bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_0397_orf1
Length=169
Score E
Sequences producing significant alignments: (Bits) Value
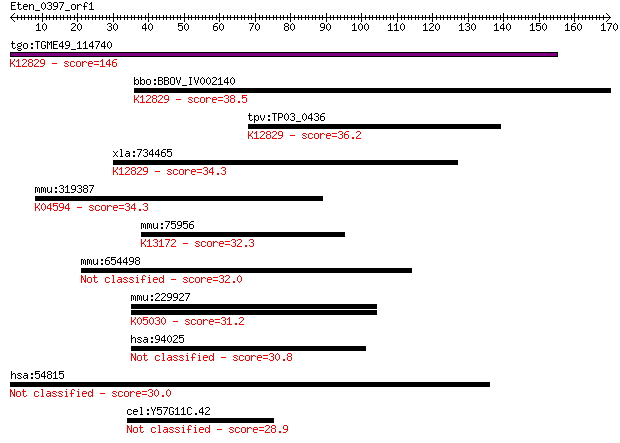
tgo:TGME49_114740 splicing factor 3B subunit 2, putative ; K12... 146 3e-35
bbo:BBOV_IV002140 21.m02783; splicing factor 3B subunit 2; K12... 38.5 0.012
tpv:TP03_0436 hypothetical protein; K12829 splicing factor 3B ... 36.2 0.047
xla:734465 sf3b2, MGC115052, cus1, sap145, sf3b145, sf3b150; s... 34.3 0.19
mmu:319387 Lphn3, 5430402I23Rik, CIRL-3, D130075K09Rik, Gm1379... 34.3 0.20
mmu:75956 Srrm2, 5033413A03Rik, AA410130, SRm300, mKIAA0324; s... 32.3 0.74
mmu:654498 Hhla1, F930104E18Rik; HERV-H LTR-associating 1 32.0
mmu:229927 Clca4; chloride channel calcium activated 4; K05030... 31.2 1.6
hsa:94025 MUC16, CA125, FLJ14303; mucin 16, cell surface assoc... 30.8 2.2
hsa:54815 GATAD2A, FLJ20085, FLJ21017, p66alpha; GATA zinc fin... 30.0 4.1
cel:Y57G11C.42 hypothetical protein 28.9 8.9
> tgo:TGME49_114740 splicing factor 3B subunit 2, putative ; K12829
splicing factor 3B subunit 2
Length=743
Score = 146 bits (369), Expect = 3e-35, Method: Compositional matrix adjust.
Identities = 98/171 (57%), Positives = 111/171 (64%), Gaps = 34/171 (19%)
Query 1 QQGQLFGVRHTYRIPAGALAGQQQQQQQQLLQQGGASGVQTPAGIATPR-LLSGRASP-- 57
QQGQLFGV+HTY+IP+ L G +G TP+G TPR LLSG A+P
Sbjct 576 QQGQLFGVKHTYKIPS-TLPGN--------------AGTATPSGHLTPRNLLSGAATPSG 620
Query 58 ----FVAGLRSPFA-PAAAAAAAAA---------SGANTPLMAQRGGAATPAGVTVSLNP 103
F G R+P P +A+ +A SGA TP A+ G TPAGVTVSLNP
Sbjct 621 VRTPFTGGARTPLVGPGGSASFGSAPGTPFLGGASGAQTP--ARGAGFRTPAGVTVSLNP 678
Query 104 NEMEQEGAFTADVIRQQLRQHEEAAARAKAAAGQIDPADQRKRKGDELRGV 154
NEMEQEG FTADVIRQQLRQHEEAAARAK AAGQ+DPAD RKRKGDE+RG
Sbjct 679 NEMEQEGVFTADVIRQQLRQHEEAAARAKQAAGQVDPADTRKRKGDEIRGT 729
> bbo:BBOV_IV002140 21.m02783; splicing factor 3B subunit 2; K12829
splicing factor 3B subunit 2
Length=552
Score = 38.5 bits (88), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 47/145 (32%), Positives = 63/145 (43%), Gaps = 29/145 (20%)
Query 36 ASGVQTP----AGIATPRLLSGRASPFV---AGLRSPFAPAAAAAAAAASGANTPLMAQR 88
A+GV TP + I PR L +A + AG S + + + TP+
Sbjct 426 ATGVDTPLEVRSNIDPPRTLPRKAYTVLEPKAGKGSTGSLFGSHVTYSMPPVATPITPIT 485
Query 89 G--GAATPAG--VTVSLNPNEMEQEGAFTADVIRQQLRQHEEAAARAKAAAGQIDPADQR 144
G G ATP G VT SL + + TA + QQL+ HE A AAGQ+ P
Sbjct 486 GKPGTATPLGGLVTPSL-----QIDSNITAHGVMQQLKFHETKAKLVHEAAGQVVP---- 536
Query 145 KRKGDELRGVSAAAAKSKRKKQFKF 169
+ E R V K+KK+FKF
Sbjct 537 --EATETRRV-------KKKKEFKF 552
> tpv:TP03_0436 hypothetical protein; K12829 splicing factor 3B
subunit 2
Length=552
Score = 36.2 bits (82), Expect = 0.047, Method: Composition-based stats.
Identities = 27/71 (38%), Positives = 32/71 (45%), Gaps = 11/71 (15%)
Query 68 PAAAAAAAAASGANTPLMAQRGGAATPAGVTVSLNPNEMEQEGAFTADVIRQQLRQHEEA 127
P A G TPL GG ATP+ T E +G T D I +QL+ HEE
Sbjct 474 PPPVATPLGMGGLTTPL----GGMATPSLTT-------EEVDGTSTTDQILRQLKYHEEK 522
Query 128 AARAKAAAGQI 138
A + AGQI
Sbjct 523 AKKVHEEAGQI 533
> xla:734465 sf3b2, MGC115052, cus1, sap145, sf3b145, sf3b150;
splicing factor 3b, subunit 2, 145kDa; K12829 splicing factor
3B subunit 2
Length=764
Score = 34.3 bits (77), Expect = 0.19, Method: Compositional matrix adjust.
Identities = 32/111 (28%), Positives = 51/111 (45%), Gaps = 16/111 (14%)
Query 30 LLQQGGASGVQTPAGIATPRLLSGRASPF---VAGLRSP--FAPAAAAAAAAASGA---N 81
L+ GG S V PAG+ TP L+ R + G +P F A GA +
Sbjct 614 LITPGGFSSV--PAGMETPELIELRKKKIEEAMDGTETPQLFTVLPEKRTATVGGAMMGS 671
Query 82 TPL------MAQRGGAATPAGVTVSLNPNEMEQEGAFTADVIRQQLRQHEE 126
T + M++RG A P GV ++L P E+E + + +++R+ +E
Sbjct 672 THIYEMATAMSRRGIATEPQGVEIALAPEELELDPSAMTQKYEERVREAQE 722
> mmu:319387 Lphn3, 5430402I23Rik, CIRL-3, D130075K09Rik, Gm1379,
LEC3, MGC99439, mKIAA0768; latrophilin 3; K04594 latrophilin
3
Length=1543
Score = 34.3 bits (77), Expect = 0.20, Method: Composition-based stats.
Identities = 27/81 (33%), Positives = 32/81 (39%), Gaps = 14/81 (17%)
Query 8 VRHTYRIPAGALAGQQQQQQQQLLQQGGASGVQTPAGIATPRLLSGRASPFVAGLRSPFA 67
+RH R P GAL + QQ + G Q P G A G+R P A
Sbjct 33 LRHAERSPGGALPPRHLLQQPAAERSTAHRG-QGPRGAAR-------------GVRGPGA 78
Query 68 PAAAAAAAAASGANTPLMAQR 88
P A AA A S A P+ R
Sbjct 79 PGAQIAAQAFSRAPIPMAVVR 99
> mmu:75956 Srrm2, 5033413A03Rik, AA410130, SRm300, mKIAA0324;
serine/arginine repetitive matrix 2; K13172 serine/arginine
repetitive matrix protein 2
Length=2607
Score = 32.3 bits (72), Expect = 0.74, Method: Compositional matrix adjust.
Identities = 24/59 (40%), Positives = 32/59 (54%), Gaps = 4/59 (6%)
Query 38 GVQTPAGIATPRLLSGRASPFVAG--LRSPFAPAAAAAAAAASGANTPLMAQRGGAATP 94
G +TPAG+A L S R +P ++G L SP P +A SG +PLM R + TP
Sbjct 2211 GSRTPAGLAPTNLSSSRMAPALSGANLTSPRVP--LSAYDRVSGRTSPLMLDRARSRTP 2267
> mmu:654498 Hhla1, F930104E18Rik; HERV-H LTR-associating 1
Length=514
Score = 32.0 bits (71), Expect = 1.0, Method: Composition-based stats.
Identities = 27/93 (29%), Positives = 35/93 (37%), Gaps = 2/93 (2%)
Query 21 GQQQQQQQQLLQQGGASGVQTPAGIATPRLLSGRASPFVAGLRSPFAPAAAAAAAAASGA 80
QQ + LL G + TP+ +A P SG P APA A
Sbjct 369 AQQLRSTGNLLHPTGI--LTTPSRLAQPSRASGTLMPGTQTTNPTQAPAPRVPQTDGIPA 426
Query 81 NTPLMAQRGGAATPAGVTVSLNPNEMEQEGAFT 113
P + ++ A PA VS P + QE A T
Sbjct 427 EWPFIPEKEPARDPAAHQVSKCPRPLLQEEAIT 459
> mmu:229927 Clca4; chloride channel calcium activated 4; K05030
calcium-activated chloride channel family member 4
Length=1044
Score = 31.2 bits (69), Expect = 1.6, Method: Composition-based stats.
Identities = 20/70 (28%), Positives = 31/70 (44%), Gaps = 9/70 (12%)
Query 35 GASGVQTPAGIATPRLLSGRASPFV-AGLRSPFAPAAAAAAAAASGANTPLMAQRGGAAT 93
G S TP G++TP G ++P GL +P P + + G +TP +T
Sbjct 887 GLSTPSTPPGLSTPSTPPGLSTPSTPPGLSTPSTPPGLSTPSTPPGLSTP--------ST 938
Query 94 PAGVTVSLNP 103
P G++ P
Sbjct 939 PPGLSTPSTP 948
Score = 31.2 bits (69), Expect = 1.6, Method: Composition-based stats.
Identities = 20/70 (28%), Positives = 31/70 (44%), Gaps = 9/70 (12%)
Query 35 GASGVQTPAGIATPRLLSGRASPFV-AGLRSPFAPAAAAAAAAASGANTPLMAQRGGAAT 93
G S TP G++TP G ++P GL +P P + + G +TP +T
Sbjct 932 GLSTPSTPPGLSTPSTPPGLSTPSTPPGLSTPSTPPGLSTPSTPPGLSTP--------ST 983
Query 94 PAGVTVSLNP 103
P G++ P
Sbjct 984 PPGLSTPSTP 993
> hsa:94025 MUC16, CA125, FLJ14303; mucin 16, cell surface associated
Length=14507
Score = 30.8 bits (68), Expect = 2.2, Method: Composition-based stats.
Identities = 22/66 (33%), Positives = 28/66 (42%), Gaps = 0/66 (0%)
Query 35 GASGVQTPAGIATPRLLSGRASPFVAGLRSPFAPAAAAAAAAASGANTPLMAQRGGAATP 94
G SGV P+ T L AS A R+P AAS NT + G + TP
Sbjct 2076 GHSGVSNPSSTTTEFPLFSAASTSAAKQRNPETETHGPQNTAASTLNTDASSVTGLSETP 2135
Query 95 AGVTVS 100
G ++S
Sbjct 2136 VGASIS 2141
> hsa:54815 GATAD2A, FLJ20085, FLJ21017, p66alpha; GATA zinc finger
domain containing 2A
Length=633
Score = 30.0 bits (66), Expect = 4.1, Method: Compositional matrix adjust.
Identities = 37/142 (26%), Positives = 57/142 (40%), Gaps = 14/142 (9%)
Query 1 QQGQLFGVRHTYRIPAGALAGQQQQQQ--QQLLQQGGASGVQTPAGIATPRLLSGRASPF 58
Q + V HT R+ A + QQ+Q+ Q+LLQQG A A P P
Sbjct 445 NQKKALKVEHTSRLKAAFVKALQQEQEIEQRLLQQGTAPAQAKAEPTAAPH-------PV 497
Query 59 VAGLRSPFAPAAAAAAAAASGANTPLM-----AQRGGAATPAGVTVSLNPNEMEQEGAFT 113
+ + P A + A +N ++ RG A TP GV + +P+ Q A
Sbjct 498 LKQVIKPRRKLAFRSGEARDWSNGAVLQASSQLSRGSATTPRGVLHTFSPSPKLQNSASA 557
Query 114 ADVIRQQLRQHEEAAARAKAAA 135
++ + R E + K +A
Sbjct 558 TALVSRTGRHSERTVSAGKGSA 579
> cel:Y57G11C.42 hypothetical protein
Length=661
Score = 28.9 bits (63), Expect = 8.9, Method: Compositional matrix adjust.
Identities = 14/41 (34%), Positives = 21/41 (51%), Gaps = 0/41 (0%)
Query 34 GGASGVQTPAGIATPRLLSGRASPFVAGLRSPFAPAAAAAA 74
G TPA P L+ + P+ G ++P APAA++A
Sbjct 118 GAQKATTTPAPTGFPTLIPFSSEPWTTGTQTPLAPAASSAV 158
Lambda K H
0.313 0.125 0.344
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4222647260
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40