TG + ++CL E M+
Sbjct 209 YEITQLTITGEWTTPNINEAMQLCLDASSKLGEIMR 244
> dre:751632 exosc5, MGC153707, si:ch211-194n6.2, zgc:153707;
exosome component 5; K12590 exosome complex component RRP46
Length=218
Score = 54.3 bits (129), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 59/232 (25%), Positives = 94/232 (40%), Gaps = 50/232 (21%)
Query 104 PAIATAGASGSLASC-DGRAVVSLGCTXVMALVYGPQPSSNMSTLSTSSSNTWSVSCTDN 162
P + G+ SL S DG + G T ++A VYGP VS
Sbjct 8 PVLREYGSEQSLLSRPDGSSTFVQGDTSILAGVYGP--------------AEVKVSKEIY 53
Query 163 TSAAMQMMHEGKQGLARGELAVPVSVVCSVGLVDSCVRTRSRYTPDAAEIAAAVRTAAEG 222
A ++++ + K GL VR R+R VR E
Sbjct 54 DRATVEVLIQPKMGLPS-------------------VRERAR--------EQCVRETCEA 86
Query 223 IILRRLYTQTRITISILVLADDGSILSASLIAASLALADAGVAMRDLLPSCTVLLLPQQF 282
+L L+ ++ +T+ + V+ DDGS+LS L AA +AL DAG+ M L S T +
Sbjct 87 ALLLTLHPRSSLTVILQVVHDDGSLLSCCLNAACMALMDAGLPMSRLFCSVTCAI----- 141
Query 283 PQQQQEPLLLVDPTSDETRSGGPCLTLGVTAETNNIVALQLTGRVDRQTYEK 334
+E ++ DPT+ + + LT + + N++ TG Q ++
Sbjct 142 ---SKEGQIITDPTARQEKVSRALLTFAIDSNERNVLMSSTTGSFSVQELQQ 190
> mmu:27998 Exosc5, D7Wsu180e; exosome component 5; K12590 exosome
complex component RRP46
Length=235
Score = 50.8 bits (120), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 33/104 (31%), Positives = 54/104 (51%), Gaps = 8/104 (7%)
Query 216 VRTAAEGIILRRLYTQTRITISILVLADDGSILSASLIAASLALADAGVAMRDLLPSCTV 275
VR E ++L L+ +T IT+ + V++D GS+L+ L AA +AL DAGV MR L T
Sbjct 98 VRNTCEAVVLGALHPRTSITVVLQVVSDAGSLLACCLNAACMALVDAGVPMRALFCGVTC 157
Query 276 LLLPQQFPQQQQEPLLLVDPTSDETRSGGPCLTLGVTAETNNIV 319
L + L++DPT+ + + LT + + ++
Sbjct 158 AL--------DSDGNLVLDPTTKQEKEARAILTFALDSAEQKLL 193
> tpv:TP01_0398 exosome complex exonuclease Rrp41; K11600 exosome
complex component RRP41
Length=259
Score = 50.4 bits (119), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 33/99 (33%), Positives = 54/99 (54%), Gaps = 6/99 (6%)
Query 180 GELAVPVSVVCSVGLVDSCVRTRSRYTPDA---AEIAAAVRTAAEGIILRRLYTQTRITI 236
G L V + C + L + + +P+ ++A AV++ + +I+ Y + I+I
Sbjct 98 GILEEKVDIHCEILL---STEKKVKNSPNERLIKDLAMAVKSTYQEMIISHCYKTSSISI 154
Query 237 SILVLADDGSILSASLIAASLALADAGVAMRDLLPSCTV 275
I V+ DGSI S L A LAL DAGV+++D++ S TV
Sbjct 155 FINVIEYDGSIKSTVLNAVGLALIDAGVSLKDIVSSSTV 193
> hsa:56915 EXOSC5, MGC111224, MGC12901, RRP41B, RRP46, Rrp46p,
hRrp46p, p12B; exosome component 5; K12590 exosome complex
component RRP46
Length=235
Score = 50.1 bits (118), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 32/96 (33%), Positives = 50/96 (52%), Gaps = 8/96 (8%)
Query 216 VRTAAEGIILRRLYTQTRITISILVLADDGSILSASLIAASLALADAGVAMRDLLPSCTV 275
+R E ++L L+ +T IT+ + V++D GS+L+ L AA +AL DAGV MR L
Sbjct 98 IRNTCEAVVLGTLHPRTSITVVLQVVSDAGSLLACCLNAACMALVDAGVPMRALFCGVAC 157
Query 276 LLLPQQFPQQQQEPLLLVDPTSDETRSGGPCLTLGV 311
L + L++DPTS + + LT +
Sbjct 158 AL--------DSDGTLVLDPTSKQEKEARAVLTFAL 185
> sce:YGR195W SKI6, ECM20, RRP41; Exosome non-catalytic core component;
involved in 3'-5' RNA processing and degradation in
both the nucleus and the cytoplasm; has similarity to E. coli
RNase PH and to human hRrp41p (EXOSC4) (EC:3.1.13.-); K11600
exosome complex component RRP41
Length=246
Score = 50.1 bits (118), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 61/297 (20%), Positives = 106/297 (35%), Gaps = 71/297 (23%)
Query 56 MFKAEAVSPCGFRADGRLVYECRQLRCRTSVPSAAASDLTTSAEVPDGPAIATAGASGSL 115
M + E SP G R DGR E R+ + AA
Sbjct 1 MSRLEIYSPEGLRLDGRRWNELRRFESSINTHPHAA------------------------ 36
Query 116 ASCDGRAVVSLGCTXVMALVYGPQPSSNMSTLSTSSSNTWSVSCTDNTSAAMQMMHEGKQ 175
DG + + G ++ LV GP+ S + TS +
Sbjct 37 ---DGSSYMEQGNNKIITLVKGPKEPRLKSQMDTSKA----------------------- 70
Query 176 GLARGELAVPVSVVCSVGLVDSCVRTRSRYTPD--AAEIAAAVRTAAEGIILRRLYTQTR 233
++V ++ R++S + + EI ++ E ++ +Y +T
Sbjct 71 ---------LLNVSVNITKFSKFERSKSSHKNERRVLEIQTSLVRMFEKNVMLNIYPRTV 121
Query 234 ITISILVLADDGSILSASLIAASLALADAGVAMRDLLPSCTVLLLPQQFPQQQQEPLLLV 293
I I I VL DG I+ + + +LAL DAG++M D + +V L PLL
Sbjct 122 IDIEIHVLEQDGGIMGSLINGITLALIDAGISMFDYISGISVGLYDTT-------PLLDT 174
Query 294 DPTSDETRSGGPCLTLGVTAETNNIVALQLTGRVDRQTYEKMFEVCLRGCLATAECM 350
+ + S +TLGV ++ + L + ++ E + + + G + M
Sbjct 175 NSLEENAMS---TVTLGVVGKSEKLSLLLVEDKIPLDRLENVLAIGIAGAHRVRDLM 228
> pfa:PFB0415c 3' exoribonuclease, putative
Length=275
Score = 49.3 bits (116), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 39/144 (27%), Positives = 59/144 (40%), Gaps = 37/144 (25%)
Query 125 SLGCTXVMALVYGPQPSSNMSTLSTSSSNTWSVSCTDNTSAAMQMMHEGKQGLARGELAV 184
SLG T ++ VYGP P S +T S +G++ +
Sbjct 61 SLGNTKILTTVYGPNPDSKYATYS------------------------------KGKVFL 90
Query 185 PVSVVCSVGLVDSCVRTRSRYTPDAAEIAAAVRTAAEGIILRRLYTQTRITISILVLADD 244
V + ++ + + R R +I + + IIL Y Q I I L++ DD
Sbjct 91 DVKSL-NINTIGASDRQRD------DDIKSLLIECISNIILLEKYPQCSIKIKCLIIQDD 143
Query 245 GSILSASLIAASLALADAGVAMRD 268
G LSA+L SLAL A + M+D
Sbjct 144 GGCLSATLTCISLALIKAQIQMKD 167
> cel:Y6D11A.1 exos-4.2; EXOSome (multiexonuclease complex) component
family member (exos-4.2)
Length=241
Score = 45.1 bits (105), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 32/128 (25%), Positives = 56/128 (43%), Gaps = 8/128 (6%)
Query 210 AEIAAAVRTAAEGIILRRLYTQTRITISILVLADDGSILSASLIAASLALADAGVAMRDL 269
AE A + +A +I Y I I I VL+DDG +LS +L A +LA++ +G+ L
Sbjct 97 AEYRAQLASAVSAVIFASKYPGKVIEIEITVLSDDGGVLSTALSAVTLAISHSGIENMGL 156
Query 270 LPSCTVLLLPQQFPQQQQEPLLLVDPTSDETRSGGPCLTLGVTAETNNIVALQLTGRVDR 329
+ S V + + L DP++ E+ +T + + GR+
Sbjct 157 MASVHVAM--------NSDGECLTDPSTSESEGAIGGVTFAFVPNLGQTTCVDIYGRIPL 208
Query 330 QTYEKMFE 337
++ + E
Sbjct 209 KSTSSLLE 216
> cpv:cgd4_3930 archeo-eukaryotic exosomal RNAse ; K11600 exosome
complex component RRP41
Length=351
Score = 43.5 bits (101), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 62/268 (23%), Positives = 103/268 (38%), Gaps = 82/268 (30%)
Query 60 EAVSPCGFRADGRLVYECRQLRCRTSVPSAAASDLTTSAEVPDGPAIATAGASGSLASCD 119
E V+ G R DGR + E R+++C I G + D
Sbjct 91 EIVNKHGIRQDGRKLTEIRRIKC-----------------------IINEGNQRGIE--D 125
Query 120 GRAVVSLGCTXVMALVYGPQPSSNMSTLSTSSSNTWSVSCTDNTSAAMQMMHEGKQGLAR 179
G G ++ + GP P S +++ T+N S
Sbjct 126 GNVYFEQGQNKLIVSIVGPVPISG------------NINYTNNNSG-------------- 159
Query 180 GELAVPVSVVCSVGLVDSCVRTRSRYTPDAAEIAAAVRTAAE-GIILRRL--------YT 230
V + C+ R + D + R E G+I+ R Y+
Sbjct 160 ------VQINCNF-------RVSPFSSQDRRKRGKNDRFCIESGLIISRTFSSVICDQYS 206
Query 231 QTRITISILVLADDGSILSASLIAASLALADAGVAMRDLLPSCTVLLLPQQFPQQQQEPL 290
+++I I+I++L DGS+ SA++ A S+ALA +G++M+DL+ S T L +Q
Sbjct 207 KSQIIINIIILEGDGSVRSAAINATSIALAISGISMKDLIVSATCGLYGKQ--------- 257
Query 291 LLVDPTSDETRSGGPCLTLGVTAETNNI 318
+L D T E S L + + + N+I
Sbjct 258 VLYDLTQSEMESLKGTLLMAIHSTDNDI 285
> cpv:cgd4_1950 RPR46-like RNAse PH domain ; K12590 exosome complex
component RRP46
Length=244
Score = 42.4 bits (98), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 27/100 (27%), Positives = 49/100 (49%), Gaps = 1/100 (1%)
Query 221 EGIILRRLYTQTRITISILVLADDGSILSASLIAASLALADAGVAMRDLLPSCTVLLLPQ 280
E +I +++ I+I++ ++++DG IL + AA LAL D G+ M + P +
Sbjct 90 EKVIDFNSFSRCVISITLQIVSEDGPILPVCINAAVLALIDLGIPM-EFFPLAVSIAESS 148
Query 281 QFPQQQQEPLLLVDPTSDETRSGGPCLTLGVTAETNNIVA 320
F ++ L++DPT E C T+ + NI +
Sbjct 149 HFYGERDISHLMLDPTQSEFEQCISCSTIVINTTEKNIFS 188
> tgo:TGME49_106610 3' exoribonuclease family, domain 1 containing
protein (EC:2.7.7.56)
Length=392
Score = 38.9 bits (89), Expect = 0.033, Method: Compositional matrix adjust.
Identities = 42/159 (26%), Positives = 68/159 (42%), Gaps = 27/159 (16%)
Query 120 GRAVVSLGCTXVMALVYGPQPSSNMSTLSTSSSNTWSVSCTDNTSAAMQMMHEGKQGLAR 179
G A VS G T V+ALV GP + S+S H + LA
Sbjct 53 GSARVSQGLTSVIALVVGPVEAPPGPKYSSSGC----------------FFHVILKPLAG 96
Query 180 GELAVPVSVVCSVGLVDSCV---------RTRSRYTPDAAE--IAAAVRTAAEGIILRRL 228
+ ++G DS V R R + ++ E + ++T + ++L
Sbjct 97 PPMLSAARTAAAIGGCDSAVASGTARGTRRGRGAFGSESFERWMECQLQTLLQHMVLPGE 156
Query 229 YTQTRITISILVLADDGSILSASLIAASLALADAGVAMR 267
Y + + +++++L DDG SA + AA AL DAGV++R
Sbjct 157 YPRCLVQLTLMILQDDGGAESACINAALAALIDAGVSLR 195
> mmu:69944 2810021J22Rik, AI449137; RIKEN cDNA 2810021J22 gene;
K09228 KRAB domain-containing zinc finger protein
Length=539
Score = 37.7 bits (86), Expect = 0.065, Method: Compositional matrix adjust.
Identities = 35/107 (32%), Positives = 46/107 (42%), Gaps = 10/107 (9%)
Query 98 AEVPDG-PAI-ATAGASGSLASCDGRAVVSLGCTXVMALVYG-PQPSSNMSTLSTSSSNT 154
+ PDG PA A AG +L S G+ GC V+ P+P SN S S+ SN
Sbjct 270 GKCPDGEPAFRAKAGLQHTLHSW-GKP---RGCNDNKKCVHQMPKPRSNQSVFSSKESNC 325
Query 155 WSVSCTDNTSAAMQMMHEGKQGLARGELAVPVSVVCSVGLVDSCVRT 201
C D+T + Q H GK+ E + PV + C RT
Sbjct 326 VKTFCPDSTLSVQQRPHTGKKPQ---ESSTPVKINSQPRRRHRCGRT 369
> mmu:72544 Exosc6, 2610510N21Rik, C76919, MGC151362, Mtr3; exosome
component 6 (EC:3.1.13.-); K12587 exosome complex component
MTR3, animal type
Length=273
Score = 37.4 bits (85), Expect = 0.078, Method: Compositional matrix adjust.
Identities = 46/148 (31%), Positives = 70/148 (47%), Gaps = 12/148 (8%)
Query 202 RSRYTPDAA-----EIAAAVRTAAEGIILRRLYTQTRITISILVLADDGSILSASLIAAS 256
R R P E+ A++ A E + Y + ++ +S L+L D G L+A+L AA+
Sbjct 108 RRRRAPQGGGGEDRELGLALQEALEPAVRLGRYPRAQLEVSALLLEDGGCALAAALTAAA 167
Query 257 LALADAGVAMRDLLPSCTVLLLPQQFPQQQQEPLLLVDPTSDETRSGGPCLTLGVTAETN 316
LALADAGV M DL+ C + L P P L+DPT E LT+ + N
Sbjct 168 LALADAGVEMYDLVVGCGLSLTP------GPSPTWLLDPTRLEEEHSAAGLTVALMPVLN 221
Query 317 NIVALQLTGRVDR-QTYEKMFEVCLRGC 343
+ L +G + +++ + L GC
Sbjct 222 QVAGLLGSGEGGQTESWTDAVRLGLEGC 249
> tpv:TP01_1052 hypothetical protein
Length=148
Score = 37.0 bits (84), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 22/73 (30%), Positives = 39/73 (53%), Gaps = 1/73 (1%)
Query 205 YTPDAAE-IAAAVRTAAEGIILRRLYTQTRITISILVLADDGSILSASLIAASLALADAG 263
+TP E + + + E IL + Y I I+V DDG +L A ++ L+L D G
Sbjct 75 HTPKYLETLKSHIIETFERHILLQNYPSQIIEAYIIVSNDDGGLLPALIMGMCLSLIDCG 134
Query 264 VAMRDLLPSCTVL 276
+ + D++ +C+V+
Sbjct 135 IQVYDVISACSVV 147
> hsa:118460 EXOSC6, EAP4, MTR3, Mtr3p, hMtr3p, p11; exosome component
6; K12587 exosome complex component MTR3, animal type
Length=272
Score = 37.0 bits (84), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 40/108 (37%), Positives = 58/108 (53%), Gaps = 6/108 (5%)
Query 211 EIAAAVRTAAEGIILRRLYTQTRITISILVLADDGSILSASLIAASLALADAGVAMRDLL 270
E+A A++ A E + Y + ++ +S L+L D GS L+A+L AA+LALADAGV M DL+
Sbjct 121 ELALALQEALEPAVRLGRYPRAQLEVSALLLEDGGSALAAALTAAALALADAGVEMYDLV 180
Query 271 PSCTVLLLPQQFPQQQQEPLLLVDPTSDETRSGGPCLTLGVTAETNNI 318
C + L P P L+DPT E LT+ + N +
Sbjct 181 VGCGLSLAPGPAPT------WLLDPTRLEEERAAAGLTVALMPVLNQV 222
> ath:AT3G46210 3' exoribonuclease family domain 1-containing
protein; K12590 exosome complex component RRP46
Length=239
Score = 35.0 bits (79), Expect = 0.40, Method: Compositional matrix adjust.
Identities = 17/39 (43%), Positives = 25/39 (64%), Gaps = 0/39 (0%)
Query 231 QTRITISILVLADDGSILSASLIAASLALADAGVAMRDL 269
T ++ I V+ DDGS+L ++ AA AL DAG+ M+ L
Sbjct 99 NTTTSVIIQVVHDDGSLLPCAINAACAALVDAGIPMKHL 137
> cel:C14A4.5 crn-5; Cell-death-Related Nuclease family member
(crn-5); K12590 exosome complex component RRP46
Length=214
Score = 33.1 bits (74), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 19/73 (26%), Positives = 39/73 (53%), Gaps = 9/73 (12%)
Query 227 RLYTQTRITISILVLADDGSILSASLIAASLALADAGVAMRDLLPSCTVLLLPQQFPQQQ 286
L+ T I++++ + DDGS+ + ++ A AL D G+ + C VL++ +
Sbjct 84 ELFPHTTISVTVHGIQDDGSMGAVAINGACFALLDNGMPFETVF--CGVLIV-------R 134
Query 287 QEPLLLVDPTSDE 299
+ L++DPT+ +
Sbjct 135 VKDELIIDPTAKQ 147
> bbo:BBOV_III011810 17.m10653; hypothetical protein
Length=196
Score = 32.0 bits (71), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 23/87 (26%), Positives = 42/87 (48%), Gaps = 2/87 (2%)
Query 192 VGLVDSCVRTRSRYTPDA--AEIAAAVRTAAEGIILRRLYTQTRITISILVLADDGSILS 249
VG++ VR + P + A++ + E ++ Y + I + + D G +
Sbjct 38 VGILSIEVRYSKPHIPSSSDADLRHVLTELFERHVILSRYPRQLIEAWVTIEEDAGGLFG 97
Query 250 ASLIAASLALADAGVAMRDLLPSCTVL 276
A + A SLA AD G+ M D+L + +V+
Sbjct 98 ACVTALSLAFADCGIQMLDILAATSVV 124
> ath:AT5G14580 polyribonucleotide nucleotidyltransferase, putative;
K00962 polyribonucleotide nucleotidyltransferase [EC:2.7.7.8]
Length=991
Score = 31.2 bits (69), Expect = 6.0, Method: Compositional matrix adjust.
Identities = 22/69 (31%), Positives = 34/69 (49%), Gaps = 12/69 (17%)
Query 234 ITISILVLADDGSILSASLIAASLALADAGVAMRDLLPSCTVLLLPQQFPQQQQEPLLLV 293
I I+ V++ DGS AS+ S+AL DAG+ +R + +V L+ V
Sbjct 474 IRINSEVMSSDGSTSMASVCGGSMALMDAGIPLRAHVAGVSVGLITD------------V 521
Query 294 DPTSDETRS 302
DP+S E +
Sbjct 522 DPSSGEIKD 530
Lambda K H
0.319 0.129 0.380
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 15069884872
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40