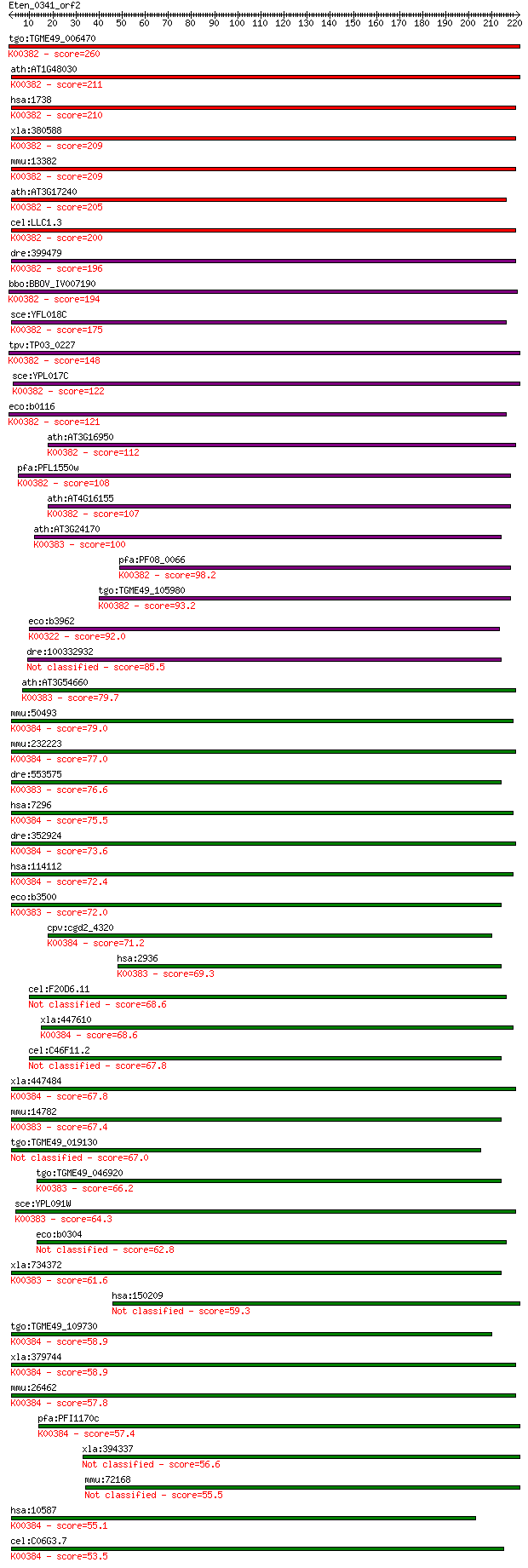
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_0341_orf2
Length=221
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_006470 dihydrolipoyl dehydrogenase, putative (EC:1.... 260 2e-69
ath:AT1G48030 mtLPD1; mtLPD1 (mitochondrial lipoamide dehydrog... 211 1e-54
hsa:1738 DLD, DLDH, E3, GCSL, LAD, PHE3; dihydrolipoamide dehy... 210 3e-54
xla:380588 dld, MGC68940; dihydrolipoamide dehydrogenase (EC:1... 209 7e-54
mmu:13382 Dld, AI315664, AI746344; dihydrolipoamide dehydrogen... 209 8e-54
ath:AT3G17240 mtLPD2; mtLPD2 (LIPOAMIDE DEHYDROGENASE 2); ATP ... 205 9e-53
cel:LLC1.3 hypothetical protein; K00382 dihydrolipoamide dehyd... 200 4e-51
dre:399479 dldh, wu:fb24b05; dihydrolipoamide dehydrogenase (E... 196 4e-50
bbo:BBOV_IV007190 23.m05858; dihydrolipoamide dehydrogenase (E... 194 2e-49
sce:YFL018C LPD1, HPD1; Dihydrolipoamide dehydrogenase, the li... 175 1e-43
tpv:TP03_0227 dihydrolipoamide dehydrogenase (EC:1.8.1.4); K00... 148 2e-35
sce:YPL017C IRC15; Irc15p; K00382 dihydrolipoamide dehydrogena... 122 9e-28
eco:b0116 lpd, dhl, ECK0115, JW0112, lpdA; lipoamide dehydroge... 121 2e-27
ath:AT3G16950 LPD1; LPD1 (LIPOAMIDE DEHYDROGENASE 1); dihydrol... 112 8e-25
pfa:PFL1550w lipoamide dehydrogenase (EC:1.8.1.4); K00382 dihy... 108 1e-23
ath:AT4G16155 dihydrolipoyl dehydrogenase; K00382 dihydrolipoa... 107 4e-23
ath:AT3G24170 ATGR1; ATGR1 (glutathione-disulfide reductase); ... 100 4e-21
pfa:PF08_0066 lipoamide dehydrogenase, putative (EC:1.8.1.4); ... 98.2 2e-20
tgo:TGME49_105980 dihydrolipoyl dehydrogenase protein, putativ... 93.2 6e-19
eco:b3962 sthA, ECK3954, JW5551, sth, udhA; pyridine nucleotid... 92.0 1e-18
dre:100332932 glutathione reductase-like 85.5 2e-16
ath:AT3G54660 GR; GR (GLUTATHIONE REDUCTASE); ATP binding / gl... 79.7 7e-15
mmu:50493 Txnrd1, TR, TR1, TrxR1; thioredoxin reductase 1 (EC:... 79.0 1e-14
mmu:232223 Txnrd3, AI196535, TR2, Tgr; thioredoxin reductase 3... 77.0 5e-14
dre:553575 MGC110010; zgc:110010 (EC:1.8.1.7); K00383 glutathi... 76.6 6e-14
hsa:7296 TXNRD1, GRIM-12, MGC9145, TR, TR1, TRXR1, TXNR; thior... 75.5 1e-13
dre:352924 txnrd1, TrxR1, cb682, fb83a08, wu:fb83a08; thioredo... 73.6 6e-13
hsa:114112 TXNRD3, TGR, TR2, TRXR3; thioredoxin reductase 3 (E... 72.4 1e-12
eco:b3500 gor, ECK3485, gorA, JW3467; glutathione oxidoreducta... 72.0 2e-12
cpv:cgd2_4320 thioredoxin reductase 1 ; K00384 thioredoxin red... 71.2 3e-12
hsa:2936 GSR, MGC78522; glutathione reductase (EC:1.8.1.7); K0... 69.3 1e-11
cel:F20D6.11 hypothetical protein 68.6 2e-11
xla:447610 txnrd1, MGC85342; thioredoxin reductase 1 (EC:1.8.1... 68.6 2e-11
cel:C46F11.2 hypothetical protein 67.8 3e-11
xla:447484 txnrd3, MGC81848; thioredoxin reductase 3 (EC:1.8.1... 67.8 3e-11
mmu:14782 Gsr, AI325518, D8Ertd238e, Gr-1, Gr1; glutathione re... 67.4 4e-11
tgo:TGME49_019130 glutathione reductase, putative (EC:1.8.1.7) 67.0 5e-11
tgo:TGME49_046920 glutathione reductase, putative (EC:1.8.1.7)... 66.2 9e-11
sce:YPL091W GLR1, LPG17; Cytosolic and mitochondrial glutathio... 64.3 3e-10
eco:b0304 ykgC, ECK0303, JW5040; predicted pyridine nucleotide... 62.8 1e-09
xla:734372 gsr, MGC84926; glutathione reductase (EC:1.8.1.7); ... 61.6 2e-09
hsa:150209 AIFM3, AIFL, FLJ30473, FLJ45137; apoptosis-inducing... 59.3 1e-08
tgo:TGME49_109730 thioredoxin reductase, putative (EC:1.8.1.7)... 58.9 1e-08
xla:379744 txnrd2, MGC69182; thioredoxin reductase 2 (EC:1.8.1... 58.9 2e-08
mmu:26462 Txnrd2, AA118373, ESTM573010, TGR, Tr3, Trxr2, Trxrd... 57.8 3e-08
pfa:PFI1170c thioredoxin reductase (EC:1.8.1.9); K00384 thiore... 57.4 4e-08
xla:394337 aifm3, MGC84340, nfrl-A; apoptosis-inducing factor,... 56.6 6e-08
mmu:72168 Aifm3, 2810401C16Rik, AI840249, Aifl; apoptosis-indu... 55.5 1e-07
hsa:10587 TXNRD2, SELZ, TR, TR-BETA, TR3, TRXR2; thioredoxin r... 55.1 2e-07
cel:C06G3.7 trxr-1; ThioRedoXin Reductase family member (trxr-... 53.5 5e-07
> tgo:TGME49_006470 dihydrolipoyl dehydrogenase, putative (EC:1.8.1.4);
K00382 dihydrolipoamide dehydrogenase [EC:1.8.1.4]
Length=519
Score = 260 bits (665), Expect = 2e-69, Method: Compositional matrix adjust.
Identities = 130/221 (58%), Positives = 165/221 (74%), Gaps = 0/221 (0%)
Query 1 IVSALSQGIEGLFKRNKVDYLKGTGRLAGPHAVQVQPIDAGNPQMLMAKNVILATGSEPA 60
+VS L+QGIE LF+RN VDY G G+L ++V+V P Q L A ++ILATGSE +
Sbjct 137 VVSTLTQGIEHLFRRNGVDYYVGEGKLTDSNSVEVTPNGKSEKQRLDAGHIILATGSEAS 196
Query 61 PLAGGALEVDEETIVSSTGALALPRVPKHLVVVGGGVIGLELGSVWRNLGAEVTVVEFCD 120
PL G + +DE+ I+SSTGALAL +VPK + V+GGGVIGLELGSVWRNLGAEVTVVEF D
Sbjct 197 PLPGNVVPIDEKVIISSTGALALDKVPKRMAVIGGGVIGLELGSVWRNLGAEVTVVEFLD 256
Query 121 KIIPALDAEIGRAFQKLLERQGIKFMFGTKVVGSQKADGGVTLSLENVKSGDASEVQCDV 180
+++P +D E+ +AFQK +E+ GIKF GTKVVG+ + TL +E K G+ E++ DV
Sbjct 257 RLLPPVDGEVAKAFQKEMEKTGIKFQLGTKVVGADVRESSATLHVEPAKGGNPFEMEADV 316
Query 181 VLVAVGRRPYTKDLGLEELGINLDNRGRVGVNEQMLVPNYP 221
VLVAVGRRPYTK+LGLEELGI D GRV V+++ VPNYP
Sbjct 317 VLVAVGRRPYTKNLGLEELGIETDRVGRVVVDDRFCVPNYP 357
> ath:AT1G48030 mtLPD1; mtLPD1 (mitochondrial lipoamide dehydrogenase
1); ATP binding / dihydrolipoyl dehydrogenase; K00382
dihydrolipoamide dehydrogenase [EC:1.8.1.4]
Length=507
Score = 211 bits (538), Expect = 1e-54, Method: Compositional matrix adjust.
Identities = 110/220 (50%), Positives = 153/220 (69%), Gaps = 4/220 (1%)
Query 2 VSALSQGIEGLFKRNKVDYLKGTGRLAGPHAVQVQPIDAGNPQMLMAKNVILATGSEPAP 61
V L++GIEGLFK+NKV Y+KG G+ P+ V V+ ID GN ++ K++I+ATGS+
Sbjct 132 VKNLTRGIEGLFKKNKVTYVKGYGKFISPNEVSVETIDGGN-TIVKGKHIIVATGSDVKS 190
Query 62 LAGGALEVDEETIVSSTGALALPRVPKHLVVVGGGVIGLELGSVWRNLGAEVTVVEFCDK 121
L G + +DE+ IVSSTGAL+L VPK L+V+G G IGLE+GSVW LG+EVTVVEF
Sbjct 191 LPG--ITIDEKKIVSSTGALSLSEVPKKLIVIGAGYIGLEMGSVWGRLGSEVTVVEFAGD 248
Query 122 IIPALDAEIGRAFQKLLERQGIKFMFGTKVVGSQKADGGVTLSLENVKSGDASEVQCDVV 181
I+P++D EI + FQ+ LE+Q +KFM TKVV + GV L++E + G+ S ++ DVV
Sbjct 249 IVPSMDGEIRKQFQRSLEKQKMKFMLKTKVVSVDSSSDGVKLTVEPAEGGEQSILEADVV 308
Query 182 LVAVGRRPYTKDLGLEELGINLDNRGRVGVNEQMLVPNYP 221
LV+ GR P+T L LE++G+ D GR+ VN++ L N P
Sbjct 309 LVSAGRTPFTSGLDLEKIGVETDKAGRILVNDRFL-SNVP 347
> hsa:1738 DLD, DLDH, E3, GCSL, LAD, PHE3; dihydrolipoamide dehydrogenase
(EC:1.8.1.4); K00382 dihydrolipoamide dehydrogenase
[EC:1.8.1.4]
Length=509
Score = 210 bits (534), Expect = 3e-54, Method: Compositional matrix adjust.
Identities = 112/222 (50%), Positives = 155/222 (69%), Gaps = 7/222 (3%)
Query 2 VSALSQGIEGLFKRNKVDYLKGTGRLAGPHAVQVQPIDAGNPQMLMAKNVILATGSEPAP 61
V AL+ GI LFK+NKV ++ G G++ G + V D G Q++ KN+++ATGSE P
Sbjct 131 VKALTGGIAHLFKQNKVVHVNGYGKITGKNQVTATKADGGT-QVIDTKNILIATGSEVTP 189
Query 62 LAGGALEVDEETIVSSTGALALPRVPKHLVVVGGGVIGLELGSVWRNLGAEVTVVEFCDK 121
G + +DE+TIVSSTGAL+L +VP+ +VV+G GVIG+ELGSVW+ LGA+VT VEF
Sbjct 190 FPG--ITIDEDTIVSSTGALSLKKVPEKMVVIGAGVIGVELGSVWQRLGADVTAVEFLGH 247
Query 122 IIP-ALDAEIGRAFQKLLERQGIKFMFGTKVVG-SQKADGGVTLSLENVKSGDASEVQCD 179
+ +D EI + FQ++L++QG KF TKV G ++K+DG + +S+E G A + CD
Sbjct 248 VGGVGIDMEISKNFQRILQKQGFKFKLNTKVTGATKKSDGKIDVSIEAASGGKAEVITCD 307
Query 180 VVLVAVGRRPYTKDLGLEELGINLDNRGRVGVNE--QMLVPN 219
V+LV +GRRP+TK+LGLEELGI LD RGR+ VN Q +PN
Sbjct 308 VLLVCIGRRPFTKNLGLEELGIELDPRGRIPVNTRFQTKIPN 349
> xla:380588 dld, MGC68940; dihydrolipoamide dehydrogenase (EC:1.8.1.4);
K00382 dihydrolipoamide dehydrogenase [EC:1.8.1.4]
Length=509
Score = 209 bits (531), Expect = 7e-54, Method: Compositional matrix adjust.
Identities = 109/222 (49%), Positives = 157/222 (70%), Gaps = 7/222 (3%)
Query 2 VSALSQGIEGLFKRNKVDYLKGTGRLAGPHAVQVQPIDAGNPQMLMAKNVILATGSEPAP 61
V +L+ GI LFK+NKV +++G G++ G + V D G+ Q++ KN+++ATGSE AP
Sbjct 131 VKSLTSGIAHLFKQNKVVHVQGFGKITGKNQVTATKAD-GSTQVVNTKNILIATGSEVAP 189
Query 62 LAGGALEVDEETIVSSTGALALPRVPKHLVVVGGGVIGLELGSVWRNLGAEVTVVEFCDK 121
G + +DEETIVSSTGAL+L +VP+ +VV+G GVIG+ELGSVW+ LGA+VT VEF
Sbjct 190 FPG--IPIDEETIVSSTGALSLKQVPEKMVVIGAGVIGVELGSVWQRLGADVTAVEFLGH 247
Query 122 I-IPALDAEIGRAFQKLLERQGIKFMFGTKVVG-SQKADGGVTLSLENVKSGDASEVQCD 179
+ +D EI + F ++L++QG+KF TKV G S++ DG + +S+E G + CD
Sbjct 248 VGGVGIDMEISKNFHRILQKQGLKFKLSTKVTGASKRPDGKIDVSIEAAAGGKEEVITCD 307
Query 180 VVLVAVGRRPYTKDLGLEELGINLDNRGRVGVNE--QMLVPN 219
V+LV +GRRP+T++LGL+ELGI LDNRGR+ +N Q +PN
Sbjct 308 VLLVCIGRRPFTENLGLQELGIELDNRGRIPINSRFQTKIPN 349
> mmu:13382 Dld, AI315664, AI746344; dihydrolipoamide dehydrogenase
(EC:1.8.1.4); K00382 dihydrolipoamide dehydrogenase [EC:1.8.1.4]
Length=509
Score = 209 bits (531), Expect = 8e-54, Method: Compositional matrix adjust.
Identities = 112/222 (50%), Positives = 156/222 (70%), Gaps = 7/222 (3%)
Query 2 VSALSQGIEGLFKRNKVDYLKGTGRLAGPHAVQVQPIDAGNPQMLMAKNVILATGSEPAP 61
V AL+ GI LFK+NKV ++ G G++ G + V D G+ Q++ KN+++ATGSE P
Sbjct 131 VKALTGGIAHLFKQNKVVHVNGFGKITGKNQVTATKAD-GSTQVIDTKNILVATGSEVTP 189
Query 62 LAGGALEVDEETIVSSTGALALPRVPKHLVVVGGGVIGLELGSVWRNLGAEVTVVEFCDK 121
G + +DE+TIVSSTGAL+L +VP+ LVV+G GVIG+ELGSVW+ LGA+VT VEF
Sbjct 190 FPG--ITIDEDTIVSSTGALSLKKVPEKLVVIGAGVIGVELGSVWQRLGADVTAVEFLGH 247
Query 122 IIP-ALDAEIGRAFQKLLERQGIKFMFGTKVVG-SQKADGGVTLSLENVKSGDASEVQCD 179
+ +D EI + FQ++L+RQG KF TKV G ++K+DG + +S+E G A + CD
Sbjct 248 VGGIGIDMEISKNFQRILQRQGFKFKLNTKVTGATKKSDGKIDVSVEAASGGKAEVITCD 307
Query 180 VVLVAVGRRPYTKDLGLEELGINLDNRGRVGVNE--QMLVPN 219
V+LV +GRRP+T++LGLEELGI LD +GR+ VN Q +PN
Sbjct 308 VLLVCIGRRPFTQNLGLEELGIELDPKGRIPVNNRFQTKIPN 349
> ath:AT3G17240 mtLPD2; mtLPD2 (LIPOAMIDE DEHYDROGENASE 2); ATP
binding / dihydrolipoyl dehydrogenase; K00382 dihydrolipoamide
dehydrogenase [EC:1.8.1.4]
Length=507
Score = 205 bits (522), Expect = 9e-53, Method: Compositional matrix adjust.
Identities = 106/214 (49%), Positives = 148/214 (69%), Gaps = 3/214 (1%)
Query 2 VSALSQGIEGLFKRNKVDYLKGTGRLAGPHAVQVQPIDAGNPQMLMAKNVILATGSEPAP 61
V L++G+EGLFK+NKV+Y+KG G+ P V V ID G ++ K++I+ATGS+
Sbjct 132 VKNLTRGVEGLFKKNKVNYVKGYGKFLSPSEVSVDTID-GENVVVKGKHIIVATGSDVKS 190
Query 62 LAGGALEVDEETIVSSTGALALPRVPKHLVVVGGGVIGLELGSVWRNLGAEVTVVEFCDK 121
L G + +DE+ IVSSTGAL+L +PK L+V+G G IGLE+GSVW LG+EVTVVEF
Sbjct 191 LPG--ITIDEKKIVSSTGALSLTEIPKKLIVIGAGYIGLEMGSVWGRLGSEVTVVEFAAD 248
Query 122 IIPALDAEIGRAFQKLLERQGIKFMFGTKVVGSQKADGGVTLSLENVKSGDASEVQCDVV 181
I+PA+D EI + FQ+ LE+Q +KFM TKVVG + GV L +E + G+ + ++ DVV
Sbjct 249 IVPAMDGEIRKQFQRSLEKQKMKFMLKTKVVGVDSSGDGVKLIVEPAEGGEQTTLEADVV 308
Query 182 LVAVGRRPYTKDLGLEELGINLDNRGRVGVNEQM 215
LV+ GR P+T L LE++G+ D GR+ VNE+
Sbjct 309 LVSAGRTPFTSGLDLEKIGVETDKGGRILVNERF 342
> cel:LLC1.3 hypothetical protein; K00382 dihydrolipoamide dehydrogenase
[EC:1.8.1.4]
Length=495
Score = 200 bits (508), Expect = 4e-51, Method: Compositional matrix adjust.
Identities = 107/221 (48%), Positives = 152/221 (68%), Gaps = 6/221 (2%)
Query 2 VSALSQGIEGLFKRNKVDYLKGTGRLAGPHAVQVQPIDAGNPQMLMAKNVILATGSEPAP 61
V L+ GI+ LFK NKV +++G + GP+ VQ + D G+ + + A+N+++A+GSE P
Sbjct 117 VKQLTGGIKQLFKANKVGHVEGFATIVGPNTVQAKKND-GSVETINARNILIASGSEVTP 175
Query 62 LAGGALEVDEETIVSSTGALALPRVPKHLVVVGGGVIGLELGSVWRNLGAEVTVVEFCDK 121
G + +DE+ IVSSTGAL+L +VPK +VV+G GVIGLELGSVW+ LGAEVT VEF
Sbjct 176 FPG--ITIDEKQIVSSTGALSLGQVPKKMVVIGAGVIGLELGSVWQRLGAEVTAVEFLGH 233
Query 122 II-PALDAEIGRAFQKLLERQGIKFMFGTKVVGSQKADGGVTLSLENVKSGDASEVQCDV 180
+ +D E+ + FQ+ L +QG KF+ TKV+G+ + +T+ +E K G ++CD
Sbjct 234 VGGMGIDGEVSKNFQRSLTKQGFKFLLNTKVMGASQNGSTITVEVEGAKDGKKQTLECDT 293
Query 181 VLVAVGRRPYTKDLGLEELGINLDNRGRVGVNE--QMLVPN 219
+LV+VGRRPYT+ LGL + I+LDNRGRV VNE Q VP+
Sbjct 294 LLVSVGRRPYTEGLGLSNVQIDLDNRGRVPVNERFQTKVPS 334
> dre:399479 dldh, wu:fb24b05; dihydrolipoamide dehydrogenase
(EC:1.8.1.4); K00382 dihydrolipoamide dehydrogenase [EC:1.8.1.4]
Length=507
Score = 196 bits (499), Expect = 4e-50, Method: Compositional matrix adjust.
Identities = 106/222 (47%), Positives = 151/222 (68%), Gaps = 8/222 (3%)
Query 2 VSALSQGIEGLFKRNKVDYLKGTGRLAGPHAVQVQPIDAGNPQMLMAKNVILATGSEPAP 61
V AL+ GI LFK+NKV ++ G G + G + V + D Q++ KN+++ATGSE P
Sbjct 130 VKALTGGIAHLFKQNKVTHVNGFGTITGKNQVTAKTADG--EQVINTKNILIATGSEVTP 187
Query 62 LAGGALEVDEETIVSSTGALALPRVPKHLVVVGGGVIGLELGSVWRNLGAEVTVVEFCDK 121
G +E+DE+++VSSTGAL+L VP+ L+V+G GVIG+ELGSVW+ LGA+VT VEF
Sbjct 188 FPG--IEIDEDSVVSSTGALSLKNVPEELIVIGAGVIGVELGSVWQRLGAKVTAVEFLGH 245
Query 122 I-IPALDAEIGRAFQKLLERQGIKFMFGTKVVGSQKA-DGGVTLSLENVKSGDASEVQCD 179
+ +D EI + FQ++L++QG+KF TKV+G+ K DG + +++E G + CD
Sbjct 246 VGGMGIDMEISKNFQRILQKQGLKFKLSTKVMGATKRPDGKIDVAVEAAAGGKNETLTCD 305
Query 180 VVLVAVGRRPYTKDLGLEELGINLDNRGRVGVNE--QMLVPN 219
V+LV +GRRP+T +LGLE +GI LD RGR+ VN Q VPN
Sbjct 306 VLLVCIGRRPFTGNLGLESVGIELDKRGRIPVNGRFQTNVPN 347
> bbo:BBOV_IV007190 23.m05858; dihydrolipoamide dehydrogenase
(EC:1.8.1.4); K00382 dihydrolipoamide dehydrogenase [EC:1.8.1.4]
Length=481
Score = 194 bits (493), Expect = 2e-49, Method: Compositional matrix adjust.
Identities = 102/220 (46%), Positives = 147/220 (66%), Gaps = 8/220 (3%)
Query 1 IVSALSQGIEGLFKRNKVDYLKGTGRLAGPHAVQVQPIDAGNPQMLMAKNVILATGSEPA 60
I+ L GI+GLFK+N VDY+ G G L + +Q++ + + AKN+I+ATGSE
Sbjct 109 ILKTLDAGIKGLFKKNGVDYISGHGTLKSANEIQIE-----GGETVSAKNIIIATGSEVT 163
Query 61 PLAGGALEVDEETIVSSTGALALPRVPKHLVVVGGGVIGLELGSVWRNLGAEVTVVEFCD 120
G AL++D + I+SS AL L VPK +VV+GGG IGLEL SVW LGA+VT+VE+ +
Sbjct 164 TFPGDALKIDGKRIISSDEALVLDEVPKEMVVIGGGAIGLELASVWSRLGAKVTIVEYAN 223
Query 121 KIIPALDAEIGRAFQKLLERQGIKFMFGTKVVGSQKADGGVTLSLENVKSGDASEVQCDV 180
+ +D ++ A +K++E+QGI + TKV+G + D ++ E K G+ E++ DV
Sbjct 224 NLCHTMDHDVSVAIKKIVEKQGINILLSTKVLGGEVKDDCAVITAE--KDGEKIELKGDV 281
Query 181 VLVAVGRRPYTKDLGLEELGINLDNRGRVGVNEQMLVPNY 220
VL+A+GRRPYTK+LGLEELGI + RG + V+E + VPNY
Sbjct 282 VLLAMGRRPYTKNLGLEELGIKTE-RGYIVVDEMLRVPNY 320
> sce:YFL018C LPD1, HPD1; Dihydrolipoamide dehydrogenase, the
lipoamide dehydrogenase component (E3) of the pyruvate dehydrogenase
and 2-oxoglutarate dehydrogenase multi-enzyme complexes
(EC:1.8.1.4); K00382 dihydrolipoamide dehydrogenase [EC:1.8.1.4]
Length=499
Score = 175 bits (444), Expect = 1e-43, Method: Compositional matrix adjust.
Identities = 99/221 (44%), Positives = 146/221 (66%), Gaps = 9/221 (4%)
Query 2 VSALSQGIEGLFKRNKVDYLKGTGRLAGPHAVQVQPID-----AGNPQMLMAKNVILATG 56
V L+ GIE LFK+NKV Y KG G ++V P+D +L KN+I+ATG
Sbjct 116 VKQLTGGIELLFKKNKVTYYKGNGSFEDETKIRVTPVDGLEGTVKEDHILDVKNIIVATG 175
Query 57 SEPAPLAGGALEVDEETIVSSTGALALPRVPKHLVVVGGGVIGLELGSVWRNLGAEVTVV 116
SE P G +E+DEE IVSSTGAL+L +PK L ++GGG+IGLE+GSV+ LG++VTVV
Sbjct 176 SEVTPFPG--IEIDEEKIVSSTGALSLKEIPKRLTIIGGGIIGLEMGSVYSRLGSKVTVV 233
Query 117 EFCDKIIPALDAEIGRAFQKLLERQGIKFMFGTKVVGSQKADGG--VTLSLENVKSGDAS 174
EF +I ++D E+ +A QK L++QG+ F TKV+ +++ D V + +E+ K+
Sbjct 234 EFQPQIGASMDGEVAKATQKFLKKQGLDFKLSTKVISAKRNDDKNVVEIVVEDTKTNKQE 293
Query 175 EVQCDVVLVAVGRRPYTKDLGLEELGINLDNRGRVGVNEQM 215
++ +V+LVAVGRRPY LG E++G+ +D RGR+ +++Q
Sbjct 294 NLEAEVLLVAVGRRPYIAGLGAEKIGLEVDKRGRLVIDDQF 334
> tpv:TP03_0227 dihydrolipoamide dehydrogenase (EC:1.8.1.4); K00382
dihydrolipoamide dehydrogenase [EC:1.8.1.4]
Length=499
Score = 148 bits (373), Expect = 2e-35, Method: Compositional matrix adjust.
Identities = 87/221 (39%), Positives = 134/221 (60%), Gaps = 9/221 (4%)
Query 1 IVSALSQGIEGLFKRNKVDYLKGTGRLAGPHAVQVQPIDAGNPQMLMAKNVILATGSEPA 60
++ L+ GI GLFK+NK+DY++GT + V V ++L+A V++ATGSE
Sbjct 109 VMRTLNMGIFGLFKKNKIDYIQGTACFKSQNEVTV------GSKVLLADKVVVATGSEVR 162
Query 61 PLAGGALEVDEETIVSSTGALALPRVPKHLVVVGGGVIGLELGSVWRNLGAEVTVVEFCD 120
P +L+VD + +SST L L +VP L+V+G G IGLEL SVW LG++V + EF +
Sbjct 163 PFPSESLKVDGKYFLSSTETLCLDKVPNRLLVIGAGAIGLELASVWSRLGSKVDIFEFNN 222
Query 121 KIIPALDAEIGRAFQKLLERQGIKFMFGTKVVGSQKADGGVTLSLENVKSGDASEVQCDV 180
+I +D ++ +K+LE+QG+ GTKV+ ++ + VTL+ E+ G D
Sbjct 223 QICSVMDTDVCVTMRKILEKQGLNIHTGTKVLNAKVTNNTVTLTTES--EGKEMSYVGDK 280
Query 181 VLVAVGRRPYTKDLGLEELGINLDNRGRVGVNEQMLVPNYP 221
VLVA+GR PYT+ LG+++LG+ LD G+V + + V P
Sbjct 281 VLVAMGRVPYTEGLGIDKLGVTLD-YGKVPTDNNLRVLKDP 320
> sce:YPL017C IRC15; Irc15p; K00382 dihydrolipoamide dehydrogenase
[EC:1.8.1.4]
Length=499
Score = 122 bits (306), Expect = 9e-28, Method: Compositional matrix adjust.
Identities = 84/230 (36%), Positives = 122/230 (53%), Gaps = 17/230 (7%)
Query 3 SALSQGIEGL---FKR----NKVDYLKGTGRLAGPHAVQVQPIDAGNPQMLMAKNVILAT 55
SAL IE L +KR N V KGT PH V++ P ++ AK +++AT
Sbjct 102 SALKHNIEELGNVYKRELSKNNVTVYKGTAAFKDPHHVEIAQ-RGMKPFIVEAKYIVVAT 160
Query 56 GSEPAPLAGGALEVDEETIVSSTGALALPRVPKHLVVVGGGVIGLELGSVWRNLGAEVTV 115
GS G A +D + I+SS AL+L +P ++GGG IGLE+ ++ NLG+ VT+
Sbjct 161 GSAVIQCPGVA--IDNDKIISSDKALSLDYIPSRFTIMGGGTIGLEIACIFNNLGSRVTI 218
Query 116 VEFCDKIIPALDAEIGRAFQKLLERQGIKFMFGTKV-VGSQKADGGVTLSLENVKSGDAS 174
VE +I +D E+ A + LL+ QGI F+ T+V + A G + ++L N S
Sbjct 219 VESQSEICQNMDNELASATKTLLQCQGIAFLLDTRVQLAEADAAGQLNITLLNKVSKKTY 278
Query 175 EVQCDVVLVAVGRRPYTKDLGLEELGINLDNRG---RVGVNEQMLVPNYP 221
CDV++V++GRRP K L + +G LD R V V Q L+ YP
Sbjct 279 VHHCDVLMVSIGRRPLLKGLDISSIG--LDERDFVENVDVQTQSLL-KYP 325
> eco:b0116 lpd, dhl, ECK0115, JW0112, lpdA; lipoamide dehydrogenase,
E3 component is part of three enzyme complexes (EC:1.8.1.4);
K00382 dihydrolipoamide dehydrogenase [EC:1.8.1.4]
Length=474
Score = 121 bits (304), Expect = 2e-27, Method: Compositional matrix adjust.
Identities = 73/215 (33%), Positives = 125/215 (58%), Gaps = 6/215 (2%)
Query 1 IVSALSQGIEGLFKRNKVDYLKGTGRLAGPHAVQVQPIDAGNPQMLMAKNVILATGSEPA 60
+++ L+ G+ G+ K KV + G G+ G + ++V+ ++ N I+A GS P
Sbjct 93 VINQLTGGLAGMAKGRKVKVVNGLGKFTGANTLEVE--GENGKTVINFDNAIIAAGSRPI 150
Query 61 PLAGGALEVDEETIVSSTGALALPRVPKHLVVVGGGVIGLELGSVWRNLGAEVTVVEFCD 120
L + ++ I ST AL L VP+ L+V+GGG+IGLE+G+V+ LG+++ VVE D
Sbjct 151 QLP--FIPHEDPRIWDSTDALELKEVPERLLVMGGGIIGLEMGTVYHALGSQIDVVEMFD 208
Query 121 KIIPALDAEIGRAFQKLLERQGIKFMFGTKVVGSQKADGGVTLSLENVKSGDASEVQCDV 180
++IPA D +I + F K + ++ M TKV + + G+ +++E K A + D
Sbjct 209 QVIPAADKDIVKVFTKRISKK-FNLMLETKVTAVEAKEDGIYVTMEG-KKAPAEPQRYDA 266
Query 181 VLVAVGRRPYTKDLGLEELGINLDNRGRVGVNEQM 215
VLVA+GR P K+L + G+ +D+RG + V++Q+
Sbjct 267 VLVAIGRVPNGKNLDAGKAGVEVDDRGFIRVDKQL 301
> ath:AT3G16950 LPD1; LPD1 (LIPOAMIDE DEHYDROGENASE 1); dihydrolipoyl
dehydrogenase; K00382 dihydrolipoamide dehydrogenase
[EC:1.8.1.4]
Length=623
Score = 112 bits (281), Expect = 8e-25, Method: Compositional matrix adjust.
Identities = 76/216 (35%), Positives = 122/216 (56%), Gaps = 24/216 (11%)
Query 18 VDYLKGTGRLAGPHAVQVQPIDAGNPQMLMAKNVILATGSEPAPLAGGALEVDEETIVSS 77
VD L G G + GP Q + G ++ AK++I+ATGS P +EVD +T+++S
Sbjct 192 VDILTGFGSVLGP-----QKVKYGKDNIITAKDIIIATGS--VPFVPKGIEVDGKTVITS 244
Query 78 TGALALPRVPKHLVVVGGGVIGLELGSVWRNLGAEVTVVEFCDKIIPALDAEIGRAFQK- 136
AL L VP+ + +VG G IGLE V+ LG+EVT +E D+++P D EI + Q+
Sbjct 245 DHALKLESVPEWIAIVGSGYIGLEFSDVYTALGSEVTFIEALDQLMPGFDPEISKLAQRV 304
Query 137 LLERQGIKF---MFGTKVVGSQKADGG-VTLSLENVKSGDASE-VQCDVVLVAVGRRPYT 191
L+ + I + +F +K+ ++ DG V + L + K+ + + ++ D L+A GR P+T
Sbjct 305 LINPRKIDYHTGVFASKITPAR--DGKPVLIELIDAKTKEPKDTLEVDAALIATGRAPFT 362
Query 192 KDLGLEELGINLDNRGRVGVNEQM--------LVPN 219
LGLE + + + RG + V+E+M LVPN
Sbjct 363 NGLGLENVNV-VTQRGFIPVDERMRVIDGKGTLVPN 397
> pfa:PFL1550w lipoamide dehydrogenase (EC:1.8.1.4); K00382 dihydrolipoamide
dehydrogenase [EC:1.8.1.4]
Length=512
Score = 108 bits (271), Expect = 1e-23, Method: Compositional matrix adjust.
Identities = 75/228 (32%), Positives = 125/228 (54%), Gaps = 17/228 (7%)
Query 5 LSQGIEGLFKRNKVDYLKGTGRLAGPHAVQVQPIDAGNPQMLMAKNVILATGSEP--APL 62
LS GI L+K+N V+++ G G L H V ++ + + A+ +++ATGS+P PL
Sbjct 116 LSDGINFLYKKNNVNHIIGHGSLVDEHTVLIKT--EKEEKKVTAERIVIATGSKPIEIPL 173
Query 63 AG-------------GALEVDEETIVSSTGALALPRVPKHLVVVGGGVIGLELGSVWRNL 109
LE D E I +S L +VP ++ ++GGGVIGLE+GSV+ L
Sbjct 174 KKLNDNNFNDADNVNDILEYDHEIIQNSDDILNFKKVPHNISIIGGGVIGLEIGSVFSKL 233
Query 110 GAEVTVVEFCDKIIPALDAEIGRAFQKLLERQGIKFMFGTKVVGSQKADGGVTLSLENVK 169
G++VTV E+ +++ LDA++ + QK LE+ +KF+F T V+G + L +N K
Sbjct 234 GSDVTVFEYNERLCGFLDADVSKVLQKTLEKIKMKFVFNTSVIGGNIENNQAALFAKNKK 293
Query 170 SGDASEVQCDVVLVAVGRRPYTKDLGLEELGINLDNRGRVGVNEQMLV 217
+ + + ++VL+ +GR+ +L L L I L+ ++ V+E V
Sbjct 294 TNEIKKTTSEIVLICIGRKANFDNLNLHLLNIELNKNKKIPVDEYFNV 341
> ath:AT4G16155 dihydrolipoyl dehydrogenase; K00382 dihydrolipoamide
dehydrogenase [EC:1.8.1.4]
Length=630
Score = 107 bits (267), Expect = 4e-23, Method: Compositional matrix adjust.
Identities = 72/206 (34%), Positives = 117/206 (56%), Gaps = 17/206 (8%)
Query 18 VDYLKGTGRLAGPHAVQVQPIDAGNPQMLMAKNVILATGSEPAPLAGGALEVDEETIVSS 77
VD L G G + GP Q + G+ ++ K++I+ATGS P +EVD +T+++S
Sbjct 253 VDILTGFGAVLGP-----QKVKYGD-NIITGKDIIIATGS--VPFVPKGIEVDGKTVITS 304
Query 78 TGALALPRVPKHLVVVGGGVIGLELGSVWRNLGAEVTVVEFCDKIIPALDAEIGRAFQK- 136
AL L VP + +VG G IGLE V+ LG+EVT +E D+++P D EI + Q+
Sbjct 305 DHALKLESVPDWIAIVGSGYIGLEFSDVYTALGSEVTFIEALDQLMPGFDPEISKLAQRV 364
Query 137 LLERQGIKF---MFGTKVVGSQKADGG-VTLSLENVKSGDASE-VQCDVVLVAVGRRPYT 191
L+ + I + +F +K+ ++ DG V + L + K+ + + ++ D L+A GR P+T
Sbjct 365 LINTRKIDYHTGVFASKITPAK--DGKPVLIELIDAKTKEPKDTLEVDAALIATGRAPFT 422
Query 192 KDLGLEELGINLDNRGRVGVNEQMLV 217
LGLE + + RG + V+E+M V
Sbjct 423 NGLGLENINVT-TQRGFIPVDERMRV 447
> ath:AT3G24170 ATGR1; ATGR1 (glutathione-disulfide reductase);
FAD binding / NADP or NADPH binding / glutathione-disulfide
reductase/ oxidoreductase (EC:1.8.1.7); K00383 glutathione
reductase (NADPH) [EC:1.8.1.7]
Length=499
Score = 100 bits (249), Expect = 4e-21, Method: Compositional matrix adjust.
Identities = 74/205 (36%), Positives = 102/205 (49%), Gaps = 16/205 (7%)
Query 12 LFKRNKVDYLKGTGRLAGPHAVQVQPIDAGNPQMLMAKNVILATGSEP-APLAGGALEVD 70
L V +G GR+ GP+ V+V+ ID G AK++++ATGS P G
Sbjct 133 LLANAAVKLYEGEGRVVGPNEVEVRQID-GTKISYTAKHILIATGSRAQKPNIPG----- 186
Query 71 EETIVSSTGALALPRVPKHLVVVGGGVIGLELGSVWRNLGAEVTVVEFCDKIIP--ALDA 128
E ++S AL+L PK +V+GGG I +E S+WR +GA V + F K +P D
Sbjct 187 HELAITSDEALSLEEFPKRAIVLGGGYIAVEFASIWRGMGATVDL--FFRKELPLRGFDD 244
Query 129 EIGRAFQKLLERQGIKFMFGTKVVGSQKADGGVTLSLENVKSGDASEVQCDVVLVAVGRR 188
E+ + LE +G+ T + K D G+ V S E DVVL A GR
Sbjct 245 EMRALVARNLEGRGVNLHPQTSLTQLTKTDQGI-----KVISSHGEEFVADVVLFATGRS 299
Query 189 PYTKDLGLEELGINLDNRGRVGVNE 213
P TK L LE +G+ LD G V V+E
Sbjct 300 PNTKRLNLEAVGVELDQAGAVKVDE 324
> pfa:PF08_0066 lipoamide dehydrogenase, putative (EC:1.8.1.4);
K00382 dihydrolipoamide dehydrogenase [EC:1.8.1.4]
Length=666
Score = 98.2 bits (243), Expect = 2e-20, Method: Composition-based stats.
Identities = 56/181 (30%), Positives = 104/181 (57%), Gaps = 15/181 (8%)
Query 49 KNVILATGSEPAPLAGGALEVDEETIVSSTGALALPRVPKHLVVVGGGVIGLELGSVWRN 108
KN+I+ATGS P +E+D++++ +S A+ L + ++ ++G G+IGLE ++
Sbjct 285 KNIIIATGS--VPNIPNNVEIDDKSVFTSDMAVKLVGLKNYMSIIGMGIIGLEFADIYTA 342
Query 109 LGAEVTVVEFCDKIIPALDAEIGRAFQKL-LERQGIKFMFGTKV--VGSQKADGGVTLSL 165
LG+E+T +E+ +++P +D ++ + F+++ L+ + + + T+V + + K + V +
Sbjct 343 LGSEITFLEYSSELLPIIDNDVAKYFERVFLKNKPVNYHLNTEVKYIKASKNNNPVIIGY 402
Query 166 ---------ENVKSGDASEVQCDVVLVAVGRRPYTKDLGLEELGINLDNRGRVGVNEQML 216
E D E+ D LVA GR P T++LGLE+L I + NRG V VN+ +
Sbjct 403 SHRTGNDDNEKKNMTDVKELYVDSCLVATGRNPNTQNLGLEKLKIQM-NRGYVSVNDNLQ 461
Query 217 V 217
V
Sbjct 462 V 462
> tgo:TGME49_105980 dihydrolipoyl dehydrogenase protein, putative
(EC:1.8.1.4); K00382 dihydrolipoamide dehydrogenase [EC:1.8.1.4]
Length=636
Score = 93.2 bits (230), Expect = 6e-19, Method: Compositional matrix adjust.
Identities = 63/187 (33%), Positives = 103/187 (55%), Gaps = 11/187 (5%)
Query 40 AGNPQMLMAKNVILATGSEPAPLAGGALEVDEETIVSSTGALALPRVPKHLVVVGGGVIG 99
A P L KNVILA GS P AG E + ++++S ++LP +P + +VG G IG
Sbjct 274 ASLPPFLRTKNVILAPGSLPFIPAGTKEE--QFSVMTSDTCVSLPWLPSEICIVGSGYIG 331
Query 100 LELGSVWRNLGAEVTVVEFCDKIIPALDAEIGRAFQKLLERQGIK--FMFGTKVVGSQ-- 155
LE V+ +LG+EV +VE +++P +D E+ + ++LL +Q + T + SQ
Sbjct 332 LEFMDVFTSLGSEVVMVEAGPRLLPGVDKEVAKLAERLLLQQFKERPVKLYTNTLASQVR 391
Query 156 ----KADGGVTLSLENVKSGDAS-EVQCDVVLVAVGRRPYTKDLGLEELGINLDNRGRVG 210
K + V + L + ++ ++ ++ D L+A GRRP T+ LGL+ LG+ L G +
Sbjct 392 PLGPKGEAPVEVQLTDAQTKESKGKIYPDACLIATGRRPNTEGLGLDSLGVTLKRGGFIP 451
Query 211 VNEQMLV 217
V+ M V
Sbjct 452 VDACMRV 458
> eco:b3962 sthA, ECK3954, JW5551, sth, udhA; pyridine nucleotide
transhydrogenase, soluble (EC:1.6.1.1); K00322 NAD(P) transhydrogenase
[EC:1.6.1.1]
Length=466
Score = 92.0 bits (227), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 55/203 (27%), Positives = 100/203 (49%), Gaps = 8/203 (3%)
Query 10 EGLFKRNKVDYLKGTGRLAGPHAVQVQPIDAGNPQMLMAKNVILATGSEPAPLAGGALEV 69
+G ++RN + L+G R H + + D G+ + L A+ ++A GS P ++
Sbjct 102 QGFYERNHCEILQGNARFVDEHTLALDCPD-GSVETLTAEKFVIACGSRPYHPTD--VDF 158
Query 70 DEETIVSSTGALALPRVPKHLVVVGGGVIGLELGSVWRNLGAEVTVVEFCDKIIPALDAE 129
I S L++ P+H+++ G GVIG E S++R + +V ++ D+++ LD E
Sbjct 159 THPRIYDSDSILSMHHEPRHVLIYGAGVIGCEYASIFRGMDVKVDLINTRDRLLAFLDQE 218
Query 130 IGRAFQKLLERQGIKFMFGTKVVGSQKADGGVTLSLENVKSGDASEVQCDVVLVAVGRRP 189
+ + G+ + + D GV + L KSG +++ D +L A GR
Sbjct 219 MSDSLSYHFWNSGVVIRHNEEYEKIEGCDDGVIMHL---KSG--KKLKADCLLYANGRTG 273
Query 190 YTKDLGLEELGINLDNRGRVGVN 212
T L L+ +G+ D+RG++ VN
Sbjct 274 NTDSLALQNIGLETDSRGQLKVN 296
> dre:100332932 glutathione reductase-like
Length=461
Score = 85.5 bits (210), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 58/209 (27%), Positives = 105/209 (50%), Gaps = 16/209 (7%)
Query 9 IEGLFKRN----KVDYLKGTGRLAGPHAVQVQPIDAGNPQMLMAKNVILATGSEPAPLAG 64
+EGL+++ K L H V++ Q + A+++++ATG P P A
Sbjct 95 LEGLYRKGLENAKAKVFDSRAELVDAHTVRLTK----TGQTVSAEHIVIATGGTPNPHAD 150
Query 65 GALEVDEETIVSSTGALALPRVPKHLVVVGGGVIGLELGSVWRNLGAEVTVVEFCDKIIP 124
E +SS A L ++PK +++ GGG I +E +++ LG E T++ +I+
Sbjct 151 LP---GHELCISSNEAFHLEKLPKSILIAGGGYIAVEFANIFHGLGVETTLIYRGKEILS 207
Query 125 ALDAEIGRAFQKLLERQGIKFMFGTKVVGSQKADGGVTLSLENVKSGDASEVQCDVVLVA 184
D ++ R + +GI+ + + + +Q + G L E K G+ ++ D V++A
Sbjct 208 RFDGDLRRGLHNAMTAKGIRII--CQDIMTQVSRDGEGLVAET-KEGET--LRVDTVMLA 262
Query 185 VGRRPYTKDLGLEELGINLDNRGRVGVNE 213
+GR P+T+ LGLE G+ D G + V+E
Sbjct 263 LGRDPHTRGLGLEAAGVATDAHGAIIVDE 291
> ath:AT3G54660 GR; GR (GLUTATHIONE REDUCTASE); ATP binding /
glutathione-disulfide reductase (EC:1.8.1.7); K00383 glutathione
reductase (NADPH) [EC:1.8.1.7]
Length=565
Score = 79.7 bits (195), Expect = 7e-15, Method: Compositional matrix adjust.
Identities = 64/221 (28%), Positives = 105/221 (47%), Gaps = 24/221 (10%)
Query 7 QGIEGLFK----RNKVDYLKGTGRLAGPHAVQVQPIDAGNPQMLMAKNVILATGSEPA-P 61
Q + G++K + V ++G G++ PH V V + ++ +N+++A G P P
Sbjct 186 QRLTGIYKNILSKANVKLIEGRGKVIDPHTVDV------DGKIYTTRNILIAVGGRPFIP 239
Query 62 LAGGALEVDEETIVSSTGALALPRVPKHLVVVGGGVIGLELGSVWRNLGAEVTVVEFCDK 121
G +E + S AL LP PK + +VGGG I LE ++ L EV V K
Sbjct 240 DIPG-----KEFAIDSDAALDLPSKPKKIAIVGGGYIALEFAGIFNGLNCEVHVFIRQKK 294
Query 122 IIPALDAEIGRAFQKLLERQGIKFMFGTKVVGSQKA-DGGVTLSLENVKSGDASEVQCDV 180
++ D ++ + + +GI+F KA DG +L K+ +
Sbjct 295 VLRGFDEDVRDFVGEQMSLRGIEFHTEESPEAIIKAGDGSFSL-----KTSKGTVEGFSH 349
Query 181 VLVAVGRRPYTKDLGLEELGINLDNRGRVGVNE--QMLVPN 219
V+ A GR+P TK+LGLE +G+ + G + V+E Q VP+
Sbjct 350 VMFATGRKPNTKNLGLENVGVKMAKNGAIEVDEYSQTSVPS 390
> mmu:50493 Txnrd1, TR, TR1, TrxR1; thioredoxin reductase 1 (EC:1.8.1.9);
K00384 thioredoxin reductase (NADPH) [EC:1.8.1.9]
Length=499
Score = 79.0 bits (193), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 62/224 (27%), Positives = 109/224 (48%), Gaps = 12/224 (5%)
Query 2 VSALSQGIEGLFKRNKVDYLKGTGRLAGPHAVQVQPIDAGNPQMLMAKNVILATGSEPAP 61
+ +L+ G + KV Y GR GPH + V + G ++ A+ ++ATG P
Sbjct 109 IGSLNWGYRVALREKKVVYENAYGRFIGPHRI-VATNNKGKEKIYSAERFLIATGERPRY 167
Query 62 LAGGALEVDEETIVSSTGALALPRVPKHLVVVGGGVIGLELGSVWRNLGAEVTVVEFCDK 121
L + D+E +SS +LP P +VVG + LE +G +VTV+
Sbjct 168 LG---IPGDKEYCISSDDLFSLPYCPGKTLVVGASYVALECAGFLAGIGLDVTVM-VRSI 223
Query 122 IIPALDAEIGRAFQKLLERQGIKFM---FGTKVVGSQKAD-GGVTLSLENVKSGDASEVQ 177
++ D ++ + +E GIKF+ TK+ + G + ++ ++ S + E +
Sbjct 224 LLRGFDQDMANKIGEHMEEHGIKFIRQFVPTKIEQIEAGTPGRLRVTAQSTNSEETIEGE 283
Query 178 CDVVLVAVGRRPYTKDLGLEELGINLDNR-GRVGVN--EQMLVP 218
+ VL+AVGR T+ +GLE +G+ ++ + G++ V EQ VP
Sbjct 284 FNTVLLAVGRDSCTRTIGLETVGVKINEKTGKIPVTDEEQTNVP 327
> mmu:232223 Txnrd3, AI196535, TR2, Tgr; thioredoxin reductase
3 (EC:1.8.1.9); K00384 thioredoxin reductase (NADPH) [EC:1.8.1.9]
Length=652
Score = 77.0 bits (188), Expect = 5e-14, Method: Compositional matrix adjust.
Identities = 62/225 (27%), Positives = 106/225 (47%), Gaps = 12/225 (5%)
Query 2 VSALSQGIEGLFKRNKVDYLKGTGRLAGPHAVQVQPIDAGNPQMLMAKNVILATGSEPAP 61
+ +L+ G + V Y+ G H ++ G A ++ATG P
Sbjct 262 IGSLNWGYRVTLREKGVTYVNSFGEFVDLHKIKATN-KKGQETFYTASKFVIATGERPRY 320
Query 62 LAGGALEVDEETIVSSTGALALPRVPKHLVVVGGGVIGLELGSVWRNLGAEVTVVEFCDK 121
L ++ D+E ++S +LP P +VVG +GLE LG +VTV+
Sbjct 321 LG---IQGDKEYCITSDDLFSLPYCPGCTLVVGASYVGLECAGFLAGLGLDVTVM-VRSV 376
Query 122 IIPALDAEIGRAFQKLLERQGIKFMFGTKVVGSQKADGGVTLSLENV-KSGDASEVQ--- 177
++ D E+ LE+QG+KF + Q+ + G+ L+ V KS + E
Sbjct 377 LLRGFDQEMAEKVGSYLEQQGVKFQRKFTPILVQQLEKGLPGKLKVVAKSTEGPETVEGI 436
Query 178 CDVVLVAVGRRPYTKDLGLEELGINLDNR-GRVGVN--EQMLVPN 219
+ VL+A+GR T+ +GLE++G+ ++ + G++ VN EQ VP+
Sbjct 437 YNTVLLAIGRDSCTRKIGLEKIGVKINEKNGKIPVNDVEQTNVPH 481
> dre:553575 MGC110010; zgc:110010 (EC:1.8.1.7); K00383 glutathione
reductase (NADPH) [EC:1.8.1.7]
Length=425
Score = 76.6 bits (187), Expect = 6e-14, Method: Compositional matrix adjust.
Identities = 58/225 (25%), Positives = 104/225 (46%), Gaps = 24/225 (10%)
Query 2 VSALSQGIEGLFKRNKVDYLKGTGRLAGPHAVQVQPIDAGNPQMLMAKNVILATGSEPAP 61
VS L+Q ++ K++++ G R +P N + A +++++TG P+
Sbjct 95 VSRLNQIYRSNLEKGKIEFIHGYARFTD----DPEPTVEVNGKKYTATHILISTGGHPST 150
Query 62 LA-----GGALEVDEETIVSSTGALALPRVPKHLVVVGGGVIGLELGSVWRNLGAEVTVV 116
++ G +L + + G L PK V+VG G I +E+ + LG++ +++
Sbjct 151 VSEDDVPGSSLGI------TCDGFFELESCPKRSVIVGAGYIAVEMAGILSTLGSKTSII 204
Query 117 EFCDKIIPALDAEIGRAFQKLLERQGIKFMFGTKVVGSQKADGGVTLSLENVKSGDASEV 176
++ DA I K L+ GI T+V +K G++++L K D +
Sbjct 205 IRQGGVLRNFDALISSNCTKELQNNGIDLRKNTQVKSVKKNGKGLSITLV-TKDPDDKDS 263
Query 177 Q--------CDVVLVAVGRRPYTKDLGLEELGINLDNRGRVGVNE 213
Q D +L A+GR P T L L ++G+ LD RG + V+E
Sbjct 264 QEKFDTINDVDCLLWAIGREPNTAGLNLSQIGVKLDERGHIVVDE 308
> hsa:7296 TXNRD1, GRIM-12, MGC9145, TR, TR1, TRXR1, TXNR; thioredoxin
reductase 1 (EC:1.8.1.9); K00384 thioredoxin reductase
(NADPH) [EC:1.8.1.9]
Length=649
Score = 75.5 bits (184), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 58/224 (25%), Positives = 109/224 (48%), Gaps = 12/224 (5%)
Query 2 VSALSQGIEGLFKRNKVDYLKGTGRLAGPHAVQVQPIDAGNPQMLMAKNVILATGSEPAP 61
+ +L+ G + KV Y G+ GPH ++ + G ++ A+ ++ATG P
Sbjct 259 IGSLNWGYRVALREKKVVYENAYGQFIGPHRIKATN-NKGKEKIYSAERFLIATGERPRY 317
Query 62 LAGGALEVDEETIVSSTGALALPRVPKHLVVVGGGVIGLELGSVWRNLGAEVTVVEFCDK 121
L + D+E +SS +LP P +VVG + LE +G +VTV+
Sbjct 318 LG---IPGDKEYCISSDDLFSLPYCPGKTLVVGASYVALECAGFLAGIGLDVTVM-VRSI 373
Query 122 IIPALDAEIGRAFQKLLERQGIKFMFGTKVVGSQKADGGVTLSLENV-KSGDASEV---Q 177
++ D ++ + +E GIKF+ + ++ + G L V +S ++ E+ +
Sbjct 374 LLRGFDQDMANKIGEHMEEHGIKFIRQFVPIKVEQIEAGTPGRLRVVAQSTNSEEIIEGE 433
Query 178 CDVVLVAVGRRPYTKDLGLEELGINLDNR-GRVGVN--EQMLVP 218
+ V++A+GR T+ +GLE +G+ ++ + G++ V EQ VP
Sbjct 434 YNTVMLAIGRDACTRKIGLETVGVKINEKTGKIPVTDEEQTNVP 477
> dre:352924 txnrd1, TrxR1, cb682, fb83a08, wu:fb83a08; thioredoxin
reductase 1 (EC:1.8.1.9); K00384 thioredoxin reductase
(NADPH) [EC:1.8.1.9]
Length=602
Score = 73.6 bits (179), Expect = 6e-13, Method: Compositional matrix adjust.
Identities = 58/225 (25%), Positives = 106/225 (47%), Gaps = 12/225 (5%)
Query 2 VSALSQGIEGLFKRNKVDYLKGTGRLAGPHAVQVQPIDAGNPQMLMAKNVILATGSEPAP 61
+ +L+ G + V+Y+ PH ++ G A +LATG P
Sbjct 212 IGSLNWGYRVSLRDKNVNYVNAYAEFVEPHKIKATN-KRGKETFYTAAQFVLATGERPRY 270
Query 62 LAGGALEVDEETIVSSTGALALPRVPKHLVVVGGGVIGLELGSVWRNLGAEVTVVEFCDK 121
L + D+E ++S +LP P +VVG + LE G LG +VT++
Sbjct 271 LG---IPGDKEFCITSDDLFSLPYCPGKTLVVGASYVALECGGFLAGLGLDVTIM-VRSI 326
Query 122 IIPALDAEIGRAFQKLLERQGIKFM---FGTKVVGSQKAD-GGVTLSLENVKSGDASEVQ 177
++ D ++ + +E G+KF+ TK+ + G + ++ ++ +S + E +
Sbjct 327 LLRGFDQDMADRAGEYMETHGVKFLRKFVPTKIEQLEAGTPGRIKVTAKSTESEEVFEGE 386
Query 178 CDVVLVAVGRRPYTKDLGLEELGINLDNR-GRVGVN--EQMLVPN 219
+ VL+AVGR T +GL++ G+ ++ + G+V VN EQ VP+
Sbjct 387 YNTVLIAVGRDACTGKIGLDKAGVKINEKNGKVPVNDEEQTNVPH 431
> hsa:114112 TXNRD3, TGR, TR2, TRXR3; thioredoxin reductase 3
(EC:1.8.1.9); K00384 thioredoxin reductase (NADPH) [EC:1.8.1.9]
Length=607
Score = 72.4 bits (176), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 59/224 (26%), Positives = 104/224 (46%), Gaps = 12/224 (5%)
Query 2 VSALSQGIEGLFKRNKVDYLKGTGRLAGPHAVQVQPIDAGNPQMLMAKNVILATGSEPAP 61
+S+L+ G + V Y+ G H ++ G A ++ATG P
Sbjct 253 ISSLNWGYRLSLREKAVAYVNSYGEFVEHHKIKATN-KKGQETYYTAAQFVIATGERPRY 311
Query 62 LAGGALEVDEETIVSSTGALALPRVPKHLVVVGGGVIGLELGSVWRNLGAEVTVVEFCDK 121
L ++ D+E ++S +LP P +VVG + LE G +VTV+
Sbjct 312 LG---IQGDKEYCITSDDLFSLPYCPGKTLVVGASYVALECAGFLAGFGLDVTVM-VRSI 367
Query 122 IIPALDAEIGRAFQKLLERQGIKFMFGTKVVGSQKADGGVTLSLENV-KSGDASEV---Q 177
++ D E+ +E+ G+KF+ V Q+ + G L+ + KS + +E
Sbjct 368 LLRGFDQEMAEKVGSYMEQHGVKFLRKFIPVMVQQLEKGSPGKLKVLAKSTEGTETIEGV 427
Query 178 CDVVLVAVGRRPYTKDLGLEELGINLDNR-GRVGVN--EQMLVP 218
+ VL+A+GR T+ +GLE++G+ ++ + G++ VN EQ VP
Sbjct 428 YNTVLLAIGRDSCTRKIGLEKIGVKINEKSGKIPVNDVEQTNVP 471
> eco:b3500 gor, ECK3485, gorA, JW3467; glutathione oxidoreductase
(EC:1.8.1.7); K00383 glutathione reductase (NADPH) [EC:1.8.1.7]
Length=450
Score = 72.0 bits (175), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 60/217 (27%), Positives = 102/217 (47%), Gaps = 24/217 (11%)
Query 2 VSALSQGIEGLFKRNKVDYLKGTGRLAGPHAVQVQPIDAGNPQMLMAKNVILATGSEPA- 60
+ + E + +N VD +KG R ++V N + + A ++++ATG P+
Sbjct 92 IDRIHTSYENVLGKNNVDVIKGFARFVDAKTLEV------NGETITADHILIATGGRPSH 145
Query 61 PLAGGALEVDEETIVSSTGALALPRVPKHLVVVGGGVIGLELGSVWRNLGAEVTVVEFCD 120
P G E + S G ALP +P+ + VVG G I +EL V LGA+ + F
Sbjct 146 PDIPGV-----EYGIDSDGFFALPALPERVAVVGAGYIAVELAGVINGLGAKTHL--FVR 198
Query 121 KIIP--ALDAEIGRAFQKLLERQGIKFMFGT--KVVGSQKADGGVTLSLENVKSGDASEV 176
K P + D I +++ +G + K V + DG +TL LE+ G + V
Sbjct 199 KHAPLRSFDPMISETLVEVMNAEGPQLHTNAIPKAV-VKNTDGSLTLELED---GRSETV 254
Query 177 QCDVVLVAVGRRPYTKDLGLEELGINLDNRGRVGVNE 213
C ++ A+GR P ++ LE G+ + +G + V++
Sbjct 255 DC--LIWAIGREPANDNINLEAAGVKTNEKGYIVVDK 289
> cpv:cgd2_4320 thioredoxin reductase 1 ; K00384 thioredoxin reductase
(NADPH) [EC:1.8.1.9]
Length=526
Score = 71.2 bits (173), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 50/196 (25%), Positives = 90/196 (45%), Gaps = 18/196 (9%)
Query 18 VDYLKGTGRLAGPHAVQVQPIDAGNPQMLMAKNVILATGSEPA--PLAGGALEVDEETIV 75
V+Y+ +L PH+V+ + D G + + ++ ++LATG P+ GA++ +
Sbjct 144 VEYINALAKLIDPHSVEYE--DNGQKKTITSRYILLATGGRPSIPETVPGAIQYS----I 197
Query 76 SSTGALALPRVPKHLVVVGGGVIGLELGSVWRNLGAEVTVVEFCDKIIP--ALDAEIGRA 133
+S L + P +V+G IGLE LG + TV + IP D +
Sbjct 198 TSDDIFFLSKSPGKTLVIGASYIGLETAGFLNELGFDTTVAM---RSIPLRGFDRQCSEK 254
Query 134 FQKLLERQGIKFMFGTKVVGSQKADGGVTLSLENVKSGDASEVQCDVVLVAVGRRPYTKD 193
+ ++ G KF+ G + +K + + +S D S + + VL A GR P K
Sbjct 255 IVEYMKATGTKFLVGVVPINIEKVNENIKVSF-----SDGSVEEFETVLYATGRNPDVKG 309
Query 194 LGLEELGINLDNRGRV 209
L L +G+ + + G++
Sbjct 310 LNLNAIGVEVSDSGKI 325
> hsa:2936 GSR, MGC78522; glutathione reductase (EC:1.8.1.7);
K00383 glutathione reductase (NADPH) [EC:1.8.1.7]
Length=522
Score = 69.3 bits (168), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 49/175 (28%), Positives = 90/175 (51%), Gaps = 15/175 (8%)
Query 48 AKNVILATGSEPAP-----LAGGALEVDEETIVSSTGALALPRVPKHLVVVGGGVIGLEL 102
A ++++ATG P+ + G +L + +S G L +P V+VG G I +E+
Sbjct 193 APHILIATGGMPSTPHESQIPGASLGI------TSDGFFQLEELPGRSVIVGAGYIAVEM 246
Query 103 GSVWRNLGAEVTVVEFCDKIIPALDAEIGRAFQKLLERQGIKFMFGTKVVGSQKADGGVT 162
+ LG++ +++ DK++ + D+ I + LE G++ + ++V +K G+
Sbjct 247 AGILSALGSKTSLMIRHDKVLRSFDSMISTNCTEELENAGVEVLKFSQVKEVKKTLSGLE 306
Query 163 LSLENVKSGDASEV----QCDVVLVAVGRRPYTKDLGLEELGINLDNRGRVGVNE 213
+S+ G + D +L A+GR P TKDL L +LGI D++G + V+E
Sbjct 307 VSMVTAVPGRLPVMTMIPDVDCLLWAIGRVPNTKDLSLNKLGIQTDDKGHIIVDE 361
> cel:F20D6.11 hypothetical protein
Length=549
Score = 68.6 bits (166), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 62/217 (28%), Positives = 101/217 (46%), Gaps = 26/217 (11%)
Query 10 EGLFKRNKVDYLKGTGRLAGPHAVQVQPIDAGNPQMLMAKNVILATGSEPAPLA--GGAL 67
+ ++ V +L T +A H + + + N + ++ +I+ATG L G L
Sbjct 204 DAFYEERNVKFLLKTSVIAVNH--KSREVSLSNGETVVYSKLIIATGGNVRKLQVPGSDL 261
Query 68 E-------VDEETIVSSTGALALPRVPKHLVVVGGGVIGLELGSVWRNLGAEVTVVEFCD 120
+ V+E I+S+ KH+V VG IG+E+ S A VTV+
Sbjct 262 KNICYLRKVEEANIISNLHP------GKHVVCVGSSFIGMEVASALAEKAASVTVISNTP 315
Query 121 KIIPALDAEIGRAFQKLLERQGIKFMFGTKVVGSQKADGG--VTLSLENVKSGDASEVQC 178
+ +P ++IG+ + E +G+KF VV + D G + LEN K E+
Sbjct 316 EPLPVFGSDIGKGIRLKFEEKGVKFELAANVVALRGNDQGEVSKVILENGK-----ELDV 370
Query 179 DVVLVAVGRRPYTKDLGLEELGINLDNRGRVGVNEQM 215
D+++ +G P TK LE GI LDNRG + V+E+
Sbjct 371 DLLVCGIGVTPATK--FLEGSGIKLDNRGFIEVDEKF 405
> xla:447610 txnrd1, MGC85342; thioredoxin reductase 1 (EC:1.8.1.9);
K00384 thioredoxin reductase (NADPH) [EC:1.8.1.9]
Length=531
Score = 68.6 bits (166), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 55/210 (26%), Positives = 100/210 (47%), Gaps = 11/210 (5%)
Query 15 RNKVDYLKGTGRLAGPHAVQVQPIDAGNPQMLMAKNVILATGSEPAPLAGGALEVDEETI 74
N V Y G G GP+ ++ G + A+ ++ATG P L + D+E
Sbjct 157 ENNVKYENGYGEFVGPNTIKSTN-SRGKSKYFTAEKFLIATGERPRYLG---IPGDKEYC 212
Query 75 VSSTGALALPRVPKHLVVVGGGVIGLELGSVWRNLGAEVTVVEFCDKIIPALDAEIGRAF 134
++S +L P +VVG + LE LG +VTV+ ++ D ++
Sbjct 213 ITSDDLFSLTYCPGKTLVVGASYVALECAGFLAGLGLDVTVM-VRSILLRGFDQQMANKI 271
Query 135 QKLLERQGIKFMFGTKVVGSQKADGGVTLSLE-NVKSGDASEV--QCDVVLVAVGRRPYT 191
+ +E G+KF+ ++ + G+ L+ ++ D +E + + VL+A+GR T
Sbjct 272 GEYMEEHGVKFIRQFVPTKIEQIEAGMPGRLKVTSQAPDGTETTDEYNTVLLAIGRDACT 331
Query 192 KDLGLEELGINLDNR-GRVGVN--EQMLVP 218
+++GLE G+ ++ + G++ VN EQ VP
Sbjct 332 RNIGLEIPGVKINEKTGKIPVNDEEQTNVP 361
> cel:C46F11.2 hypothetical protein
Length=473
Score = 67.8 bits (164), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 59/207 (28%), Positives = 97/207 (46%), Gaps = 18/207 (8%)
Query 10 EGLFKRNKVDYLKGTGRLAGPHAVQVQPIDAGNPQMLMAKNVILATGSEPA-PLAGGALE 68
E K + V+Y++G A V+V N KN ++A G +P P GA
Sbjct 115 ESGLKGSSVEYIRGRATFAEDGTVEV------NGAKYRGKNTLIAVGGKPTIPNIKGA-- 166
Query 69 VDEETIVSSTGALALPRVPKHLVVVGGGVIGLELGSVWRNLGAEVTVVEFCDKIIPALDA 128
E + S G L +P VVVG G I +E+ V NLG++ ++ DK++ D
Sbjct 167 ---EHGIDSDGFFDLEDLPSRTVVVGAGYIAVEIAGVLANLGSDTHLLIRYDKVLRTFDK 223
Query 129 EIGRAFQKLLERQG--IKFMFGTKVVGSQKADGGVTLSLENVKSGDASEVQCDVVLVAVG 186
+ ++ + + T+V K D G+ L+++ +G +VQ ++ A+G
Sbjct 224 MLSDELTADMDEETNPLHLHKNTQVTEVIKGDDGL-LTIKTT-TGVIEKVQ--TLIWAIG 279
Query 187 RRPYTKDLGLEELGINLDNRGRVGVNE 213
R P TK+L LE +G+ D G + V+E
Sbjct 280 RDPLTKELNLERVGVKTDKSGHIIVDE 306
> xla:447484 txnrd3, MGC81848; thioredoxin reductase 3 (EC:1.8.1.9);
K00384 thioredoxin reductase (NADPH) [EC:1.8.1.9]
Length=596
Score = 67.8 bits (164), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 55/225 (24%), Positives = 103/225 (45%), Gaps = 12/225 (5%)
Query 2 VSALSQGIEGLFKRNKVDYLKGTGRLAGPHAVQVQPIDAGNPQMLMAKNVILATGSEPAP 61
+ +L+ G + +V Y G H ++ G A+ ++ATG P
Sbjct 208 IGSLNWGYRVALRDKQVRYENAYGEFVESHKIKATN-KKGKESFFTAEKFVVATGERPRY 266
Query 62 LAGGALEVDEETIVSSTGALALPRVPKHLVVVGGGVIGLELGSVWRNLGAEVTVVEFCDK 121
L + D+E ++S +LP P +VVG + LE +G + TV+
Sbjct 267 LN---IPGDKEYCITSDDLFSLPYCPGKTLVVGASYVALECAGFLAGIGLDATVM-VRSI 322
Query 122 IIPALDAEIGRAFQKLLERQGIKFM---FGTKV-VGSQKADGGVTLSLENVKSGDASEVQ 177
+ D E+ +E G+KF+ KV + + G + ++ ++ + E +
Sbjct 323 FLRGFDQEMANRAGAYMETHGVKFIKQFVPIKVELLEEGTPGRIKVTAKSTQGDQIIEDE 382
Query 178 CDVVLVAVGRRPYTKDLGLEELGINLDNR-GRVGVN--EQMLVPN 219
+ VL+AVGR T+++GLE++G+ ++ R G++ V+ EQ VP+
Sbjct 383 YNTVLIAVGRDACTRNIGLEKIGVKINERNGKIPVSDEEQTSVPH 427
> mmu:14782 Gsr, AI325518, D8Ertd238e, Gr-1, Gr1; glutathione
reductase (EC:1.8.1.7); K00383 glutathione reductase (NADPH)
[EC:1.8.1.7]
Length=500
Score = 67.4 bits (163), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 57/222 (25%), Positives = 104/222 (46%), Gaps = 21/222 (9%)
Query 2 VSALSQGIEGLFKRNKVDYLKGTGRLA-GPHAVQVQPIDAGNPQMLMAKNVILATGSEP- 59
VS L+ + ++ ++ + G A GP +P N + A ++++ATG P
Sbjct 129 VSRLNTIYQNNLTKSHIEIIHGYATFADGP-----RPTVEVNGKKFTAPHILIATGGVPT 183
Query 60 ----APLAGGALEVDEETIVSSTGALALPRVPKHLVVVGGGVIGLELGSVWRNLGAEVTV 115
+ + G +L + +S G L +P V+VG G I +E+ + LG++ ++
Sbjct 184 VPHESQIPGASLGI------TSDGFFQLEDLPSRSVIVGAGYIAVEIAGILSALGSKTSL 237
Query 116 VEFCDKIIPALDAEIGRAFQKLLERQGIKFMFGTKVVGSQKADGGVTL----SLENVKSG 171
+ DK++ D+ I + LE G++ + T+V +K G+ L S+ K
Sbjct 238 MIRHDKVLRNFDSLISSNCTEELENAGVEVLKFTQVKEVKKTSSGLELQVVTSVPGRKPT 297
Query 172 DASEVQCDVVLVAVGRRPYTKDLGLEELGINLDNRGRVGVNE 213
D +L A+GR P +K L L ++GI D +G + V+E
Sbjct 298 TTMIPDVDCLLWAIGRDPNSKGLNLNKVGIQTDEKGHILVDE 339
> tgo:TGME49_019130 glutathione reductase, putative (EC:1.8.1.7)
Length=505
Score = 67.0 bits (162), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 66/243 (27%), Positives = 103/243 (42%), Gaps = 47/243 (19%)
Query 2 VSALSQGIEGLFKRNKVDYLKGTGRLAGP------------------HAVQVQPIDAGNP 43
VS L L K V +G GRL HAV +Q ++ G
Sbjct 66 VSRLRDTFARLLKEANVTVYRGVGRLDASFDRTGKSGNSSAQRCRPRHAVLIQTLE-GKV 124
Query 44 QMLMAKNVILATGSEPAPL-AGGALEVDEETIVSSTGALALPRVPKHLVVVGGGVIGLEL 102
Q + A +V++ATG+ L GA E +SS G + +P+ + ++G G + EL
Sbjct 125 QRVTANHVLIATGTRRQVLDIPGA-----EFAISSDGFFQIQHLPRRVALIGAGYVSAEL 179
Query 103 GSVWRNLGAEVTVVEFCDKIIPALDAEIGRAFQKLLERQGIKFMFGTKVV--GSQKADGG 160
G + R+LG +V++ + + D E + + GI+ G V KADG
Sbjct 180 GGILRHLGVDVSIFMRNQRQLKRFDKEAVESLEATQRASGIQLYKGVNAVEISISKADGT 239
Query 161 -------------VTLSLE------NVKSGDASEVQCDVVLVAVGRRPYTKDLGLEELGI 201
+ LE ++ +GDA D V++AV P +DLGLEE G+
Sbjct 240 KLSASYTFHDQTETSYHLEDSLLTVHLDNGDAHH-GFDHVIMAVNPAPAIEDLGLEEAGV 298
Query 202 NLD 204
++D
Sbjct 299 DID 301
> tgo:TGME49_046920 glutathione reductase, putative (EC:1.8.1.7);
K00383 glutathione reductase (NADPH) [EC:1.8.1.7]
Length=483
Score = 66.2 bits (160), Expect = 9e-11, Method: Compositional matrix adjust.
Identities = 54/211 (25%), Positives = 97/211 (45%), Gaps = 19/211 (9%)
Query 13 FKRNKVDYLKGTGRLAGP---------HAVQVQPIDAGNPQMLMAKNVILATGSEPAPLA 63
K + V + R A P HA+ ++ D GN + + A +V++A+G PA
Sbjct 111 LKNSGVTFFPAYARFAKPEAKTDGGLAHAIVLKSAD-GNEETVTADHVLIASGGRPAK-- 167
Query 64 GGALEVDEETIVSSTGALALPRVPKHLVVVGGGVIGLELGSVWRNLGAEVTVVEFCDKII 123
+E +E TI +S G L +P+ + ++G G I +E V+ + E + ++ +
Sbjct 168 -AGIEGEEHTI-NSDGFFELEEMPQKVALLGAGYIAVEFAGVFAAMKCETHLFVRHERAL 225
Query 124 PALDAEIGRAFQKLLERQGIKFM-FGTKVVGSQKADGGVTLSLENVKSGDASEVQCDVVL 182
D I + + + G++ Q+AD +TL L N +S D V+
Sbjct 226 RKFDDMISMRVDEFMRKAGVQIHPHSVAKAVRQEADKSLTLELTNGESFRGF----DSVI 281
Query 183 VAVGRRPYTKDLGLEELGINLDNRGRVGVNE 213
V+VGR P +LGL+ +G+ + G + +E
Sbjct 282 VSVGRVPEVANLGLDVVGVKQRHGGYIVADE 312
> sce:YPL091W GLR1, LPG17; Cytosolic and mitochondrial glutathione
oxidoreductase, converts oxidized glutathione to reduced
glutathione; mitochondrial but not cytosolic form has a role
in resistance to hyperoxia (EC:1.8.1.7); K00383 glutathione
reductase (NADPH) [EC:1.8.1.7]
Length=483
Score = 64.3 bits (155), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 61/227 (26%), Positives = 105/227 (46%), Gaps = 23/227 (10%)
Query 4 ALSQGIEGLFKRN----KVDYLKGTGRLAGPHAVQVQPIDAGNPQMLMAKNVILATGSE- 58
A + G++++N KVD + G R V+VQ D ++ A ++++ATG +
Sbjct 114 AYVHRLNGIYQKNLEKEKVDVVFGWARFNKDGNVEVQKRD-NTTEVYSANHILVATGGKA 172
Query 59 --PAPLAGGALEVDEETIVSSTGALALPRVPKHLVVVGGGVIGLELGSVWRNLGAEVTVV 116
P + G L D S G L PK +VVVG G IG+EL V+ LG+E +V
Sbjct 173 IFPENIPGFELGTD------SDGFFRLEEQPKKVVVVGAGYIGIELAGVFHGLGSETHLV 226
Query 117 EFCDKIIPALDAEIGRAFQKLLERQGIKFMFGTKVVGSQK--ADGGVTLSLENVKSGDAS 174
+ ++ D I ++GI +K+V +K + + + + KS D
Sbjct 227 IRGETVLRKFDECIQNTITDHYVKEGINVHKLSKIVKVEKNVETDKLKIHMNDSKSID-- 284
Query 175 EVQCDVVLVAVGRRPYTKDLGLEELGINLDNRGRVGVNE--QMLVPN 219
D ++ +GR+ + +G E +GI L++ ++ +E VPN
Sbjct 285 --DVDELIWTIGRKSHL-GMGSENVGIKLNSHDQIIADEYQNTNVPN 328
> eco:b0304 ykgC, ECK0303, JW5040; predicted pyridine nucleotide-disulfide
oxidoreductase
Length=441
Score = 62.8 bits (151), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 57/215 (26%), Positives = 103/215 (47%), Gaps = 23/215 (10%)
Query 13 FKRNK----------VDYLKGTGRLAGPHAVQVQPIDAGNPQMLMAKNVILATGSEPA-- 60
F RNK +D + G H+++V + GN + + + + + TG++
Sbjct 77 FLRNKNFHNLADMPNIDVIDGQAEFINNHSLRVHRPE-GNLE-IHGEKIFINTGAQTVVP 134
Query 61 PLAGGALEVDEETIVSSTGALALPRVPKHLVVVGGGVIGLELGSVWRNLGAEVTVVEFCD 120
P+ G + STG L L +P HL ++GGG IG+E S++ N G++VT++E
Sbjct 135 PIPGITT---TPGVYDSTGLLNLKELPGHLGILGGGYIGVEFASMFANFGSKVTILEAAS 191
Query 121 KIIPALDAEIGRAFQKLLERQGIKFMFGTKVVGSQKADGGVTLSLENVKSGDASEVQCDV 180
+P D +I +L QG+ + V + V + E+ +++ D
Sbjct 192 LFLPREDRDIADNIATILRDQGVDIILNAHVERISHHENQVQVHSEH------AQLAVDA 245
Query 181 VLVAVGRRPYTKDLGLEELGINLDNRGRVGVNEQM 215
+L+A GR+P T L E GI ++ RG + V++++
Sbjct 246 LLIASGRQPATASLHPENAGIAVNERGAIVVDKRL 280
> xla:734372 gsr, MGC84926; glutathione reductase (EC:1.8.1.7);
K00383 glutathione reductase (NADPH) [EC:1.8.1.7]
Length=476
Score = 61.6 bits (148), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 58/221 (26%), Positives = 109/221 (49%), Gaps = 19/221 (8%)
Query 2 VSALSQGIEGLFKRNKVDYLKGTGRLAGPHAVQVQPIDAGNPQMLMAKNVILATGSEP-- 59
VS L+ + ++ +++ ++G +P N Q A ++++ATG +P
Sbjct 105 VSRLNDIYQNNLQKAQIEIIRGNANFTS----DPEPTVEVNGQKYSAPHILIATGGKPSM 160
Query 60 ---APLAGGALEVDEETIVSSTGALALPRVPKHLVVVGGGVIGLELGSVWRNLGAEVTVV 116
A L G +L + +S G L +P+ +VVG G I +E+ + LG++ +++
Sbjct 161 PSDAELPGASLGI------TSDGFFELTDLPRRSIVVGAGYIAVEIAGILSALGSKASLL 214
Query 117 EFCDKIIPALDAEIGRAFQKLLERQGIKFMFGTKVVGSQKADGGVTLSLENVKSGDASEV 176
DK++ D+ I + LE G++ +V +K+ G+ ++++ G V
Sbjct 215 IRQDKVLRTFDSIISSNCTEELENAGVEVWKYAQVKSVKKSTTGLEINVQCSMPGRKPTV 274
Query 177 QC----DVVLVAVGRRPYTKDLGLEELGINLDNRGRVGVNE 213
+ D +L A+GR P T+ LGLE LG+ LD +G + V+E
Sbjct 275 RTIQDVDCLLWAIGRDPNTEYLGLENLGLELDEKGHIVVDE 315
> hsa:150209 AIFM3, AIFL, FLJ30473, FLJ45137; apoptosis-inducing
factor, mitochondrion-associated, 3
Length=598
Score = 59.3 bits (142), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 56/180 (31%), Positives = 88/180 (48%), Gaps = 11/180 (6%)
Query 46 LMAKNVILATGSEPAPLAGGALEVDE-ETIVSSTGALALPRVPK--HLVVVGGGVIGLEL 102
L ++LA GS P L+ EV+ TI + A + R+ + ++VVVG G +G+E+
Sbjct 288 LEYSKLLLAPGSSPKTLSCKGKEVENVFTIRTPEDANRVVRLARGRNVVVVGAGFLGMEV 347
Query 103 GSVWRNLGAEVTVVEFCDKIIPALDAE-IGRAFQKLLERQGIKFMFGTKVVGSQKADGGV 161
+ V+VVE + E +GRA K+ E +KF T+V + +G
Sbjct 348 AAYLTEKAHSVSVVELEETPFRRFLGERVGRALMKMFENNRVKFYMQTEVSELRGQEG-- 405
Query 162 TLSLENVKSGDASEVQCDVVLVAVGRRPYTKDLGLEELGINLDNRGRVGVNEQMLVPNYP 221
L+ V + V+ DV +V +G P T L + GI LD+RG + VN +M+ N P
Sbjct 406 --KLKEVVLKSSKVVRADVCVVGIGAVPATG--FLRQSGIGLDSRGFIPVN-KMMQTNVP 460
> tgo:TGME49_109730 thioredoxin reductase, putative (EC:1.8.1.7);
K00384 thioredoxin reductase (NADPH) [EC:1.8.1.9]
Length=662
Score = 58.9 bits (141), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 49/211 (23%), Positives = 93/211 (44%), Gaps = 10/211 (4%)
Query 2 VSALSQGIEGLFKRNKVDYLKGTGRLAGPHAVQVQPIDAGNPQMLMAKNVILATGSEPAP 61
+ +L+ G ++ V Y+ + PH ++ G ++ A+N+++A G P
Sbjct 257 IKSLNFGYRTGLRKAGVTYINAYAKFVSPH--ELAYTFRGEDKICKARNIVVAVGGRPH- 313
Query 62 LAGGALEVDEETIVSSTGALALPRVPKHLVVVGGGVIGLELGSVWRNLGAEVTVVEFCDK 121
+E +E ++S +L + P + VG I LE R LG +VTV
Sbjct 314 -IPEEVEGAKELAITSDDIFSLKQAPNKTLCVGASYISLECAGFLRELGFDVTVAVR-SI 371
Query 122 IIPALDAEIGRAFQKLLERQGIKFMFGT---KVVGSQKADGGVTLSLENVKSGDASEVQC 178
++ D + LE G++ + T K+V ++A+G + ++ + K +
Sbjct 372 LLRGFDRQCAEQVGLCLEEAGVRILRETIPAKMV--KQANGKIQVTFQVGKEKKELVEEF 429
Query 179 DVVLVAVGRRPYTKDLGLEELGINLDNRGRV 209
D VL A GR+ T +L L+ G+ G++
Sbjct 430 DTVLYATGRKADTSNLNLQAAGVETTETGKI 460
> xla:379744 txnrd2, MGC69182; thioredoxin reductase 2 (EC:1.8.1.9);
K00384 thioredoxin reductase (NADPH) [EC:1.8.1.9]
Length=504
Score = 58.9 bits (141), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 57/226 (25%), Positives = 97/226 (42%), Gaps = 16/226 (7%)
Query 2 VSALSQGIEGLFKRNKVDYLKGTGRLAGPHAVQVQPIDAGNPQMLMAKNVILATGSEPA- 60
V +L+ G + KV Y H ++ AG ++ A+N+++ATG P
Sbjct 116 VKSLNWGHRIQLQDKKVKYFNLKANFVDEHCIR-GVTKAGKETLVTAQNIVIATGGRPKY 174
Query 61 -PLAGGALEVDEETIVSSTGALALPRVPKHLVVVGGGVIGLELGSVWRNLGAEVTVVEFC 119
GALE ++S L P +VVG + LE +G T +
Sbjct 175 PTHVPGALEYG----ITSDDLFWLKESPGKTLVVGASYVSLECAGFLTGIGLNTTAMV-- 228
Query 120 DKIIP--ALDAEIGRAFQKLLERQGIKFMFGTKVVGSQK-ADGGVTLSLENVKSGDASEV 176
+ IP D ++ +E G KF++ +K +G + ++ +N +SG
Sbjct 229 -RSIPLRGFDQQMAYLVADYMESHGTKFLWKCTPSHVEKLKNGKLQVTWKNTQSGKEGVD 287
Query 177 QCDVVLVAVGRRPYTKDLGLEELGINLD-NRGRVGVN--EQMLVPN 219
D V+ AVGR T+ L LE++G+ + G++ V+ E VP+
Sbjct 288 IYDTVMWAVGRAAETQYLNLEKVGVKIKPETGKIIVDASEATSVPH 333
> mmu:26462 Txnrd2, AA118373, ESTM573010, TGR, Tr3, Trxr2, Trxrd2;
thioredoxin reductase 2 (EC:1.8.1.9); K00384 thioredoxin
reductase (NADPH) [EC:1.8.1.9]
Length=527
Score = 57.8 bits (138), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 59/227 (25%), Positives = 97/227 (42%), Gaps = 18/227 (7%)
Query 2 VSALSQGIEGLFKRNKVDYLKGTGRLAGPHAVQVQPID-AGNPQMLMAKNVILATGSEP- 59
V +L+ G + KV Y H V+ +D G +L A+++++ATG P
Sbjct 139 VKSLNWGHRVQLQDRKVKYFNIKASFVDEHTVR--GVDKGGKATLLSAEHIVIATGGRPR 196
Query 60 -APLAGGALEVDEETIVSSTGALALPRVPKHLVVVGGGVIGLELGSVWRNLGAEVTVVEF 118
GALE ++S L P +VVG + LE +G + TV+
Sbjct 197 YPTQVKGALEYG----ITSDDIFWLKESPGKTLVVGASYVALECAGFLTGIGLDTTVMM- 251
Query 119 CDKIIP--ALDAEIGRAFQKLLERQGIKFMFGTKVVGSQK-ADGGVTLSLENVKSGDASE 175
+ IP D ++ + +E G +F+ G +K + ++ E+ SG
Sbjct 252 --RSIPLRGFDQQMSSLVTEHMESHGTQFLKGCVPSHIKKLPTNQLQVTWEDHASGKEDT 309
Query 176 VQCDVVLVAVGRRPYTKDLGLEELGINLDNRGR---VGVNEQMLVPN 219
D VL A+GR P T+ L LE+ GI+ + + + V E VP+
Sbjct 310 GTFDTVLWAIGRVPETRTLNLEKAGISTNPKNQKIIVDAQEATSVPH 356
> pfa:PFI1170c thioredoxin reductase (EC:1.8.1.9); K00384 thioredoxin
reductase (NADPH) [EC:1.8.1.9]
Length=617
Score = 57.4 bits (137), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 51/209 (24%), Positives = 88/209 (42%), Gaps = 9/209 (4%)
Query 14 KRNKVDYLKGTGRLAGPHAVQVQPI-DAGNPQMLMAKNVILATGSEPAPLAGGALEVDEE 72
+ +KV Y+ G +L + V D + + K +++ATG P +E +E
Sbjct 226 RSSKVKYINGLAKLKDKNTVSYYLKGDLSKEETVTGKYILIATGCRPH--IPDDVEGAKE 283
Query 73 TIVSSTGALALPRVPKHLVVVGGGVIGLELGSVWRNLGAEVTVVEFCDKIIPALDAEIGR 132
++S +L + P +VVG + LE +LG +VTV ++ D +
Sbjct 284 LSITSDDIFSLKKDPGKTLVVGASYVALECSGFLNSLGYDVTVA-VRSIVLRGFDQQCAV 342
Query 133 AFQKLLERQGIKFMFGTKVVGSQKADGGVTLSLENVKSGDASEVQCDVVLVAVGRRPYTK 192
+ +E QG+ F G K D + + S SE+ D VL A+GR+
Sbjct 343 KVKLYMEEQGVMFKNGILPKKLTKMDDKILVEF----SDKTSELY-DTVLYAIGRKGDID 397
Query 193 DLGLEELGINLDNRGRVGVNEQMLVPNYP 221
L LE L +N++ + + + N P
Sbjct 398 GLNLESLNMNVNKSNNKIIADHLSCTNIP 426
> xla:394337 aifm3, MGC84340, nfrl-A; apoptosis-inducing factor,
mitochondrion-associated, 3
Length=605
Score = 56.6 bits (135), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 59/203 (29%), Positives = 102/203 (50%), Gaps = 24/203 (11%)
Query 33 VQVQPIDAGNPQMLMAKN--------VILATGSEPAPLAGGALEVDEE-TIVSSTGALAL 83
QV +D N +++M K+ +++ATGS P L E+D TI + A +
Sbjct 268 TQVVSVDTKN-KIVMFKDGFRMEYNKLLIATGSTPKTLTCKGKELDNVITIRTPEDANKV 326
Query 84 PRV--PKHLVVVGGGVIGLELGSVWRNLGAEVTVVEFCDKIIPA---LDAEIGRAFQKLL 138
R+ K+ V+VG +G+E+ + V+VVE + IP L ++G A K+
Sbjct 327 VRLASSKNAVIVGASFLGMEVAAYLCEKAHSVSVVELEN--IPFKKFLGEKVGLAIMKMF 384
Query 139 ERQGIKFMFGTKVVGSQKADGGVTLSLENVKSGDASEVQCDVVLVAVGRRPYTKDLGLEE 198
E +KF T+V ++ +G + + +KSG ++ DV ++ +G P T L++
Sbjct 385 ENNRVKFYMQTEVSELREQEGKLKEVV--LKSGKV--LRADVCVIGIGASPTTG--FLKQ 438
Query 199 LGINLDNRGRVGVNEQMLVPNYP 221
G+ LD+RG + VN +M+ N P
Sbjct 439 SGVALDSRGYIPVN-KMMQTNIP 460
> mmu:72168 Aifm3, 2810401C16Rik, AI840249, Aifl; apoptosis-inducing
factor, mitochondrion-associated 3
Length=598
Score = 55.5 bits (132), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 57/199 (28%), Positives = 93/199 (46%), Gaps = 18/199 (9%)
Query 34 QVQPIDAGNPQM-------LMAKNVILATGSEPAPLAGGALEVDE-ETIVSSTGALALPR 85
QV +D N ++ L ++LA GS P L +V+ TI + A + R
Sbjct 269 QVVTVDVRNKKVVFKDGFKLEYSKLLLAPGSSPKTLTCKGKDVENVFTIRTPEDANRVLR 328
Query 86 VPK--HLVVVGGGVIGLELGSVWRNLGAEVTVVEFCDKIIPALDAE-IGRAFQKLLERQG 142
+ + + VVVG G +G+E+ + V+VVE + E +GRA K+ E
Sbjct 329 LARGRNAVVVGAGFLGMEVAAYLTEKAHSVSVVELEETPFRRFLGERVGRALMKMFENNR 388
Query 143 IKFMFGTKVVGSQKADGGVTLSLENVKSGDASEVQCDVVLVAVGRRPYTKDLGLEELGIN 202
+KF T+V + +G L+ V + ++ DV ++ +G P T L + GI
Sbjct 389 VKFYMQTEVSELRAQEG----KLQEVVLKSSKVLRADVCVLGIGAVPATG--FLRQSGIG 442
Query 203 LDNRGRVGVNEQMLVPNYP 221
LD+RG + VN +M+ N P
Sbjct 443 LDSRGFIPVN-KMMQTNVP 460
> hsa:10587 TXNRD2, SELZ, TR, TR-BETA, TR3, TRXR2; thioredoxin
reductase 2 (EC:1.8.1.9); K00384 thioredoxin reductase (NADPH)
[EC:1.8.1.9]
Length=524
Score = 55.1 bits (131), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 52/206 (25%), Positives = 86/206 (41%), Gaps = 13/206 (6%)
Query 2 VSALSQGIEGLFKRNKVDYLKGTGRLAGPHAVQVQPIDAGNPQMLMAKNVILATGSEPAP 61
V +L+ G + KV Y H V G +L A ++I+ATG P
Sbjct 136 VKSLNWGHRVQLQDRKVKYFNIKASFVDEHTV-CGVAKGGKEILLSADHIIIATGGRPRY 194
Query 62 LAG--GALEVDEETIVSSTGALALPRVPKHLVVVGGGVIGLELGSVWRNLGAEVTVVEFC 119
GALE ++S L P +VVG + LE +G + T++
Sbjct 195 PTHIEGALEYG----ITSDDIFWLKESPGKTLVVGASYVALECAGFLTGIGLDTTIMM-- 248
Query 120 DKIIP--ALDAEIGRAFQKLLERQGIKFMFGTKVVGSQK-ADGGVTLSLENVKSGDASEV 176
+ IP D ++ + + G +F+ G ++ DG + ++ E+ +G
Sbjct 249 -RSIPLRGFDQQMSSMVIEHMASHGTRFLRGCAPSRVRRLPDGQLQVTWEDSTTGKEDTG 307
Query 177 QCDVVLVAVGRRPYTKDLGLEELGIN 202
D VL A+GR P T+ L LE+ G++
Sbjct 308 TFDTVLWAIGRVPDTRSLNLEKAGVD 333
> cel:C06G3.7 trxr-1; ThioRedoXin Reductase family member (trxr-1);
K00384 thioredoxin reductase (NADPH) [EC:1.8.1.9]
Length=667
Score = 53.5 bits (127), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 54/225 (24%), Positives = 92/225 (40%), Gaps = 18/225 (8%)
Query 2 VSALSQGIEGLFKRNKVDYLKGTGRLAGPHAVQVQPIDAGNPQMLMAKNVILATGSEPA- 60
+++L+ G + V Y+ G GP + + L A +++TG P
Sbjct 270 IASLNWGYRVQLREKTVTYINSYGEFTGPFEISATN-KKKKVEKLTADRFLISTGLRPKY 328
Query 61 PLAGGALEVDEETIVSSTGALALPRVPKHLVVVGGGVIGLELGSVWRNLGAEVTVVEFCD 120
P G +E ++S LP P + VG + LE G +VTV+
Sbjct 329 PEIPGV----KEYTITSDDLFQLPYSPGKTLCVGASYVSLECAGFLHGFGFDVTVM-VRS 383
Query 121 KIIPALDAEIGRAFQKLLERQGIKFMFGTKVVGSQ---KADGG-----VTLSLENVKSGD 172
++ D ++ +K + G+KF G Q K D V +N ++G+
Sbjct 384 ILLRGFDQDMAERIRKHMIAYGMKFEAGVPTRIEQIDEKTDEKAGKYRVFWPKKNEETGE 443
Query 173 ASEV--QCDVVLVAVGRRPYTKDLGLEELGINLDNRGRV-GVNEQ 214
EV + + +L+A+GR T D+GL +G+ +V G EQ
Sbjct 444 MQEVSEEYNTILMAIGREAVTDDVGLTTIGVERAKSKKVLGRREQ 488
Lambda K H
0.316 0.138 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 7266557660
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40