bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_0393_orf1
Length=124
Score E
Sequences producing significant alignments: (Bits) Value
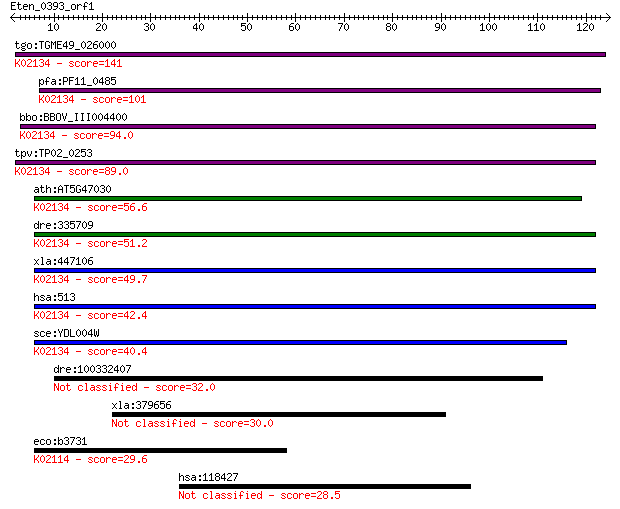
tgo:TGME49_026000 ATP synthase, putative (EC:3.6.3.14); K02134... 141 5e-34
pfa:PF11_0485 mitochondrial ATP synthase delta subunit, putati... 101 6e-22
bbo:BBOV_III004400 17.m07397; ATP synthase, Delta/Epsilon chai... 94.0 1e-19
tpv:TP02_0253 ATP synthase F1 subunit delta (EC:3.6.3.14); K02... 89.0 3e-18
ath:AT5G47030 ATP synthase delta' chain, mitochondrial; K02134... 56.6 2e-08
dre:335709 atp5d, fk58f09, wu:fk58f09, zgc:73303; ATP synthase... 51.2 8e-07
xla:447106 atp5d, MGC85306; ATP synthase, H+ transporting, mit... 49.7 2e-06
hsa:513 ATP5D; ATP synthase, H+ transporting, mitochondrial F1... 42.4 3e-04
sce:YDL004W ATP16; Atp16p (EC:3.6.3.14); K02134 F-type H+-tran... 40.4 0.001
dre:100332407 hypothetical protein LOC100332407 32.0 0.57
xla:379656 stard3, MGC68989; StAR-related lipid transfer (STAR... 30.0 2.2
eco:b3731 atpC, ECK3724, JW3709, papG, uncC; F1 sector of memb... 29.6 2.5
hsa:118427 OLFM3, NOE3, NOELIN3, NOELIN3_V1, NOELIN3_V2, NOELI... 28.5 5.9
> tgo:TGME49_026000 ATP synthase, putative (EC:3.6.3.14); K02134
F-type H+-transporting ATPase subunit delta [EC:3.6.3.14]
Length=183
Score = 141 bits (355), Expect = 5e-34, Method: Compositional matrix adjust.
Identities = 67/123 (54%), Positives = 88/123 (71%), Gaps = 1/123 (0%)
Query 2 PAAAVSLPGSTGHFTVTNGHGPLVTRLRAGPLAVFAANNNNPTT-FFVADGFLVLSPPQD 60
P +V++PGS G T+TNGH V RL+AG + V + FF++DGF++ P+D
Sbjct 61 PVRSVTVPGSEGAMTMTNGHSQTVARLKAGEIIVRKGETGDEVERFFLSDGFVLFKSPED 120
Query 61 DSNCTLAEVVATEIVPLELLDKEAAAQQLQQLLQEAAAATDQWTKARALLGQELLSAVIR 120
DS C AEV+ E+VP+ +LDKE+AA LQ+LLQ+ A ATD+WTKAR LLGQELLS+VIR
Sbjct 121 DSGCCTAEVLGVEVVPVSMLDKESAATALQELLQQGAGATDEWTKARTLLGQELLSSVIR 180
Query 121 AAP 123
AAP
Sbjct 181 AAP 183
> pfa:PF11_0485 mitochondrial ATP synthase delta subunit, putative
(EC:3.6.3.14); K02134 F-type H+-transporting ATPase subunit
delta [EC:3.6.3.14]
Length=154
Score = 101 bits (252), Expect = 6e-22, Method: Compositional matrix adjust.
Identities = 49/116 (42%), Positives = 69/116 (59%), Gaps = 1/116 (0%)
Query 7 SLPGSTGHFTVTNGHGPLVTRLRAGPLAVFAANNNNPTTFFVADGFLVLSPPQDDSNCTL 66
S PG G+FTVTN H PLVT LR G + V ++ FF++DG + D+++
Sbjct 39 SFPGIEGYFTVTNNHSPLVTLLRNGIITV-EFDDKEKKQFFISDGIFIYKKSNDNNSSNN 97
Query 67 AEVVATEIVPLELLDKEAAAQQLQQLLQEAAAATDQWTKARALLGQELLSAVIRAA 122
AE+V EIVPLE LDK + LQ++ A D+W K + LLG+EL S+++R A
Sbjct 98 AEIVGVEIVPLEYLDKNKTIKVLQEMCAINDATDDKWRKIKTLLGKELCSSILRVA 153
> bbo:BBOV_III004400 17.m07397; ATP synthase, Delta/Epsilon chain,
beta-sandwich domain containing protein; K02134 F-type
H+-transporting ATPase subunit delta [EC:3.6.3.14]
Length=154
Score = 94.0 bits (232), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 46/119 (38%), Positives = 73/119 (61%), Gaps = 3/119 (2%)
Query 3 AAAVSLPGSTGHFTVTNGHGPLVTRLRAGPLAVFAANNNNPTTFFVADGFLVLSPPQDDS 62
A ++PGS G+FTVT GH ++T L+ G ++V A + +F++ GF ++ +S
Sbjct 38 AKQATVPGSEGYFTVTKGHAAMLTVLKPGVVSVVAEETGEVSKYFLSSGFFKIAHVDGNS 97
Query 63 NCTLAEVVATEIVPLELLDKEAAAQQLQQLLQEAAAATDQWTKARALLGQELLSAVIRA 121
+AEV E VPL+ LDKE Q LQ+LL E + D W KA+ +LGQ+L +++++A
Sbjct 98 ---VAEVSGVEAVPLDHLDKERTTQVLQELLAEGHGSNDPWIKAKTMLGQDLCASILKA 153
> tpv:TP02_0253 ATP synthase F1 subunit delta (EC:3.6.3.14); K02134
F-type H+-transporting ATPase subunit delta [EC:3.6.3.14]
Length=164
Score = 89.0 bits (219), Expect = 3e-18, Method: Compositional matrix adjust.
Identities = 45/130 (34%), Positives = 72/130 (55%), Gaps = 13/130 (10%)
Query 2 PAAAVSLPGSTGHFTVTNGHGPLVTRLRAGPLAVFAANNNNPTTFFVADGFLVLSPPQDD 61
P ++++PG G+FTVT GH P+++ L+ G ++ ++N FF++ GF V ++
Sbjct 36 PVKSLTVPGYDGYFTVTPGHSPMLSTLKPGVVSFSLKDSNEVHKFFISSGFFVYRQSEES 95
Query 62 SNCTLAEVVATEIVPLELLDKEAAAQQLQQLLQEAAAATDQWTKARALLG---------- 111
AEV+ E+VPL+ LDKE A LQ++L E + QW K + L
Sbjct 96 HT---AEVMGVEVVPLDQLDKERATSVLQEVLTETQGDSSQWAKVKTFLTLVISNTNIHR 152
Query 112 QELLSAVIRA 121
Q+L S+VI+A
Sbjct 153 QDLCSSVIKA 162
> ath:AT5G47030 ATP synthase delta' chain, mitochondrial; K02134
F-type H+-transporting ATPase subunit delta [EC:3.6.3.14]
Length=203
Score = 56.6 bits (135), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 35/113 (30%), Positives = 58/113 (51%), Gaps = 8/113 (7%)
Query 6 VSLPGSTGHFTVTNGHGPLVTRLRAGPLAVFAANNNNPTTFFVADGFLVLSPPQDDSNCT 65
V +P STG V GH P + L+ G ++V + +F++ GF L +
Sbjct 93 VIIPASTGQMGVLPGHVPTIAELKPGIMSVHEGTDVK--KYFLSSGFAFL------HANS 144
Query 66 LAEVVATEIVPLELLDKEAAAQQLQQLLQEAAAATDQWTKARALLGQELLSAV 118
+A+++A E VPL+ +D + L + Q+ A+AT KA A +G E+ SA+
Sbjct 145 VADIIAVEAVPLDHIDPSQVQKGLAEFQQKLASATTDLEKAEAQIGVEVHSAI 197
> dre:335709 atp5d, fk58f09, wu:fk58f09, zgc:73303; ATP synthase,
H+ transporting, mitochondrial F1 complex, delta subunit
(EC:3.6.1.14); K02134 F-type H+-transporting ATPase subunit
delta [EC:3.6.3.14]
Length=159
Score = 51.2 bits (121), Expect = 8e-07, Method: Compositional matrix adjust.
Identities = 36/116 (31%), Positives = 61/116 (52%), Gaps = 7/116 (6%)
Query 6 VSLPGSTGHFTVTNGHGPLVTRLRAGPLAVFAANNNNPTTFFVADGFLVLSPPQDDSNCT 65
+ +P TG F + H P + LR G + VF ++ + +FV+ G + ++ DS+
Sbjct 49 IDVPTLTGAFGILPAHVPTLQVLRPGVVTVFN-DDGSSKKYFVSSGSVTVNA---DSSVQ 104
Query 66 LAEVVATEIVPLELLDKEAAAQQLQQLLQEAAAATDQWTKARALLGQELLSAVIRA 121
L +A E PLE LD AA L++ E +A+D+ T+A L+ E A+++A
Sbjct 105 L---LAEEAFPLESLDVAAAKANLEKAQSELVSASDEATRAEVLISIEANEAIVKA 157
> xla:447106 atp5d, MGC85306; ATP synthase, H+ transporting, mitochondrial
F1 complex, delta subunit (EC:3.6.1.14); K02134
F-type H+-transporting ATPase subunit delta [EC:3.6.3.14]
Length=162
Score = 49.7 bits (117), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 37/116 (31%), Positives = 61/116 (52%), Gaps = 7/116 (6%)
Query 6 VSLPGSTGHFTVTNGHGPLVTRLRAGPLAVFAANNNNPTTFFVADGFLVLSPPQDDSNCT 65
V +P TG F + H P + LR G + VF +++ T +FV+ G + ++ DS+
Sbjct 52 VDVPTLTGMFGILPAHVPTLQVLRPGLVTVF-SDDGVATKYFVSSGSVTVNA---DSSVQ 107
Query 66 LAEVVATEIVPLELLDKEAAAQQLQQLLQEAAAATDQWTKARALLGQELLSAVIRA 121
L +A E V L++LD A L++ E +A D+ KA AL+ E A+++A
Sbjct 108 L---LAEEAVTLDMLDLSTAKSNLEKAQAELQSAGDEAAKAEALINVEASEAIVKA 160
> hsa:513 ATP5D; ATP synthase, H+ transporting, mitochondrial
F1 complex, delta subunit (EC:3.6.1.14); K02134 F-type H+-transporting
ATPase subunit delta [EC:3.6.3.14]
Length=168
Score = 42.4 bits (98), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 35/116 (30%), Positives = 57/116 (49%), Gaps = 7/116 (6%)
Query 6 VSLPGSTGHFTVTNGHGPLVTRLRAGPLAVFAANNNNPTTFFVADGFLVLSPPQDDSNCT 65
V +P TG F + H P + LR G L V A + + +FV+ G + ++ DS+
Sbjct 58 VDVPTLTGAFGILAAHVPTLQVLRPG-LVVVHAEDGTTSKYFVSSGSIAVNA---DSSVQ 113
Query 66 LAEVVATEIVPLELLDKEAAAQQLQQLLQEAAAATDQWTKARALLGQELLSAVIRA 121
L +A E V L++LD AA L++ E D+ T+A + E A+++A
Sbjct 114 L---LAEEAVTLDMLDLGAAKANLEKAQAELVGTADEATRAEIQIRIEANEALVKA 166
> sce:YDL004W ATP16; Atp16p (EC:3.6.3.14); K02134 F-type H+-transporting
ATPase subunit delta [EC:3.6.3.14]
Length=160
Score = 40.4 bits (93), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 35/113 (30%), Positives = 52/113 (46%), Gaps = 15/113 (13%)
Query 6 VSLPGSTGHFTVTNGHGPLVTRLRAGPLAVFAANNNNPTTFFVADGFLVLSPPQDDSNCT 65
V+LP +G V H P V +L G + V +N+ FF++ GF + P D C
Sbjct 52 VNLPAKSGRIGVLANHVPTVEQLLPGVVEVMEGSNSK--KFFISGGFATVQP--DSQLC- 106
Query 66 LAEVVATEIVPLELLDKEAAAQQLQQLLQEA---AAATDQWTKARALLGQELL 115
V A E PLE +E ++ LL EA +++D A A + E+L
Sbjct 107 ---VTAIEAFPLESFSQE----NIKNLLAEAKKNVSSSDAREAAEAAIQVEVL 152
> dre:100332407 hypothetical protein LOC100332407
Length=756
Score = 32.0 bits (71), Expect = 0.57, Method: Compositional matrix adjust.
Identities = 35/107 (32%), Positives = 48/107 (44%), Gaps = 16/107 (14%)
Query 10 GSTGHFTVTNGHGPLVTRLRA--GPLAVFAANNNNPTTFFVADGFLVLSPPQDDSNCTLA 67
GS H V GPL T A P ++NNN+ T LVL+PP ++ T
Sbjct 46 GSPRHKLVKQPSGPLETAFSAFNPPKDAASSNNNSEAT----GPSLVLAPPSVNAQTT-- 99
Query 68 EVVATEIVPLELLDKEAAA----QQLQQLLQEAAAATDQWTKARALL 110
E +PLE K+ + QQ Q++ QEA A Q + R L+
Sbjct 100 ----KEQLPLETRIKQDKSNFHEQQRQKVAQEARAERRQKKQERELV 142
> xla:379656 stard3, MGC68989; StAR-related lipid transfer (START)
domain containing 3
Length=444
Score = 30.0 bits (66), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 20/72 (27%), Positives = 31/72 (43%), Gaps = 9/72 (12%)
Query 22 GPLVTRLRAGPLAVFAANNNNPTTFFVADGFLVLSPPQDDSNCTLAEVVATEI---VPLE 78
G T + PL + N P GFLVL P++ S CT V+ T++ +P
Sbjct 362 GISTTHISRPPLTKYVRGENGP------GGFLVLKYPKNPSMCTFIWVLNTDLKGRLPRS 415
Query 79 LLDKEAAAQQLQ 90
L+ + A +
Sbjct 416 LIHQSLGATMFE 427
> eco:b3731 atpC, ECK3724, JW3709, papG, uncC; F1 sector of membrane-bound
ATP synthase, epsilon subunit (EC:3.6.3.14); K02114
F-type H+-transporting ATPase subunit epsilon [EC:3.6.3.14]
Length=139
Score = 29.6 bits (65), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 12/52 (23%), Positives = 26/52 (50%), Gaps = 1/52 (1%)
Query 6 VSLPGSTGHFTVTNGHGPLVTRLRAGPLAVFAANNNNPTTFFVADGFLVLSP 57
+ + GS G + GH PL+T ++ G + + + + +++ G L + P
Sbjct 24 IQVTGSEGELGIYPGHAPLLTAIKPGMIRIV-KQHGHEEFIYLSGGILEVQP 74
> hsa:118427 OLFM3, NOE3, NOELIN3, NOELIN3_V1, NOELIN3_V2, NOELIN3_V3,
NOELIN3_V4, NOELIN3_V5, NOELIN3_V6, OPTIMEDIN; olfactomedin
3
Length=458
Score = 28.5 bits (62), Expect = 5.9, Method: Composition-based stats.
Identities = 19/60 (31%), Positives = 31/60 (51%), Gaps = 3/60 (5%)
Query 36 FAANNNNPTTFFVADGFLVLSPPQDDSNCTLAEVVATEIVPLELLDKEAAAQQLQQLLQE 95
FA + + T +G+ V S QD + VVA E L ++A ++QL+QLL++
Sbjct 16 FAGLDPSKTQISPKEGWQVYSSAQDPDGRCICTVVAPE---QNLCSRDAKSRQLRQLLEK 72
Lambda K H
0.316 0.129 0.367
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2069971060
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40