bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_0361_orf2
Length=257
Score E
Sequences producing significant alignments: (Bits) Value
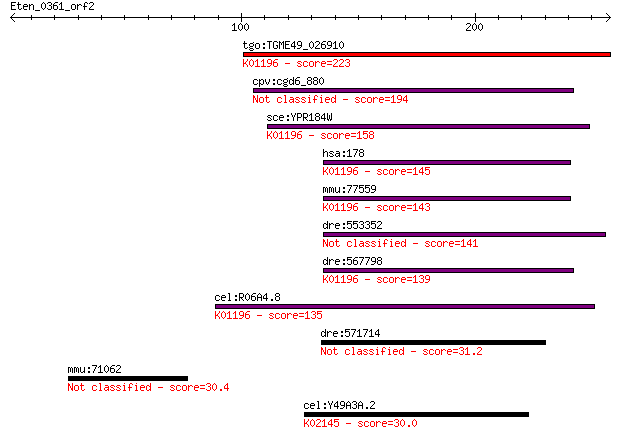
tgo:TGME49_026910 glycogen debranching enzyme, putative (EC:3.... 223 6e-58
cpv:cgd6_880 Gdb1p; glycogen debranching enzyme 194 2e-49
sce:YPR184W GDB1; Gdb1p (EC:3.2.1.33 2.4.1.25); K01196 glycoge... 158 2e-38
hsa:178 AGL, GDE; amylo-alpha-1, 6-glucosidase, 4-alpha-glucan... 145 1e-34
mmu:77559 Agl, 1110061O17Rik, 9430004C13Rik, 9630046L06Rik, AI... 143 6e-34
dre:553352 im:7145503 141 2e-33
dre:567798 agl; amylo-1, 6-glucosidase, 4-alpha-glucanotransfe... 139 8e-33
cel:R06A4.8 hypothetical protein; K01196 glycogen debranching ... 135 2e-31
dre:571714 C11orf66 homolog 31.2 3.7
mmu:71062 Tekt3, 4933407G07Rik; tektin 3 30.4
cel:Y49A3A.2 vha-13; Vacuolar H ATPase family member (vha-13);... 30.0 7.8
> tgo:TGME49_026910 glycogen debranching enzyme, putative (EC:3.2.1.33);
K01196 glycogen debranching enzyme [EC:2.4.1.25 3.2.1.33]
Length=1882
Score = 223 bits (567), Expect = 6e-58, Method: Compositional matrix adjust.
Identities = 107/157 (68%), Positives = 124/157 (78%), Gaps = 0/157 (0%)
Query 101 AAAADPLLLHLALASLQCYGAVASAPLIWGSNDPSLAAGLPHFATGFMRNWGRDTFIAIR 160
AD LL L +A+LQ Y V SAPLI G+ PSL+AGLP F++GFMR WGRDTFIA+R
Sbjct 1365 GVVADKLLAELQVATLQFYSFVPSAPLISGTTRPSLSAGLPFFSSGFMRAWGRDTFIALR 1424
Query 161 GILIATGRYAEARREILGYARVLRHGLIPNLLDSGNNPRYNARDATWFFMQAIQDYCLAA 220
GIL+ TGRY A+ EILG+ARV+RHGLIPNLLDSGNNPRYNARDATWFF QAIQDY L A
Sbjct 1425 GILLTTGRYEAAKAEILGFARVMRHGLIPNLLDSGNNPRYNARDATWFFCQAIQDYSLCA 1484
Query 221 PEGLDFLLCPVEMKYGKNASQLPEEEIRNIGDVMHHI 257
P+GL L PVEMKY NA+Q P+ I+N+ DV+HHI
Sbjct 1485 PDGLQILKEPVEMKYEGNATQAPDLPIKNLADVLHHI 1521
> cpv:cgd6_880 Gdb1p; glycogen debranching enzyme
Length=1891
Score = 194 bits (493), Expect = 2e-49, Method: Composition-based stats.
Identities = 84/137 (61%), Positives = 105/137 (76%), Gaps = 0/137 (0%)
Query 105 DPLLLHLALASLQCYGAVASAPLIWGSNDPSLAAGLPHFATGFMRNWGRDTFIAIRGILI 164
DPL +HL LA+ Q Y V S+PLIW S PS++AGLPHF+TGFMR+WGRDTFIA GILI
Sbjct 1380 DPLYIHLQLATYQVYSYVPSSPLIWNSKIPSISAGLPHFSTGFMRSWGRDTFIAFNGILI 1439
Query 165 ATGRYAEARREILGYARVLRHGLIPNLLDSGNNPRYNARDATWFFMQAIQDYCLAAPEGL 224
++ RY EA++EILG AR++RHGLIPNL+DSGN PRYNARDATWFF+ A DYC+ P G
Sbjct 1440 SSKRYLEAKQEILGVARLMRHGLIPNLIDSGNRPRYNARDATWFFLNAALDYCVNIPNGY 1499
Query 225 DFLLCPVEMKYGKNASQ 241
L +E+++ N +
Sbjct 1500 QILNEDIELRFTLNLEE 1516
> sce:YPR184W GDB1; Gdb1p (EC:3.2.1.33 2.4.1.25); K01196 glycogen
debranching enzyme [EC:2.4.1.25 3.2.1.33]
Length=1536
Score = 158 bits (399), Expect = 2e-38, Method: Composition-based stats.
Identities = 68/138 (49%), Positives = 97/138 (70%), Gaps = 0/138 (0%)
Query 111 LALASLQCYGAVASAPLIWGSNDPSLAAGLPHFATGFMRNWGRDTFIAIRGILIATGRYA 170
L++ S+Q + S ++ G N PS+AAGLPHF+ +MR WGRD FI++RG+L+ TGR+
Sbjct 1043 LSMTSIQMVSRMKSTSILPGENVPSMAAGLPHFSVNYMRCWGRDVFISLRGMLLTTGRFD 1102
Query 171 EARREILGYARVLRHGLIPNLLDSGNNPRYNARDATWFFMQAIQDYCLAAPEGLDFLLCP 230
EA+ IL +A+ L+HGLIPNLLD+G NPRYNARDA WFF+QA+QDY P+G L
Sbjct 1103 EAKAHILAFAKTLKHGLIPNLLDAGRNPRYNARDAAWFFLQAVQDYVYIVPDGEKILQEQ 1162
Query 231 VEMKYGKNASQLPEEEIR 248
V ++ + + +P ++ R
Sbjct 1163 VTRRFPLDDTYIPVDDPR 1180
> hsa:178 AGL, GDE; amylo-alpha-1, 6-glucosidase, 4-alpha-glucanotransferase
(EC:2.4.1.25 3.2.1.33); K01196 glycogen debranching
enzyme [EC:2.4.1.25 3.2.1.33]
Length=1532
Score = 145 bits (367), Expect = 1e-34, Method: Composition-based stats.
Identities = 67/106 (63%), Positives = 76/106 (71%), Gaps = 0/106 (0%)
Query 135 SLAAGLPHFATGFMRNWGRDTFIAIRGILIATGRYAEARREILGYARVLRHGLIPNLLDS 194
SLAAGLPHF++G R WGRDTFIA+RGIL+ TGRY EAR IL +A LRHGLIPNLL
Sbjct 1083 SLAAGLPHFSSGIFRCWGRDTFIALRGILLITGRYVEARNIILAFAGTLRHGLIPNLLGE 1142
Query 195 GNNPRYNARDATWFFMQAIQDYCLAAPEGLDFLLCPVEMKYGKNAS 240
G RYN RDA W+++Q IQDYC P GLD L CPV Y + S
Sbjct 1143 GIYARYNCRDAVWWWLQCIQDYCKMVPNGLDILKCPVSRMYPTDDS 1188
> mmu:77559 Agl, 1110061O17Rik, 9430004C13Rik, 9630046L06Rik,
AI850929, C77197; amylo-1,6-glucosidase, 4-alpha-glucanotransferase
(EC:3.2.1.33); K01196 glycogen debranching enzyme [EC:2.4.1.25
3.2.1.33]
Length=1532
Score = 143 bits (361), Expect = 6e-34, Method: Compositional matrix adjust.
Identities = 66/106 (62%), Positives = 76/106 (71%), Gaps = 0/106 (0%)
Query 135 SLAAGLPHFATGFMRNWGRDTFIAIRGILIATGRYAEARREILGYARVLRHGLIPNLLDS 194
SLAAGLPHF++G R WGRDTFIA+RG+L+ TGRY EAR IL +A LRHGLIPNLL
Sbjct 1083 SLAAGLPHFSSGLFRCWGRDTFIALRGMLLVTGRYLEARNIILAFASTLRHGLIPNLLGE 1142
Query 195 GNNPRYNARDATWFFMQAIQDYCLAAPEGLDFLLCPVEMKYGKNAS 240
G RYN RDA W+++Q IQDYC P GLD L CPV Y + S
Sbjct 1143 GTYARYNCRDAVWWWLQCIQDYCRTVPNGLDILKCPVSRMYPTDDS 1188
> dre:553352 im:7145503
Length=996
Score = 141 bits (356), Expect = 2e-33, Method: Composition-based stats.
Identities = 66/121 (54%), Positives = 82/121 (67%), Gaps = 5/121 (4%)
Query 135 SLAAGLPHFATGFMRNWGRDTFIAIRGILIATGRYAEARREILGYARVLRHGLIPNLLDS 194
S+AAGLPHF+TG R WGRDTFIA+RG+++ TGR+ EAR IL +A +RHGLIPNLL
Sbjct 545 SIAAGLPHFSTGIFRCWGRDTFIALRGLMLLTGRHLEARDIILAFAGTMRHGLIPNLLGQ 604
Query 195 GNNPRYNARDATWFFMQAIQDYCLAAPEGLDFLLCPVEMKYGKNASQLPEEEIRNIGDVM 254
G RYN RDA W+++Q IQDYC P G D L+CPV Y + S E + +G V
Sbjct 605 GVAARYNCRDAVWWWLQCIQDYCTIVPIGTDILMCPVSRMYPTDDS-----EAQALGTVA 659
Query 255 H 255
H
Sbjct 660 H 660
> dre:567798 agl; amylo-1, 6-glucosidase, 4-alpha-glucanotransferase;
K01196 glycogen debranching enzyme [EC:2.4.1.25 3.2.1.33]
Length=1532
Score = 139 bits (351), Expect = 8e-33, Method: Composition-based stats.
Identities = 62/107 (57%), Positives = 75/107 (70%), Gaps = 0/107 (0%)
Query 135 SLAAGLPHFATGFMRNWGRDTFIAIRGILIATGRYAEARREILGYARVLRHGLIPNLLDS 194
S+AAGLPHF+ G R WGRDTFIA+RG+L+ TGR+ EAR IL +A LRHGLIPNLL
Sbjct 1083 SMAAGLPHFSAGIFRCWGRDTFIALRGLLLVTGRHLEARNIILAFAGTLRHGLIPNLLGE 1142
Query 195 GNNPRYNARDATWFFMQAIQDYCLAAPEGLDFLLCPVEMKYGKNASQ 241
G R+N RDA W+++Q IQDYC P G D L CPV Y + S+
Sbjct 1143 GVGARFNCRDAVWWWLQCIQDYCTMVPNGTDILQCPVSRMYPTDDSK 1189
> cel:R06A4.8 hypothetical protein; K01196 glycogen debranching
enzyme [EC:2.4.1.25 3.2.1.33]
Length=1467
Score = 135 bits (340), Expect = 2e-31, Method: Composition-based stats.
Identities = 73/170 (42%), Positives = 99/170 (58%), Gaps = 8/170 (4%)
Query 89 LRGQTAAAAAAAAAAADPLLLHLALASLQCYGAVASAPL--------IWGSNDPSLAAGL 140
+R + A +++ L+ HLA+++L G + A L I SLAAGL
Sbjct 974 IRKEALKRMAPQISSSSALVRHLAISTLSFLGYIPGAGLAPIPTSLQIEDQYPSSLAAGL 1033
Query 141 PHFATGFMRNWGRDTFIAIRGILIATGRYAEARREILGYARVLRHGLIPNLLDSGNNPRY 200
HFA G RNWGRDTFIA+ G L++TGR+ EAR+ IL +A +RHGLIPNLL G RY
Sbjct 1034 SHFAVGIWRNWGRDTFIALPGCLLSTGRFQEARQIILSFAGAIRHGLIPNLLAEGIGARY 1093
Query 201 NARDATWFFMQAIQDYCLAAPEGLDFLLCPVEMKYGKNASQLPEEEIRNI 250
N RDATWF++ +I Y +AP G+ L PV Y + S E E++ +
Sbjct 1094 NCRDATWFWLVSIVKYVESAPNGVGILEDPVRRIYPNDDSVYGEGEVQQM 1143
> dre:571714 C11orf66 homolog
Length=331
Score = 31.2 bits (69), Expect = 3.7, Method: Compositional matrix adjust.
Identities = 27/107 (25%), Positives = 45/107 (42%), Gaps = 21/107 (19%)
Query 134 PSLAAGLP----------HFATGFMRNWGRDTFIAIRGILIATGRYAEARREILGYARVL 183
PS G P H+ T + +++G++ F G TG Y+ R +L Y+
Sbjct 10 PSFGTGRPAGRPANISIDHYCTSYRQSYGKELFQPCLGYHHGTG-YSANHRPVLYYS--- 65
Query 184 RHGLIPNLLDSGNNPRYNARDATWFFMQAIQDYC-LAAPEGLDFLLC 229
+ LD +NP++ F Q+ + Y L P+G + L C
Sbjct 66 ------SRLDDYDNPQFGFSLLDSFESQSKRHYQRLVQPDGTEPLAC 106
> mmu:71062 Tekt3, 4933407G07Rik; tektin 3
Length=490
Score = 30.4 bits (67), Expect = 6.4, Method: Compositional matrix adjust.
Identities = 20/51 (39%), Positives = 26/51 (50%), Gaps = 3/51 (5%)
Query 26 GQTLARVYIHRPPRKPSAARKLFRLSGQSDFLSPRRPPRSLQHTLVLPWGP 76
G TL Y H PP P++A L + + R P R+L H+L LPW P
Sbjct 5 GSTLTATYAH-PP--PASASFLPAIGTITSSYKDRFPHRNLTHSLSLPWRP 52
> cel:Y49A3A.2 vha-13; Vacuolar H ATPase family member (vha-13);
K02145 V-type H+-transporting ATPase subunit A [EC:3.6.3.14]
Length=606
Score = 30.0 bits (66), Expect = 7.8, Method: Compositional matrix adjust.
Identities = 31/97 (31%), Positives = 48/97 (49%), Gaps = 9/97 (9%)
Query 127 LIWGSNDPSLAAGLPHFATGF-MRNWGRDTFIAIRGILIATGRYAEARREILGYARVLRH 185
L+ +++ +AA TG + + RD + + + +T R+AEA REI G R
Sbjct 300 LVANTSNMPVAAREASIYTGITLAEYFRDMGLNVAMMADSTSRWAEALREISG-----RL 354
Query 186 GLIPNLLDSGNNPRYNARDATWFFMQAIQDYCLAAPE 222
G +P DSG P Y A F+ +A + CL +PE
Sbjct 355 GEMP--ADSG-YPAYLAARLASFYERAGRVKCLGSPE 388
Lambda K H
0.325 0.139 0.445
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 9404593716
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40