bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_0377_orf2
Length=211
Score E
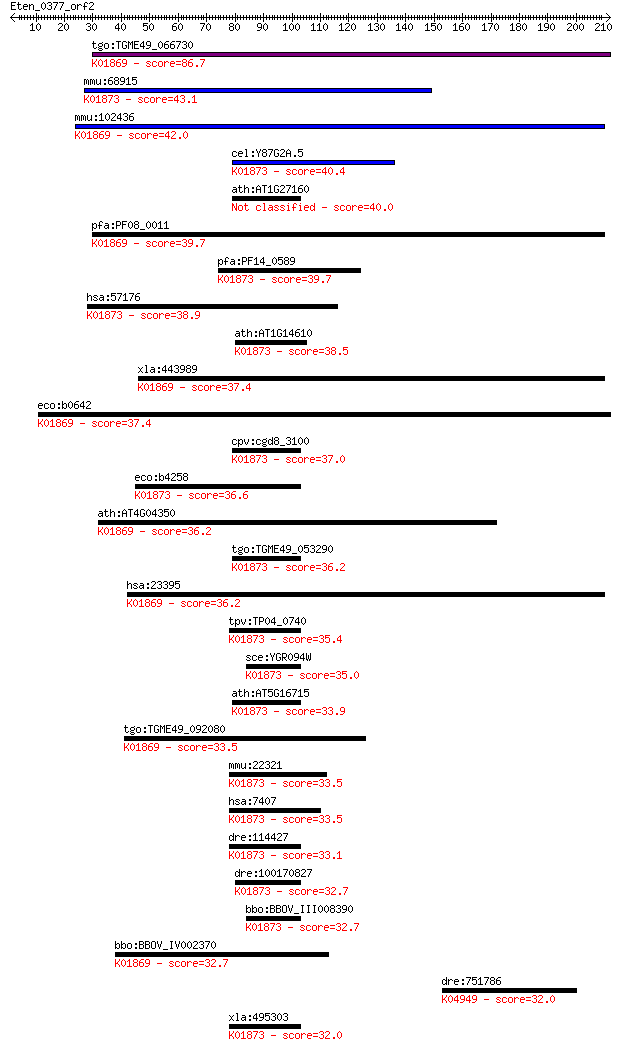
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_066730 leucyl-tRNA synthetase, putative (EC:6.1.1.4... 86.7 5e-17
mmu:68915 Vars2, 1190004I24Rik, Vars2l, mKIAA1885; valyl-tRNA ... 43.1 7e-04
mmu:102436 Lars2, AI035546, Kiaa0028, LEURS; leucyl-tRNA synth... 42.0 0.002
cel:Y87G2A.5 vrs-2; Valyl tRNA Synthetase family member (vrs-2... 40.4 0.004
ath:AT1G27160 valyl-tRNA synthetase / valine--tRNA ligase-related 40.0 0.007
pfa:PF08_0011 leucine-tRNA ligase; K01869 leucyl-tRNA syntheta... 39.7 0.007
pfa:PF14_0589 valine-tRNA ligase, putative; K01873 valyl-tRNA ... 39.7 0.009
hsa:57176 VARS2, MGC138259, MGC142165, VARS2L, VARSL; valyl-tR... 38.9 0.014
ath:AT1G14610 TWN2; TWN2 (TWIN 2); ATP binding / aminoacyl-tRN... 38.5 0.018
xla:443989 lars2, MGC80460; leucyl-tRNA synthetase 2, mitochon... 37.4 0.034
eco:b0642 leuS, ECK0635, JW0637; leucyl-tRNA synthetase (EC:6.... 37.4 0.039
cpv:cgd8_3100 valyl-tRNA synthetase ; K01873 valyl-tRNA synthe... 37.0 0.047
eco:b4258 valS, ECK4251, JW4215; valyl-tRNA synthetase (EC:6.1... 36.6 0.071
ath:AT4G04350 EMB2369 (EMBRYO DEFECTIVE 2369); ATP binding / a... 36.2 0.078
tgo:TGME49_053290 valyl-tRNA synthetase, putative (EC:6.1.1.9)... 36.2 0.078
hsa:23395 LARS2, KIAA0028, LEURS, MGC26121; leucyl-tRNA synthe... 36.2 0.089
tpv:TP04_0740 valyl-tRNA synthetase (EC:6.1.1.9); K01873 valyl... 35.4 0.15
sce:YGR094W VAS1; Mitochondrial and cytoplasmic valyl-tRNA syn... 35.0 0.17
ath:AT5G16715 EMB2247 (embryo defective 2247); ATP binding / a... 33.9 0.39
tgo:TGME49_092080 leucyl-tRNA synthetase, putative (EC:6.1.1.4... 33.5 0.53
mmu:22321 Vars, Bat6, D17H6S56E, G7a, Vars2; valyl-tRNA synthe... 33.5 0.59
hsa:7407 VARS, G7A, VARS1, VARS2; valyl-tRNA synthetase (EC:6.... 33.5 0.59
dre:114427 vars, CHUNP6879, im:7137378, vars1, vars2, wu:fb07a... 33.1 0.73
dre:100170827 vars2, si:ch211-152p11.2; valyl-tRNA synthetase ... 32.7 0.82
bbo:BBOV_III008390 17.m07734; valyl-tRNA synthetase (EC:6.1.1.... 32.7 0.93
bbo:BBOV_IV002370 21.m02840; leucyl-tRNA synthetase (EC:6.1.1.... 32.7 1.0
dre:751786 cnga5; cyclic nucleotide gated channel alpha 5; K04... 32.0 1.6
xla:495303 vars, vars2; valyl-tRNA synthetase (EC:6.1.1.9); K0... 32.0 1.6
> tgo:TGME49_066730 leucyl-tRNA synthetase, putative (EC:6.1.1.4);
K01869 leucyl-tRNA synthetase [EC:6.1.1.4]
Length=1465
Score = 86.7 bits (213), Expect = 5e-17, Method: Compositional matrix adjust.
Identities = 60/193 (31%), Positives = 95/193 (49%), Gaps = 15/193 (7%)
Query 30 ERKLLAPLVANVTRSLERMQVNRAVAALMAGSRHLQQQQQRQGGTLSVSCVKTLLTLLHP 89
ER+L+ LVA VT +ERM +N A+AALMA R L + G LSV L+ LLHP
Sbjct 1256 ERELMKLLVARVTADIERMSLNTAIAALMANVRKLTAWTAK-GNRLSVGTAIDLVKLLHP 1314
Query 90 VAPFITEELWKRLAAEYPAAFAFPGW-TLAASGEWPTTEGEE----------FEKEKTAE 138
APF+TEELW+ + A +P + +L AS WPT EE E E A+
Sbjct 1315 FAPFVTEELWQMIHASFPDVSLYSTTPSLLASATWPTPSKEESRVHPRSGRVAESENEAD 1374
Query 139 NSRKVQVSVQFNGRHRLSVPLSVTEMSTPSDLFETVRRFSLVQQRLQLENDRGKALSRVI 198
+ + S+ +G+ + +V + + M + +++ L G+ ++++
Sbjct 1375 S---IVFSIHVDGKLQGTVHVPRSSMHDEGTVSALATTSPCLKKWEILLKSGGRGFTKIV 1431
Query 199 SKPEACLVNFIFG 211
P ++NF+ G
Sbjct 1432 CIPARRVINFVTG 1444
> mmu:68915 Vars2, 1190004I24Rik, Vars2l, mKIAA1885; valyl-tRNA
synthetase 2, mitochondrial (putative) (EC:6.1.1.9); K01873
valyl-tRNA synthetase [EC:6.1.1.9]
Length=1060
Score = 43.1 bits (100), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 39/128 (30%), Positives = 56/128 (43%), Gaps = 13/128 (10%)
Query 27 SEGERKLLAPLVANVTRSLERMQVNRAVAALMAGSRHLQQQQQRQGGTLSV--SCVKTLL 84
SE ER L+ ++ VT +L ++ + + + R G V SC L
Sbjct 792 SECERGFLSRELSLVTHTLYHFWLHNLCDVYLEAVKPVLSSVPRPPGPPQVLFSCADVGL 851
Query 85 TLLHPVAPFITEELWKRLAAEYPAAFAFPGWTLAAS---GEWPTTEGEEFEKEKTAENS- 140
LL P+ PF+ EELW+RL PG LA S +P+T EF ++ E
Sbjct 852 RLLAPLMPFLAEELWQRLPPR-------PGGPLAPSICVAPYPSTRSLEFWRQPELERCF 904
Query 141 RKVQVSVQ 148
+VQ VQ
Sbjct 905 SRVQEVVQ 912
> mmu:102436 Lars2, AI035546, Kiaa0028, LEURS; leucyl-tRNA synthetase,
mitochondrial (EC:6.1.1.4); K01869 leucyl-tRNA synthetase
[EC:6.1.1.4]
Length=902
Score = 42.0 bits (97), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 44/189 (23%), Positives = 75/189 (39%), Gaps = 17/189 (8%)
Query 24 KNMSEGERKLLAPLVANVTRSLERMQVNRAVAALMAGSRHLQQQQQR--QGGTLSVSCVK 81
+N+ E + ++A + + T E +N V+ LM S L Q QR +
Sbjct 725 QNLWEYKNAVIAQVTTHFT---EDFALNSVVSQLMGLSSALSQASQRVVLHSPEFEDALC 781
Query 82 TLLTLLHPVAPFITEELWKRLAAEYPAAFAFPGWTLAASGE-WPTTEGEEFEKEKTAENS 140
LL + P+AP +T ELW L W + WPT + + +K
Sbjct 782 ALLVMAAPLAPHVTSELWAGLTLVPSKLCDHYAWDSGVMLQAWPTVDSQFLQKPDM---- 837
Query 141 RKVQVSVQFNGRHRLSVPLSVTEMSTPSDLFETVRRFSLVQQRLQLENDRGKALSRVISK 200
VQ++V N + +P+ + E V + L + LQ G+++ +
Sbjct 838 --VQMAVLINNKACGKIPVPQHVAQDQDKVHELVLQSELGMKLLQ-----GRSIKKAFLS 890
Query 201 PEACLVNFI 209
P L+NF+
Sbjct 891 PRTALINFL 899
> cel:Y87G2A.5 vrs-2; Valyl tRNA Synthetase family member (vrs-2);
K01873 valyl-tRNA synthetase [EC:6.1.1.9]
Length=1050
Score = 40.4 bits (93), Expect = 0.004, Method: Composition-based stats.
Identities = 20/58 (34%), Positives = 35/58 (60%), Gaps = 4/58 (6%)
Query 79 CVKTLLTLLHPVAPFITEELWKRLAAEYPAAFAFPGWTLAASGEWPTTEG-EEFEKEK 135
C+ T L L+ P+ PFI+EELW+R+ + + P +A ++P T+ E+++ EK
Sbjct 833 CIDTGLRLISPLMPFISEELWQRMPRLDDSDYTSPSIIVA---QYPLTQKYEKYQNEK 887
> ath:AT1G27160 valyl-tRNA synthetase / valine--tRNA ligase-related
Length=200
Score = 40.0 bits (92), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 16/24 (66%), Positives = 20/24 (83%), Gaps = 0/24 (0%)
Query 79 CVKTLLTLLHPVAPFITEELWKRL 102
C++T L LLHP P+ITEELW+RL
Sbjct 48 CLETGLRLLHPFMPYITEELWQRL 71
> pfa:PF08_0011 leucine-tRNA ligase; K01869 leucyl-tRNA synthetase
[EC:6.1.1.4]
Length=1481
Score = 39.7 bits (91), Expect = 0.007, Method: Composition-based stats.
Identities = 37/185 (20%), Positives = 82/185 (44%), Gaps = 24/185 (12%)
Query 30 ERKLLAPLVANVTRSLERMQVNRAVAALMAGSRHLQQQQQRQGGTLSVSCVKTLLTLLHP 89
++K + + +T + +++N AV+ M ++ + + + LL+P
Sbjct 1314 KKKKVNYYIEKITNCINDIKLNTAVSFFMKFYNEIKTWD-----IVPLKIFIIFVKLLYP 1368
Query 90 VAPFITEELW-----KRLAAEYPAAFAFPGWTLAASGEWPTTEGEEFEKEKTAENSRKVQ 144
P I EE W + + F L G+WP+ FE + ++ V
Sbjct 1369 FCPHICEEFWFYYLKRYKIKKRKKLCYFCNSNLLYYGKWPSL----FE----IKQNKLVN 1420
Query 145 VSVQFNGRHRLSVPLSVTEMSTPSDLFETVRRFSLVQQRLQLENDRGKALSRVISKPEAC 204
+S++ N +H + ++S+ SD+ E +L++ +++ E +GK + +++ P
Sbjct 1421 ISIKLNNKH---ITFLQRDVSSTSDITEEAT--NLIKHKIENEMKKGKKIVNIVNIPNK- 1474
Query 205 LVNFI 209
++NFI
Sbjct 1475 VINFI 1479
> pfa:PF14_0589 valine-tRNA ligase, putative; K01873 valyl-tRNA
synthetase [EC:6.1.1.9]
Length=1090
Score = 39.7 bits (91), Expect = 0.009, Method: Composition-based stats.
Identities = 21/50 (42%), Positives = 33/50 (66%), Gaps = 4/50 (8%)
Query 74 TLSVSCVKTLLTLLHPVAPFITEELWKRLAAEYPAAFAFPGWTLAASGEW 123
TL V C+ L LLHP++PFITEEL++++++E + F +LA E+
Sbjct 865 TLHV-CLDYGLRLLHPISPFITEELYQKISSE---PYKFNSISLANYPEY 910
> hsa:57176 VARS2, MGC138259, MGC142165, VARS2L, VARSL; valyl-tRNA
synthetase 2, mitochondrial (putative) (EC:6.1.1.9); K01873
valyl-tRNA synthetase [EC:6.1.1.9]
Length=923
Score = 38.9 bits (89), Expect = 0.014, Method: Compositional matrix adjust.
Identities = 29/103 (28%), Positives = 42/103 (40%), Gaps = 15/103 (14%)
Query 28 EGERKLLAPLVANVTRSLERMQVNRAVAALMAGSRHLQQQQQRQGGTLSV--SCVKTLLT 85
E ER L ++ VT +L ++ + + + R G V SC L
Sbjct 652 ECERGFLTRELSLVTHALHHFWLHNLCDVYLEAVKPVLWHSPRPLGPPQVLFSCADLGLR 711
Query 86 LLHPVAPFITEELWKRL-------------AAEYPAAFAFPGW 115
LL P+ PF+ EELW+RL A YP+A + W
Sbjct 712 LLAPLMPFLAEELWQRLPPRPGCPPAPSISVAPYPSACSLEHW 754
> ath:AT1G14610 TWN2; TWN2 (TWIN 2); ATP binding / aminoacyl-tRNA
ligase/ nucleotide binding / valine-tRNA ligase (EC:6.1.1.9);
K01873 valyl-tRNA synthetase [EC:6.1.1.9]
Length=1108
Score = 38.5 bits (88), Expect = 0.018, Method: Compositional matrix adjust.
Identities = 16/25 (64%), Positives = 20/25 (80%), Gaps = 0/25 (0%)
Query 80 VKTLLTLLHPVAPFITEELWKRLAA 104
++T L LLHP PF+TEELW+RL A
Sbjct 888 LETGLRLLHPFMPFVTEELWQRLPA 912
> xla:443989 lars2, MGC80460; leucyl-tRNA synthetase 2, mitochondrial
(EC:6.1.1.4); K01869 leucyl-tRNA synthetase [EC:6.1.1.4]
Length=900
Score = 37.4 bits (85), Expect = 0.034, Method: Compositional matrix adjust.
Identities = 37/173 (21%), Positives = 72/173 (41%), Gaps = 26/173 (15%)
Query 46 ERMQVNRAVAALMAGSRHLQQQQQRQ--GGTLSVSCVKTLLTLLHPVAPFITEELWKRLA 103
E N +++ LM+ S L Q R + + +L ++ P+AP + ELWK ++
Sbjct 742 EDYLFNASISQLMSLSNTLSQASSRVILHSSEFEDALASLCIMVAPMAPHLASELWKGMS 801
Query 104 -AEYPAAFAFPGWTLAASGEWPTTEGEEFEKEKTAENSRKVQVSVQFNGRHRLSVPLSVT 162
++ + + +WP+ + + + E VQV + + VP
Sbjct 802 HVQFKLCTQYKWDEDVLAQQWPSVDPDYLRQPDIVE----VQVLINNKTFGTVGVP---- 853
Query 163 EMSTPSDLFETVRRFSLVQQRLQLEND------RGKALSRVISKPEACLVNFI 209
+ V R S V Q + L++D +G+ +++V P L+NF+
Sbjct 854 ---------QEVSRDSKVVQEMVLQSDLGVTHLQGRTITKVFLSPRTALINFL 897
> eco:b0642 leuS, ECK0635, JW0637; leucyl-tRNA synthetase (EC:6.1.1.4);
K01869 leucyl-tRNA synthetase [EC:6.1.1.4]
Length=860
Score = 37.4 bits (85), Expect = 0.039, Method: Composition-based stats.
Identities = 50/207 (24%), Positives = 85/207 (41%), Gaps = 31/207 (14%)
Query 11 EAVYAGRLRGVSMKNMSEGERKL---LAPLVANVTRSLERMQV-NRAVAALMAGSRHLQQ 66
E G + +++ ++E ++ L + +A VT + R Q N A+AA+M L +
Sbjct 679 EHTAKGDVAALNVDALTENQKALRRDVHKTIAKVTDDIGRRQTFNTAIAAIMELMNKLAK 738
Query 67 QQQ--RQGGTLSVSCVKTLLTLLHPVAPFITEELWKRLAAEYPAAFAFPGWTLAASGEWP 124
Q L + ++ +L+P P I LW+ L E A WP
Sbjct 739 APTDGEQDRALMQEALLAVVRMLNPFTPHICFTLWQELKGEGDIDNA----------PWP 788
Query 125 TTEGEEFEKEKTAENSRKVQVSVQFNGRHRLSVPLSVTEMSTPSDLFETVRRFSLVQQRL 184
+++ E+S V V V R +++VP+ TE E VR + Q+ L
Sbjct 789 VA-----DEKAMVEDSTLVVVQVNGKVRAKITVPVDATE--------EQVRERA-GQEHL 834
Query 185 QLENDRGKALSRVISKPEACLVNFIFG 211
+ G + +VI P L+N + G
Sbjct 835 VAKYLDGVTVRKVIYVPGK-LLNLVVG 860
> cpv:cgd8_3100 valyl-tRNA synthetase ; K01873 valyl-tRNA synthetase
[EC:6.1.1.9]
Length=1042
Score = 37.0 bits (84), Expect = 0.047, Method: Composition-based stats.
Identities = 13/24 (54%), Positives = 19/24 (79%), Gaps = 0/24 (0%)
Query 79 CVKTLLTLLHPVAPFITEELWKRL 102
C++ L LLHP+ PF+TEEL++R
Sbjct 817 CIEYGLVLLHPLTPFVTEELYQRF 840
> eco:b4258 valS, ECK4251, JW4215; valyl-tRNA synthetase (EC:6.1.1.9);
K01873 valyl-tRNA synthetase [EC:6.1.1.9]
Length=951
Score = 36.6 bits (83), Expect = 0.071, Method: Composition-based stats.
Identities = 21/58 (36%), Positives = 32/58 (55%), Gaps = 11/58 (18%)
Query 45 LERMQVNRAVAALMAGSRHLQQQQQRQGGTLSVSCVKTLLTLLHPVAPFITEELWKRL 102
L + +N A + G+RH TL V+ ++ LL L HP+ PFITE +W+R+
Sbjct 719 LTKPVMNGGTEAELRGTRH----------TL-VTVLEGLLRLAHPIIPFITETIWQRV 765
> ath:AT4G04350 EMB2369 (EMBRYO DEFECTIVE 2369); ATP binding /
aminoacyl-tRNA ligase/ leucine-tRNA ligase/ nucleotide binding
(EC:6.1.1.4); K01869 leucyl-tRNA synthetase [EC:6.1.1.4]
Length=973
Score = 36.2 bits (82), Expect = 0.078, Method: Composition-based stats.
Identities = 33/140 (23%), Positives = 58/140 (41%), Gaps = 24/140 (17%)
Query 32 KLLAPLVANVTRSLERMQVNRAVAALMAGSRHLQQQQQRQGGTLSVSCVKTLLTLLHPVA 91
+ L +A VT +E + N ++ +M + + G ++ + LL P A
Sbjct 815 RTLHKCIAKVTEEIESTRFNTGISGMMEFVNAAYKWNNQPRGI-----IEPFVLLLSPYA 869
Query 92 PFITEELWKRLAAEYPAAFAFPGWTLAASGEWPTTEGEEFEKEKTAENSRKVQVSVQFNG 151
P + EELW RL +P + A+ + A ++ K T + + VQ NG
Sbjct 870 PHMAEELWSRLG--HPNSLAYESFPKA---------NPDYLKNTT------IVLPVQING 912
Query 152 RHRLSVPLSVTEMSTPSDLF 171
+ R ++ V E + D F
Sbjct 913 KTRGTI--EVEEGCSEDDAF 930
> tgo:TGME49_053290 valyl-tRNA synthetase, putative (EC:6.1.1.9);
K01873 valyl-tRNA synthetase [EC:6.1.1.9]
Length=1042
Score = 36.2 bits (82), Expect = 0.078, Method: Compositional matrix adjust.
Identities = 15/24 (62%), Positives = 19/24 (79%), Gaps = 0/24 (0%)
Query 79 CVKTLLTLLHPVAPFITEELWKRL 102
C+ L LLHP+ PFITEEL++RL
Sbjct 818 CLDRGLRLLHPLCPFITEELFQRL 841
> hsa:23395 LARS2, KIAA0028, LEURS, MGC26121; leucyl-tRNA synthetase
2, mitochondrial (EC:6.1.1.4); K01869 leucyl-tRNA synthetase
[EC:6.1.1.4]
Length=903
Score = 36.2 bits (82), Expect = 0.089, Method: Compositional matrix adjust.
Identities = 43/179 (24%), Positives = 72/179 (40%), Gaps = 30/179 (16%)
Query 42 TRSLERMQVNRAVAALMAGSRHLQQQQQRQGGTLSV--------SCVKTLLTLLHPVAPF 93
T E +N A++ LM S L Q Q SV + L+ + P+AP
Sbjct 741 THFTEDFSLNSAISQLMGLSNALSQASQ------SVILHSPEFEDALCALMVMAAPLAPH 794
Query 94 ITEELWKRLAAEYPAAFAFPGWTLAASGE-WPTTEGEEFEKEKTAENSRKVQVSVQFNGR 152
+T E+W LA A W + + WP + E ++ + VQ++V N +
Sbjct 795 VTSEIWAGLALVPRKLCAHYTWDASVLLQAWPAVDPEFLQQPEV------VQMAVLINNK 848
Query 153 H--RLSVPLSVTEMSTPSDLFETVRRFSLVQQRLQLENDRGKALSRVISKPEACLVNFI 209
++ VP V + V F L Q L + +G+++ + P L+NF+
Sbjct 849 ACGKIPVPQQVARDQ------DKVHEFVL-QSELGVRLLQGRSIKKSFLSPRTALINFL 900
> tpv:TP04_0740 valyl-tRNA synthetase (EC:6.1.1.9); K01873 valyl-tRNA
synthetase [EC:6.1.1.9]
Length=1010
Score = 35.4 bits (80), Expect = 0.15, Method: Composition-based stats.
Identities = 14/25 (56%), Positives = 17/25 (68%), Gaps = 0/25 (0%)
Query 78 SCVKTLLTLLHPVAPFITEELWKRL 102
+C L LLHP+ PFITEEL+ L
Sbjct 794 TCFSESLKLLHPIMPFITEELYHHL 818
> sce:YGR094W VAS1; Mitochondrial and cytoplasmic valyl-tRNA synthetase
(EC:6.1.1.9); K01873 valyl-tRNA synthetase [EC:6.1.1.9]
Length=1104
Score = 35.0 bits (79), Expect = 0.17, Method: Composition-based stats.
Identities = 12/19 (63%), Positives = 16/19 (84%), Gaps = 0/19 (0%)
Query 84 LTLLHPVAPFITEELWKRL 102
L L+HP PFI+EE+W+RL
Sbjct 897 LKLIHPFMPFISEEMWQRL 915
> ath:AT5G16715 EMB2247 (embryo defective 2247); ATP binding /
aminoacyl-tRNA ligase/ nucleotide binding / valine-tRNA ligase
(EC:6.1.1.9); K01873 valyl-tRNA synthetase [EC:6.1.1.9]
Length=970
Score = 33.9 bits (76), Expect = 0.39, Method: Compositional matrix adjust.
Identities = 12/24 (50%), Positives = 17/24 (70%), Gaps = 0/24 (0%)
Query 79 CVKTLLTLLHPVAPFITEELWKRL 102
+ +L LLHP PF+TE+LW+ L
Sbjct 757 VFENILKLLHPFMPFVTEDLWQAL 780
> tgo:TGME49_092080 leucyl-tRNA synthetase, putative (EC:6.1.1.4);
K01869 leucyl-tRNA synthetase [EC:6.1.1.4]
Length=1162
Score = 33.5 bits (75), Expect = 0.53, Method: Composition-based stats.
Identities = 22/85 (25%), Positives = 34/85 (40%), Gaps = 16/85 (18%)
Query 41 VTRSLERMQVNRAVAALMAGSRHLQQQQQRQGGTLSVSCVKTLLTLLHPVAPFITEELWK 100
+ + L M R L+ G H+ + + V+ +KT L P+AP I E +W
Sbjct 897 LKKGLYEMHTRRDQYRLLCGEDHMHKD-------MVVTWLKTQCQTLAPIAPHICEHIWS 949
Query 101 RLAAEYPAAFAFPGWTLAASGEWPT 125
+ E +L S WPT
Sbjct 950 EILKEP---------SLIVSSAWPT 965
> mmu:22321 Vars, Bat6, D17H6S56E, G7a, Vars2; valyl-tRNA synthetase
(EC:6.1.1.9); K01873 valyl-tRNA synthetase [EC:6.1.1.9]
Length=1263
Score = 33.5 bits (75), Expect = 0.59, Method: Compositional matrix adjust.
Identities = 16/34 (47%), Positives = 21/34 (61%), Gaps = 0/34 (0%)
Query 78 SCVKTLLTLLHPVAPFITEELWKRLAAEYPAAFA 111
+C+ L LL P PF+TEEL++RL P A A
Sbjct 1050 TCLDVGLRLLSPFMPFVTEELFQRLPRRTPKAPA 1083
> hsa:7407 VARS, G7A, VARS1, VARS2; valyl-tRNA synthetase (EC:6.1.1.9);
K01873 valyl-tRNA synthetase [EC:6.1.1.9]
Length=1264
Score = 33.5 bits (75), Expect = 0.59, Method: Compositional matrix adjust.
Identities = 15/32 (46%), Positives = 20/32 (62%), Gaps = 0/32 (0%)
Query 78 SCVKTLLTLLHPVAPFITEELWKRLAAEYPAA 109
+C+ L LL P PF+TEEL++RL P A
Sbjct 1051 TCLDVGLRLLSPFMPFVTEELFQRLPRRMPQA 1082
> dre:114427 vars, CHUNP6879, im:7137378, vars1, vars2, wu:fb07a11,
wu:fb07d10; valyl-tRNA synthetase; K01873 valyl-tRNA synthetase
[EC:6.1.1.9]
Length=1111
Score = 33.1 bits (74), Expect = 0.73, Method: Composition-based stats.
Identities = 12/25 (48%), Positives = 20/25 (80%), Gaps = 0/25 (0%)
Query 78 SCVKTLLTLLHPVAPFITEELWKRL 102
+C++ L LL P+ PF++EEL++RL
Sbjct 897 TCLEVGLRLLSPIMPFVSEELFQRL 921
> dre:100170827 vars2, si:ch211-152p11.2; valyl-tRNA synthetase
2, mitochondrial (putative) (EC:6.1.1.9); K01873 valyl-tRNA
synthetase [EC:6.1.1.9]
Length=1057
Score = 32.7 bits (73), Expect = 0.82, Method: Compositional matrix adjust.
Identities = 14/23 (60%), Positives = 16/23 (69%), Gaps = 0/23 (0%)
Query 80 VKTLLTLLHPVAPFITEELWKRL 102
V L LL P PF+TEELW+RL
Sbjct 839 VSVSLALLSPFMPFLTEELWQRL 861
> bbo:BBOV_III008390 17.m07734; valyl-tRNA synthetase (EC:6.1.1.9);
K01873 valyl-tRNA synthetase [EC:6.1.1.9]
Length=972
Score = 32.7 bits (73), Expect = 0.93, Method: Composition-based stats.
Identities = 13/19 (68%), Positives = 15/19 (78%), Gaps = 0/19 (0%)
Query 84 LTLLHPVAPFITEELWKRL 102
L LLHP+ PFITEEL+ L
Sbjct 766 LRLLHPIMPFITEELYHHL 784
> bbo:BBOV_IV002370 21.m02840; leucyl-tRNA synthetase (EC:6.1.1.4);
K01869 leucyl-tRNA synthetase [EC:6.1.1.4]
Length=782
Score = 32.7 bits (73), Expect = 1.0, Method: Composition-based stats.
Identities = 21/75 (28%), Positives = 33/75 (44%), Gaps = 2/75 (2%)
Query 38 VANVTRSLERMQVNRAVAALMAGSRHLQQQQQRQGGTLSVSCVKTLLTLLHPVAPFITEE 97
VA +TR++E N A+A ++ G + T + LL P AP I E+
Sbjct 704 VARITRAIEEYSFNVAIADFFKLFDYIYDIS--NGRAIDRGVGGTYIKLLAPFAPHIAED 761
Query 98 LWKRLAAEYPAAFAF 112
LW L + + A+
Sbjct 762 LWHILYPDRTGSVAY 776
> dre:751786 cnga5; cyclic nucleotide gated channel alpha 5; K04949
cyclic nucleotide gated channel alpha 2
Length=673
Score = 32.0 bits (71), Expect = 1.6, Method: Composition-based stats.
Identities = 23/60 (38%), Positives = 30/60 (50%), Gaps = 14/60 (23%)
Query 153 HRLSVPLSVTEMSTPSDLF-------------ETVRRFSLVQQRLQLENDRGKALSRVIS 199
HRLSV S+ + S P+D E R +L QQRL DRG ALSR+++
Sbjct 15 HRLSVKTSIEDESEPADCTFSRDQSICDDTSSELQRVATLEQQRLNSFRDRG-ALSRLVN 73
> xla:495303 vars, vars2; valyl-tRNA synthetase (EC:6.1.1.9);
K01873 valyl-tRNA synthetase [EC:6.1.1.9]
Length=1243
Score = 32.0 bits (71), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 13/25 (52%), Positives = 18/25 (72%), Gaps = 0/25 (0%)
Query 78 SCVKTLLTLLHPVAPFITEELWKRL 102
+C+ L LL P PF+TEEL++RL
Sbjct 1030 TCLDVGLRLLSPFMPFLTEELYQRL 1054
Lambda K H
0.316 0.130 0.372
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 6623499460
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40