bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_0384_orf9
Length=539
Score E
Sequences producing significant alignments: (Bits) Value
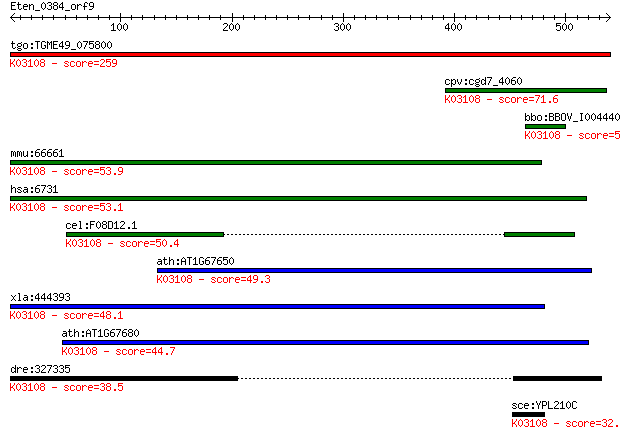
tgo:TGME49_075800 microneme protein, putative (EC:3.4.21.69); ... 259 2e-68
cpv:cgd7_4060 signal recognition particle SPR72 ; K03108 signa... 71.6 7e-12
bbo:BBOV_I004440 19.m02337; hypothetical protein; K03108 signa... 57.0 2e-07
mmu:66661 Srp72, 5730576P14Rik, 72kDa, AI132477, BC019196, C77... 53.9 2e-06
hsa:6731 SRP72; signal recognition particle 72kDa; K03108 sign... 53.1 2e-06
cel:F08D12.1 hypothetical protein; K03108 signal recognition p... 50.4 2e-05
ath:AT1G67650 7S RNA binding; K03108 signal recognition partic... 49.3 4e-05
xla:444393 srp72, MGC82921; signal recognition particle 72kDa;... 48.1 8e-05
ath:AT1G67680 7S RNA binding; K03108 signal recognition partic... 44.7 0.001
dre:327335 srp72, wu:fi03d11; signal recognition particle 72; ... 38.5 0.062
sce:YPL210C SRP72; Core component of the signal recognition pa... 32.0 7.0
> tgo:TGME49_075800 microneme protein, putative (EC:3.4.21.69);
K03108 signal recognition particle subunit SRP72
Length=1122
Score = 259 bits (661), Expect = 2e-68, Method: Compositional matrix adjust.
Identities = 208/558 (37%), Positives = 316/558 (56%), Gaps = 51/558 (9%)
Query 1 LPFNAAALAMQQQQPQAAAALLQRSLELCAAEAGVAVGDALRCIDSGELAGVLVQQAVLQ 60
LPFNAA + + + + A LLQ++ ELCA E GV +AL G+LAGVLVQ+A L+
Sbjct 596 LPFNAACVRLLEGRLDDADVLLQQARELCAEECGVEE-EAL---SQGDLAGVLVQEAFLR 651
Query 61 QQVGNAAAAEDIYSAVLAAVEQQQT-------IDAGVAAVAVTNAFAAALNRGQQQQQQE 113
G A +YS +L V +++T +D V VA TNAF R + +
Sbjct 652 DLQGRGEEASALYSRLLQKVAEERTAGEGPEEVDVAVLTVAQTNAFVHDARRRLTETGER 711
Query 114 EQQQQH------DLDALIRRATKGATETLERKLTVPQALGIGINRCIGLLKGGRVEECRR 167
+ LD I+R +G + L++KLT QAL + +NRC+ + GR+EE RR
Sbjct 712 SEDSSFARSGGCTLDDAIKRLARGTGQALDQKLTTAQALTLAVNRCVAFILAGRLEEARR 771
Query 168 VLASLTSRFGSS-VSLLRLKAAAQFAASRPTKADELLKLLLSSPALSSSSSSNTAAAAAT 226
+LA+ ++RFG L+ L+AA Q A+ RP KA+E L+ L SSSS ++ + +
Sbjct 772 LLAASSARFGPRQPKLILLRAALQLASGRPAKAEEQLR------HLLSSSSPSSPSVSLP 825
Query 227 AAQRLQLQICRLALLLQQGNRQGFRTLLRDVQQQLLQHLQQQQQQQQGSSREYLALLSAA 286
A+ +++ + AL L+QG+ G ++L + +L + ++ + R L+LL
Sbjct 826 PAEEIRVNLALAALRLEQGDSAGASSVLAAARARLQSMSEDCPEKSAATDR--LSLLRDL 883
Query 287 AELQQQAGDVEGVLASLDLQRRALEE-TQGSAAAY--VDLCLTAAEIASSLSRWEYAAAQ 343
+QQ+AGD EG + SL+ ++ A ++ + G + A V + LT+A ++L W
Sbjct 884 VAMQQRAGDAEGAIQSLEAEQTAWKDKSSGDSGARFAVSVLLTSASACAALGHWA----- 938
Query 344 SEAALQRLQQQQQQQQQEPAAAAAAAAAELRATLTLAEALARLNL--PSAQVYVHRLMRW 401
AA Q+ ++ Q Q A + LRA + A + + P A+ ++ L +
Sbjct 939 --AAGQKCREVLAQTQ-------ATSPEYLRALVGAVHASSFSSSSEPDAK-FLRELRQR 988
Query 402 LPMSLDLFDSEDVEKQLLPPSSRRAPAAAAAAAAAGKPAADKKKKRKKIKYPKGWDPSKP 461
+P + L DS++VEKQ + + R + +AG A+KKKK++K ++PKG+DP++P
Sbjct 989 IPQRILLLDSDEVEKQDVTACAPRK--TKESRPSAGDAEAEKKKKKRKPRWPKGFDPNEP 1046
Query 462 QMPPDPERWKPKHERSGYKKMLRKRKECLRGGAQGAVAPGIAEAVTGFRDSGPTTAKTKA 521
+PPDPERW PK ER+GYKKMLR+RKE RGGAQGAVAPG AEA TGFR++GP+TAKT+A
Sbjct 1047 HLPPDPERWLPKCERTGYKKMLRRRKEAGRGGAQGAVAPGTAEATTGFRNAGPSTAKTEA 1106
Query 522 ATDEGTMRAITRKKSGRR 539
A D+ +RKK G R
Sbjct 1107 AKDKNVG---SRKKRGTR 1121
> cpv:cgd7_4060 signal recognition particle SPR72 ; K03108 signal
recognition particle subunit SRP72
Length=680
Score = 71.6 bits (174), Expect = 7e-12, Method: Compositional matrix adjust.
Identities = 51/150 (34%), Positives = 80/150 (53%), Gaps = 10/150 (6%)
Query 392 QVYVHRLMRWLPMSLDLFDSEDVEKQLLPP--SSRRAPAAAAAAAAAGKPAADKKKKRKK 449
+VY+ +L + LP S+ D+E +EK P S + + + + + A K K++K
Sbjct 533 KVYLRQLDKKLPSSVFSIDAEVLEKMEAPTRASEKDSHDSKSMSHANLTIKTKSKHKKRK 592
Query 450 IKYPKGWDPSKPQMPPDPERWKPKHERSGYKKMLRKRK----ECLRGGAQGAVAPGIAEA 505
+YPKG+DP P PDPERW PK +RS +KK+ + R + +GG QGA+ EA
Sbjct 593 PRYPKGFDPLNPGQEPDPERWLPKEQRSSFKKLNKNRSSKKGQIGKGGHQGAIPTSNIEA 652
Query 506 VTGFRDSGPTTAKTKAATDEGTMRAITRKK 535
+ + P+TAK A + +R +KK
Sbjct 653 ----KPTVPSTAKQAALSSSSGIRRSHKKK 678
> bbo:BBOV_I004440 19.m02337; hypothetical protein; K03108 signal
recognition particle subunit SRP72
Length=713
Score = 57.0 bits (136), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 21/36 (58%), Positives = 30/36 (83%), Gaps = 0/36 (0%)
Query 464 PPDPERWKPKHERSGYKKMLRKRKECLRGGAQGAVA 499
PPDPERW PK+ERS +KK L+++KE ++G +QGA +
Sbjct 651 PPDPERWLPKYERSAFKKQLKRKKEMVKGHSQGATS 686
> mmu:66661 Srp72, 5730576P14Rik, 72kDa, AI132477, BC019196, C77589;
signal recognition particle 72; K03108 signal recognition
particle subunit SRP72
Length=671
Score = 53.9 bits (128), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 112/489 (22%), Positives = 186/489 (38%), Gaps = 94/489 (19%)
Query 1 LPFNAAALAMQQQQPQAAAALLQRSLELCAAEAGVAVGDALRCIDSGELAGVLVQQAVLQ 60
L +NAA + Q Q A +LQ++ +LC + D ELA + Q A +
Sbjct 178 LCYNAACALIGQGQLTQAMKILQKAEDLCR-RSFSEDSDGAEEDPQAELAIIHGQMAYIM 236
Query 61 QQVGNAAAAEDIYSAVLAAVEQQQTIDAGVAAVAVTNAFAAALNRGQQQQQQEEQQQQHD 120
Q G A +Y+ ++ + + D + AV N +N+ Q +
Sbjct 237 QLQGRTEEALQLYNQII----KLKPTDVALLAVIANNIIT--INKDQNVFDSK------- 283
Query 121 LDALIRRATKGATETLERKLTVPQALGIGINRCIGLLKGGRVEECRRVLASLTSRFGSSV 180
++ E +E KL+ Q I N+ + + + E+CR++ ASL S+ +
Sbjct 284 -----KKVKLTNAEGVEFKLSKRQLQAIEFNKALLAMYTNQAEQCRKIAASLQSQSPEYL 338
Query 181 SLLRLKAAAQFAASRPTKADELLKLLLSSPALSSSSSSNTAAAAATAAQRLQLQICRLAL 240
+ ++AA + TKA ELL+ S N A T AQ L
Sbjct 339 LPVLIQAAQLCREKQHTKAIELLQ------EFSDQHPENAAEIKLTMAQ----------L 382
Query 241 LLQQGNRQGFRTLLRDVQQQLLQHLQQQQQQQQGSSREYLALLSAAAELQQQAGDVEGVL 300
+ QGN +LR +++ L+H ++SA + D++ +
Sbjct 383 KISQGNISKACLILRSIEE--LRHKP--------------GMVSALVTMYSHEEDIDSAI 426
Query 301 ASLDLQRRALEETQGSAAAYVDLCLTAAEIASSLSRWEYAAAQSEAALQRLQQQQQQQQQ 360
+ + Q + A++ L AA R + A+ L+Q +Q +
Sbjct 427 EVFTQAIQWYQSHQPKSPAHLSLIREAANFKLKYGR-------KKEAVSDLEQLWKQNSK 479
Query 361 EPAAAAAAAAAELRATLTLAEALARLNLPSAQVYVHRLMRWLPMSLDLFDSEDVEKQLLP 420
+ A +A A+AL++ +LPS+ MSL + DVE
Sbjct 480 DIHTLAQLISAYSLVDPEKAKALSK-HLPSSD----------SMSLKV----DVE----- 519
Query 421 PSSRRAPAAAAAAAAAGKPAADKKKKRK------------KIKYPKGWDPSKPQMPPDPE 468
+ +P A GK D + K + K K PK +DP ++ PDPE
Sbjct 520 -ALENSPGATYIRKKGGKVTGDNQPKEQGQGDLKKKKKKKKGKLPKNYDP---KVTPDPE 575
Query 469 RWKPKHERS 477
RW P ERS
Sbjct 576 RWLPMRERS 584
> hsa:6731 SRP72; signal recognition particle 72kDa; K03108 signal
recognition particle subunit SRP72
Length=671
Score = 53.1 bits (126), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 124/530 (23%), Positives = 202/530 (38%), Gaps = 95/530 (17%)
Query 1 LPFNAAALAMQQQQPQAAAALLQRSLELCAAEAGVAVGDALRCIDSGELAGVLVQQAVLQ 60
L +N A + Q Q A +LQ++ +LC D ELA + Q A +
Sbjct 178 LCYNTACALIGQGQLNQAMKILQKAEDLCRRSLSEDT-DGTEEDPQAELAIIHGQMAYIL 236
Query 61 QQVGNAAAAEDIYSAVLAAVEQQQTIDAGVAAVAVTNAFAAALNRGQQQQQQEEQQQQHD 120
Q G A +Y+ ++ + + D G+ AV N +N+ Q +
Sbjct 237 QLQGRTEEALQLYNQII----KLKPTDVGLLAVIANNIIT--INKDQNVFDSK------- 283
Query 121 LDALIRRATKGATETLERKLTVPQALGIGINRCIGLLKGGRVEECRRVLASLTSRFGSSV 180
++ E +E KL+ Q I N+ + + + E+CR++ ASL S+ +
Sbjct 284 -----KKVKLTNAEGVEFKLSKKQLQAIEFNKALLAMYTNQAEQCRKISASLQSQSPEHL 338
Query 181 SLLRLKAAAQFAASRPTKADELLKLLLSSPALSSSSSSNTAAAAATAAQRLQLQICRLAL 240
+ ++AA + TKA ELL+ S N A T AQ L
Sbjct 339 LPVLIQAAQLCREKQHTKAIELLQ------EFSDQHPENAAEIKLTMAQ----------L 382
Query 241 LLQQGNRQGFRTLLRDVQQQLLQHLQQQQQQQQGSSREYLALLSAAAELQQQAGDVEGVL 300
+ QGN +LR +++ L+H ++SA + D++ +
Sbjct 383 KISQGNISKACLILRSIEE--LKHKP--------------GMVSALVTMYSHEEDIDSAI 426
Query 301 ASLDLQRRALEETQGSAAAYVDLCLTAAEIASSLSRWEYAAAQSEAALQRLQQQQQQQQQ 360
+ + Q + A++ L AA R + A+ LQQ +Q +
Sbjct 427 EVFTQAIQWYQNHQPKSPAHLSLIREAANFKLKYGR-------KKEAISDLQQLWKQNPK 479
Query 361 EPAAAAAAAAAELRATLTLAEALARLNLPSAQVYVHRLMRWLPMSLDLFDSEDVEKQLLP 420
+ A +A A+AL++ +LPS+ MSL + DVE
Sbjct 480 DIHTLAQLISAYSLVDPEKAKALSK-HLPSSD----------SMSLKV----DVE----- 519
Query 421 PSSRRAPAAAAAAAAAGKPAADKKKKRK------------KIKYPKGWDPSKPQMPPDPE 468
+ + A GK D + K + K K PK +DP ++ PDPE
Sbjct 520 -ALENSAGATYIRKKGGKVTGDSQPKEQGQGDLKKKKKKKKGKLPKNYDP---KVTPDPE 575
Query 469 RWKPKHERSGYKKMLRKRKECLRG-GAQGAVAPGIAEAVTGFRDSGPTTA 517
RW P ERS Y+ + +K+ G G QGA A +E S P T+
Sbjct 576 RWLPMRERSYYRGRKKGKKKDQIGKGTQGATAGASSELDASKTVSSPPTS 625
> cel:F08D12.1 hypothetical protein; K03108 signal recognition
particle subunit SRP72
Length=635
Score = 50.4 bits (119), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 25/65 (38%), Positives = 41/65 (63%), Gaps = 6/65 (9%)
Query 445 KKRKKIKYPKGWDPSKPQMPPDPERWKPKHERSGYKKMLRKRKECLRGGAQGAVA--PGI 502
K+++KI+ PK ++ + + PDPERW P+ ERS YK+ + R+ + G QG+ + P +
Sbjct 539 KRKRKIRLPKNYNSA---VTPDPERWLPRQERSTYKRKRKNREREIGRGTQGSSSANPNV 595
Query 503 AEAVT 507
E VT
Sbjct 596 -EYVT 599
Score = 45.1 bits (105), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 40/141 (28%), Positives = 62/141 (43%), Gaps = 19/141 (13%)
Query 51 GVLVQQAVLQQQVGNAAAAEDIYSAVLAAVEQQQTIDAGVAAVAVTNAFAAALNRGQQQQ 110
+ VQ+A + Q++G A A IY V AA D+ V A N AA+ + +
Sbjct 221 SIRVQKAYVLQRMGQKAEALAIYEKVQAA----NHPDSSVKATITNNIPAASSDFALPES 276
Query 111 QQEEQQQQHDLDALIRRATKGATETLERKLTVPQALGIGINRCIGLLKGGRVEECRRVLA 170
R+ K A + + KLT Q L + +N + LL + E C+R L
Sbjct 277 ---------------RKRFKAALQIDQTKLTRRQRLTLMLNNALVLLLSNQREPCKRALE 321
Query 171 SLTSRFGSSVSLLRLKAAAQF 191
L ++FGSS + ++A F
Sbjct 322 ELVAKFGSSKDVALIEATLHF 342
> ath:AT1G67650 7S RNA binding; K03108 signal recognition particle
subunit SRP72
Length=651
Score = 49.3 bits (116), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 97/401 (24%), Positives = 169/401 (42%), Gaps = 85/401 (21%)
Query 133 TETLERKLTVPQALGIGINRCIGLLKGGRVEECRRVLASLTSRFGSSVSLLRLKAAAQFA 192
++ L+ KL+ I NR + LL ++++ R + A+L F SV L+AA
Sbjct 303 SQELDAKLSHKHKEAIYANRVLLLLHANKMDQARELCATLPGMFPESVIPTLLQAAVLVR 362
Query 193 ASRPTKADELL-----------KLLLSSPALSSSSSSNTAAAAATAAQRLQLQICRLALL 241
++ KA+ELL KL+L + A ++S+S+ AA + ++
Sbjct 363 ENKAAKAEELLGQCAENFPEKSKLVLLARAQIAASASHPHVAAESLSK------------ 410
Query 242 LQQGNRQGFRTLLRDVQQQLLQHLQQQQQQQQGSSREYLALLSAAAELQQQAGDVEGVLA 301
+ D+Q HL A ++ L+++AGD +G A
Sbjct 411 ------------IPDIQ-----HLP--------------ATVATIVALRERAGDNDGATA 439
Query 302 SLDLQRRALEETQGSAAAYVDLCLTAAEIASSLSRWEYAAAQSEAALQRLQQQQQQQQQE 361
LD SA + +T + + L AA +L+ Q+++
Sbjct 440 VLD-----------SAIRWWSDSMTDSNMLRIL--------MPVAAAFKLRHGQEEEASR 480
Query 362 PAAAAAAAAAELRATLTLAEALARLNLPSAQVYVHRLMRWLPMSLDLFDSEDVEKQLLPP 421
A + L LAR+N+ A+ Y + ++ LP L D +++EK
Sbjct 481 LYEEIVKNHNSTDALVGLVTTLARVNVEKAEAY-EKQLKPLP-GLKAVDVDNLEKT---S 535
Query 422 SSRRAPAAAAAAAAAGKPAADKKKKRKKIKYPKGWDPSKPQMPPDPERWKPKHERSGYKK 481
++ +AA+ + + +K K+++K KYPKG+D PDPERW P+ ERS Y+
Sbjct 536 GAKPIEGISAASLSQEEVKKEKVKRKRKPKYPKGFDLENSGPTPDPERWLPRRERSSYRP 595
Query 482 MLRKRKECLRGGAQGAVAPGIAEAVTGFRDSGPTTAKTKAA 522
+ ++ G+QGA+ E P+T+K+K A
Sbjct 596 KRKDKRAAQIRGSQGALTKVKQE-------EAPSTSKSKQA 629
> xla:444393 srp72, MGC82921; signal recognition particle 72kDa;
K03108 signal recognition particle subunit SRP72
Length=673
Score = 48.1 bits (113), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 109/484 (22%), Positives = 189/484 (39%), Gaps = 78/484 (16%)
Query 1 LPFNAAALAMQQQQPQAAAALLQRSLELC----AAEAGVAVGDALRCIDSGELAGVLVQQ 56
L +N+A + Q Q + L+ + ELC + +A + D ELA + Q
Sbjct 179 LCYNSACTLIGQGQLREGMVKLREAEELCRQSISEDADLTEEDI-----EAELAIIHGQM 233
Query 57 AVLQQQVGNAAAAEDIYSAVLAAVEQQQTIDAGVAAVAVTNAFAAALNRGQQQQQQEEQQ 116
A + Q GN A +Y+ ++ + + D G+ AV N +
Sbjct 234 AYVMQLQGNTDNALQLYNQII----KLKPTDVGLLAVVANNIITV-------------NK 276
Query 117 QQHDLDALIRRATKGATETLERKLTVPQALGIGINRCIGLLKGGRVEECRRVLASLTSRF 176
Q+ D+ + A E +E KL Q I N+ + + + ++CR++ ASL S+
Sbjct 277 DQNVFDSKKKVKLTNA-EGVEHKLCKKQLQAIEFNKALLSMYTNQADQCRKLSASLQSQD 335
Query 177 GSSVSLLRLKAAAQFAASRPTKADELLKLLLSSPALSSSSSSNTAAAAATAAQRLQLQIC 236
+ + ++AA + + TKA ELL+ + N A T AQ
Sbjct 336 PEDLLPVLIQAAQLYKEKQHTKAVELLQ------EFAEQHPDNAAQIKLTMAQ------- 382
Query 237 RLALLLQQGNRQGFRTLLRDVQQQLLQHLQQQQQQQQGSSREYLALLSAAAELQQQAGDV 296
L L QGN +L+ +++ LQH ++SA + D+
Sbjct 383 ---LKLAQGNVTKACMILKSIKE--LQHTP--------------GMVSALVTMHSHDEDI 423
Query 297 EGVLASLDLQRRALEETQGSAAAYVDLCLTAAEIASSLSRWEYAAAQSEAALQRLQQQQQ 356
+ + +E Q ++ ++ L AA ++ + + A+ L+Q +
Sbjct 424 DSAIEVFSNAISWYQENQPTSPLHLSLVREAA-------NFKLKHGRKKDAISDLEQLWR 476
Query 357 QQQQEPAAAAAAAAAELRATLTLAEALARLNLPSAQVYVHRLMRWLPMSLDLFDSEDVEK 416
Q ++ A +A A+ L++ +LPS+ M L + D + +E
Sbjct 477 QNPKDIHTLAQLISAYSLVDAEKAKILSK-HLPSSDT----------MKLKV-DVDALEN 524
Query 417 QLLPPSSRRAPAAAAAAAAAGKPAADKKKKRKKIKYPKGWDPSKPQMPPDPERWKPKHER 476
R+ A A A + D KKK+ K K K P++ PD ERW P ER
Sbjct 525 STGATFIRKKAAKVAGEQAKEQGPTDIKKKKTKKKKGKLPKHYDPKVTPDAERWLPMRER 584
Query 477 SGYK 480
S Y+
Sbjct 585 SYYR 588
> ath:AT1G67680 7S RNA binding; K03108 signal recognition particle
subunit SRP72
Length=664
Score = 44.7 bits (104), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 123/491 (25%), Positives = 204/491 (41%), Gaps = 109/491 (22%)
Query 48 ELAGVLVQQAVLQQQVGNAAAAEDIYSAVLAAVEQQQTIDAGVAAVAVTNAFAAALNRGQ 107
ELA + VQ A +QQ +G + Y + ++ D AVAV N A +G
Sbjct 240 ELAPIAVQLAYVQQILGQTQESTSSYVDFI----KRNLADEPSLAVAVNNLVAL---KGF 292
Query 108 QQQQQEEQQQQHDLDALIRRATKGATETLERKLTVPQALGIGINRCIGLLKGGRVEECRR 167
+ + ++ DL T ++ L+ KL+ I NR + LL ++++ R
Sbjct 293 KDIS--DGLRKFDLLKDKDSQTFQLSQALDAKLSQKHKEAIYANRVLLLLHANKMDQARE 350
Query 168 VLASLTSRFGSSVSLLRLKAAAQFAASRPTKADELL-----------KLLLSSPALSSSS 216
+ A+L F S+ L+AA ++ KA+ELL KL+L + A ++S
Sbjct 351 LCAALPGMFPESIIPTLLQAAVLVRENKAAKAEELLGQCAEKFPEKSKLVLLARAQIAAS 410
Query 217 SSNTAAAAATAAQRLQLQICRLALLLQQGNRQGFRTLLRDVQQQLLQHLQQQQQQQQGSS 276
+S+ AA + ++ + D+Q HL
Sbjct 411 ASHPHVAAESLSK------------------------IPDIQ-----HLP---------- 431
Query 277 REYLALLSAAAELQQQAGDVEGVLASLDLQRRALEETQGSAAAYVDLCLTAAEIASSLSR 336
A ++ L+++AGD +G A LD S++
Sbjct 432 ----ATVATIVALKERAGDNDGAAAVLD---------------------------SAIKW 460
Query 337 WEYAAAQS--------EAALQRLQQQQQQQQQEPAAAAAAAAAELRATLTLAEALARLNL 388
W + +S EAA +L+ Q+++ A + L LAR+N+
Sbjct 461 WSNSMTESSKLRVLMPEAAAFKLRHGQEEEASRLYEEIVKNHNSTDALVGLVTTLARVNV 520
Query 389 PSAQVYVHRLMRWLPMSLDLFDSEDVEKQLLPPSSRRAPAAAAAAAAAGKPAADKKKKRK 448
A+ Y + ++ LP L D + +EK + AAA+++ K K+K+++
Sbjct 521 EKAESY-EKQLKPLP-GLKAVDVDKLEKTY--GAKPIEGAAASSSQEEVKKEKAKRKRKR 576
Query 449 KIKYPKGWDPSKPQMPPDPERWKPKHERSGYKKMLRKRKECLRGGAQGAVAPGIAEAVTG 508
K KYPKG+DP+ P PPDPERW P+ ERS YK + ++ G+QGAV EA
Sbjct 577 KPKYPKGFDPANPGPPPDPERWLPRRERSSYKPKRKDKRAAQIRGSQGAVTKDKQEA--- 633
Query 509 FRDSGPTTAKT 519
P+T+K+
Sbjct 634 ----APSTSKS 640
> dre:327335 srp72, wu:fi03d11; signal recognition particle 72;
K03108 signal recognition particle subunit SRP72
Length=555
Score = 38.5 bits (88), Expect = 0.062, Method: Compositional matrix adjust.
Identities = 31/85 (36%), Positives = 41/85 (48%), Gaps = 9/85 (10%)
Query 453 PKGWDPSKPQMPPDPERWKPKHERSGYK-KMLRKRKECLRGGAQGAVAPGIAEAVTGFRD 511
PK +DP+ PDPERW P ERS Y+ K K+KE + G QG + AE
Sbjct 445 PKNYDPNSS---PDPERWLPMRERSYYRGKKKGKKKEQVGRGTQGTTSGATAELDASKTA 501
Query 512 SGPTTAK-----TKAATDEGTMRAI 531
+ P T+ T A + G+ AI
Sbjct 502 TSPPTSPRPGSGTGAPSSSGSSTAI 526
Score = 36.6 bits (83), Expect = 0.28, Method: Compositional matrix adjust.
Identities = 53/208 (25%), Positives = 92/208 (44%), Gaps = 27/208 (12%)
Query 1 LPFNAAALAMQQQQPQAAAALLQRSLELC----AAEAGVAVGDALRCIDSGELAGVLVQQ 56
L +NAA + Q + A LQ++ ELC A ++ + D ID+ ELA + Q
Sbjct 60 LCYNAACCLIGQGELGQAMKKLQKAEELCRKSLAEDSDMTEED----IDA-ELAVIHSQM 114
Query 57 AVLQQQVGNAAAAEDIYSAVLAAVEQQQTIDAGVAAVAVTNAFAAALNRGQQQQQQEEQQ 116
A + Q G A +YS V+ + + D G+ AV N +N+ Q
Sbjct 115 AYVIQLQGRTEDALQLYSQVI----KLKPSDVGLLAVTANNIIT--INKDQNV------- 161
Query 117 QQHDLDALIRRATKGATETLERKLTVPQALGIGINRCIGLLKGGRVEECRRVLASLTSRF 176
D+ + A E +E KL Q I IN+ + + + ++C ++L+SL ++
Sbjct 162 ----FDSKKKVKLTNA-EGVEHKLAKKQLQAIEINKALLAMYTNQADQCNKLLSSLQTQN 216
Query 177 GSSVSLLRLKAAAQFAASRPTKADELLK 204
+ + ++ A + +KA ELL+
Sbjct 217 PTHPRPVLIQVAQLCREKQHSKAIELLQ 244
> sce:YPL210C SRP72; Core component of the signal recognition
particle (SRP) ribonucleoprotein (RNP) complex that functions
in targeting nascent secretory proteins to the endoplasmic
reticulum (ER) membrane; K03108 signal recognition particle
subunit SRP72
Length=640
Score = 32.0 bits (71), Expect = 7.0, Method: Compositional matrix adjust.
Identities = 14/29 (48%), Positives = 18/29 (62%), Gaps = 3/29 (10%)
Query 452 YPKGWDPSKPQMPPDPERWKPKHERSGYK 480
+ +G D SK PDPERW P +RS Y+
Sbjct 569 FLQGRDTSKL---PDPERWLPLRDRSTYR 594
Lambda K H
0.314 0.125 0.338
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 26380222060
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40