bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_0327_orf2
Length=168
Score E
Sequences producing significant alignments: (Bits) Value
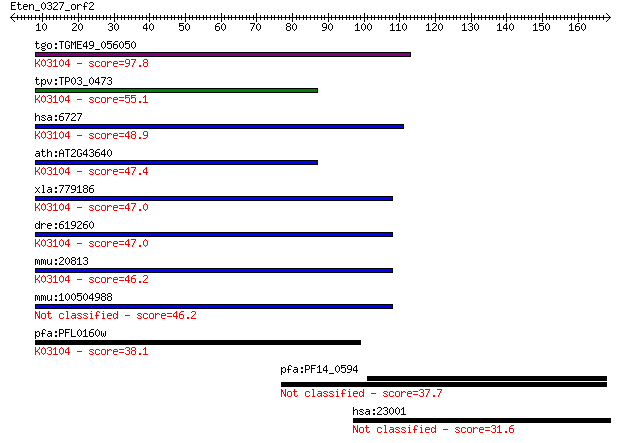
tgo:TGME49_056050 signal recognition particle 14 kDa protein, ... 97.8 2e-20
tpv:TP03_0473 signal recognition particle; K03104 signal recog... 55.1 1e-07
hsa:6727 SRP14, ALURBP, MGC14326; signal recognition particle ... 48.9 7e-06
ath:AT2G43640 signal recognition particle 14 kDa family protei... 47.4 2e-05
xla:779186 srp14, MGC115450; signal recognition particle 14kDa... 47.0 3e-05
dre:619260 srp14, MGC112290, zgc:112290; signal recognition pa... 47.0 3e-05
mmu:20813 Srp14, 14kDa, AW536328; signal recognition particle ... 46.2 5e-05
mmu:100504988 signal recognition particle 14 kDa protein-like 46.2 5e-05
pfa:PFL0160w signal recognition particle SRP14, putative; K031... 38.1 0.014
pfa:PF14_0594 conserved Plasmodium protein, unknown function 37.7 0.020
hsa:23001 WDFY3, ALFY, KIAA0993, MGC16461, ZFYVE25; WD repeat ... 31.6 1.4
> tgo:TGME49_056050 signal recognition particle 14 kDa protein,
putative ; K03104 signal recognition particle subunit SRP14
Length=172
Score = 97.8 bits (242), Expect = 2e-20, Method: Compositional matrix adjust.
Identities = 49/105 (46%), Positives = 66/105 (62%), Gaps = 2/105 (1%)
Query 8 MGLLGEAEFLEALRRLYEESRSKNSGTVWITFKRTFPEVGGSRGAKRRRKLLTCEEKPEA 67
M L FLE L RLYE++R G+VW+T KRTFP + G RG KR+R L +E+
Sbjct 1 MVLADNNVFLEELGRLYEQNRKTGCGSVWVTLKRTFPALNGVRGLKRKRALQ--DEEKLQ 58
Query 68 TPVCLVRATDGKKRISVQVEGDRALAFSQRLWGMSRIHMDQVRPA 112
T VCL+RAT+G+K+IS QV FS L G+ R++ D +RP+
Sbjct 59 TSVCLIRATNGRKKISTQVSLKEVTQFSSALMGVVRLNADDLRPS 103
> tpv:TP03_0473 signal recognition particle; K03104 signal recognition
particle subunit SRP14
Length=91
Score = 55.1 bits (131), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 29/79 (36%), Positives = 46/79 (58%), Gaps = 6/79 (7%)
Query 8 MGLLGEAEFLEALRRLYEESRSKNSGTVWITFKRTFPEVGGSRGAKRRRKLLTCEEKPEA 67
M LL EFL L ++ +E + S ++WIT+KR P +R + ++++ E P+
Sbjct 1 MVLLDCEEFLVELTKMIQEDSTNTSKSLWITYKRYIP---NTRTWRHKKEV---EIDPDE 54
Query 68 TPVCLVRATDGKKRISVQV 86
PVCL+RA G+K+IS V
Sbjct 55 DPVCLIRAKIGRKKISTYV 73
> hsa:6727 SRP14, ALURBP, MGC14326; signal recognition particle
14kDa (homologous Alu RNA binding protein); K03104 signal
recognition particle subunit SRP14
Length=136
Score = 48.9 bits (115), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 36/109 (33%), Positives = 51/109 (46%), Gaps = 20/109 (18%)
Query 8 MGLLGEAEFLEALRRLYEESRSKNSGTVWITFKRT------FPEVGGSRGAKRRRKLLTC 61
M LL +FL L RL+++ R+ SG+V+IT K+ P+ G T
Sbjct 1 MVLLESEQFLTELTRLFQKCRT--SGSVYITLKKYDGRTKPIPKKG------------TV 46
Query 62 EEKPEATPVCLVRATDGKKRISVQVEGDRALAFSQRLWGMSRIHMDQVR 110
E A CL+RATDGKK+IS V F + R +MD ++
Sbjct 47 EGFEPADNKCLLRATDGKKKISTVVSSKEVNKFQMAYSNLLRANMDGLK 95
> ath:AT2G43640 signal recognition particle 14 kDa family protein
/ SRP14 family protein; K03104 signal recognition particle
subunit SRP14
Length=121
Score = 47.4 bits (111), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 31/79 (39%), Positives = 44/79 (55%), Gaps = 9/79 (11%)
Query 8 MGLLGEAEFLEALRRLYEESRSKNSGTVWITFKRTFPEVGGSRGAKRRRKLLTCEEKPEA 67
M LL FL L ++E+S+ K G+VW+T KR+ + ++RKL + E E
Sbjct 1 MVLLQLDPFLNELTSMFEKSKEK--GSVWVTLKRS-----SLKSKVQKRKLSSVGESIEY 53
Query 68 TPVCLVRATDGKKRISVQV 86
CL+RATDGKK +S V
Sbjct 54 R--CLIRATDGKKTVSTSV 70
> xla:779186 srp14, MGC115450; signal recognition particle 14kDa
(homologous Alu RNA binding protein); K03104 signal recognition
particle subunit SRP14
Length=110
Score = 47.0 bits (110), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 35/100 (35%), Positives = 51/100 (51%), Gaps = 8/100 (8%)
Query 8 MGLLGEAEFLEALRRLYEESRSKNSGTVWITFKRTFPEVGGSRGAKRRRKLLTCEEKPEA 67
M LL +FL L RL+++ R+ SG+V+IT K+ G ++ R+ + E A
Sbjct 1 MVLLESEQFLTELTRLFQKCRT--SGSVYITLKKY---DGRTKPVPRKGQ---SESFEPA 52
Query 68 TPVCLVRATDGKKRISVQVEGDRALAFSQRLWGMSRIHMD 107
CL+RATDGKK+IS V F + R +MD
Sbjct 53 DNKCLLRATDGKKKISTVVSSKEVNKFQMAYSNLLRANMD 92
> dre:619260 srp14, MGC112290, zgc:112290; signal recognition
particle 14; K03104 signal recognition particle subunit SRP14
Length=110
Score = 47.0 bits (110), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 36/100 (36%), Positives = 50/100 (50%), Gaps = 8/100 (8%)
Query 8 MGLLGEAEFLEALRRLYEESRSKNSGTVWITFKRTFPEVGGSRGAKRRRKLLTCEEKPEA 67
M LL FL L RL+++ RS SG+V IT K+ G ++ R+ T E A
Sbjct 1 MVLLENDAFLTELTRLFQKCRS--SGSVLITLKK---YDGRTKPVPRKGHPETFEP---A 52
Query 68 TPVCLVRATDGKKRISVQVEGDRALAFSQRLWGMSRIHMD 107
CL+RA+DGK++IS V + F + R HMD
Sbjct 53 DNKCLLRASDGKRKISTVVSTKEVIKFQMAYSNLLRAHMD 92
> mmu:20813 Srp14, 14kDa, AW536328; signal recognition particle
14; K03104 signal recognition particle subunit SRP14
Length=110
Score = 46.2 bits (108), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 35/100 (35%), Positives = 51/100 (51%), Gaps = 8/100 (8%)
Query 8 MGLLGEAEFLEALRRLYEESRSKNSGTVWITFKRTFPEVGGSRGAKRRRKLLTCEEKPEA 67
M LL +FL L RL+++ RS SG+V+IT K+ G ++ R+ + E A
Sbjct 1 MVLLESEQFLTELTRLFQKCRS--SGSVFITLKKY---DGRTKPIPRKS---SVEGLEPA 52
Query 68 TPVCLVRATDGKKRISVQVEGDRALAFSQRLWGMSRIHMD 107
CL+RATDGK++IS V F + R +MD
Sbjct 53 ENKCLLRATDGKRKISTVVSSKEVNKFQMAYSNLLRANMD 92
> mmu:100504988 signal recognition particle 14 kDa protein-like
Length=110
Score = 46.2 bits (108), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 35/100 (35%), Positives = 51/100 (51%), Gaps = 8/100 (8%)
Query 8 MGLLGEAEFLEALRRLYEESRSKNSGTVWITFKRTFPEVGGSRGAKRRRKLLTCEEKPEA 67
M LL +FL L RL+++ RS SG+V+IT K+ G ++ R+ + E A
Sbjct 1 MVLLESEQFLTELTRLFQKCRS--SGSVFITLKKY---DGRTKPIPRKS---SVEGLEPA 52
Query 68 TPVCLVRATDGKKRISVQVEGDRALAFSQRLWGMSRIHMD 107
CL+RATDGK++IS V F + R +MD
Sbjct 53 ENKCLLRATDGKRKISTVVSSKEVNKFQMAYSNLLRANMD 92
> pfa:PFL0160w signal recognition particle SRP14, putative; K03104
signal recognition particle subunit SRP14
Length=104
Score = 38.1 bits (87), Expect = 0.014, Method: Compositional matrix adjust.
Identities = 31/99 (31%), Positives = 47/99 (47%), Gaps = 9/99 (9%)
Query 8 MGLLGEAEFLEALRRLYEESRSKNSGTVWITFKR-------TFPEVGGSRGAKRRRKLLT 60
M LL F++ L +L K ++WIT KR T + K +K+
Sbjct 1 MVLLSNDVFIQELNKLCGPVEGKKKTSIWITMKRVKRNDIKTLTSNKDAGDKKNTKKVNK 60
Query 61 CEEKPEATPVCLVRATDGK-KRISVQVEGDRALAFSQRL 98
+ K +CL+RATDGK K+IS V D ++F+Q +
Sbjct 61 NDNKGNKEYMCLIRATDGKNKKISTHVY-DNVISFTQNI 98
> pfa:PF14_0594 conserved Plasmodium protein, unknown function
Length=3322
Score = 37.7 bits (86), Expect = 0.020, Method: Composition-based stats.
Identities = 18/67 (26%), Positives = 37/67 (55%), Gaps = 0/67 (0%)
Query 101 MSRIHMDQVRPAPPNMEKSRQQQRHPQGQQQSSQHEQRQDEQKQQQSLREQRNNKNYRPS 160
M I D+ N+E + +QR + Q+ +++QR D+ KQ+ +QR++KN +
Sbjct 2393 MDNIINDKHEEIKINIEGDKNKQRDDKNNQRDDKNKQRDDKNKQRDDKNKQRDDKNKQRD 2452
Query 161 NASCKRE 167
+ + +R+
Sbjct 2453 DKNKQRD 2459
Score = 37.4 bits (85), Expect = 0.024, Method: Composition-based stats.
Identities = 20/91 (21%), Positives = 44/91 (48%), Gaps = 16/91 (17%)
Query 77 DGKKRISVQVEGDRALAFSQRLWGMSRIHMDQVRPAPPNMEKSRQQQRHPQGQQQSSQHE 136
D + I + +EGD+ ++ R N + +QR + +Q+ +++
Sbjct 2399 DKHEEIKINIEGDK----------------NKQRDDKNNQRDDKNKQRDDKNKQRDDKNK 2442
Query 137 QRQDEQKQQQSLREQRNNKNYRPSNASCKRE 167
QR D+ KQ+ +QR++KN + + + +R+
Sbjct 2443 QRDDKNKQRDDKNKQRDDKNKQRDDKNKQRD 2473
> hsa:23001 WDFY3, ALFY, KIAA0993, MGC16461, ZFYVE25; WD repeat
and FYVE domain containing 3
Length=3526
Score = 31.6 bits (70), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 23/80 (28%), Positives = 38/80 (47%), Gaps = 9/80 (11%)
Query 97 RLWGMSRIHMDQVRPAPPNMEKSRQQQRHPQGQQ-QSSQHEQRQDEQKQQQSLRE----- 150
R W M + + + PAP E Q+ P+ Q Q +Q E D + +QS+ +
Sbjct 3242 RFWRMEFLQVPET-PAPEPAEVLEMQEDCPEAQIGQEAQDEDSSDSEADEQSISQDPKDT 3300
Query 151 --QRNNKNYRPSNASCKREA 168
Q ++ ++RP ASC+ A
Sbjct 3301 PSQPSSTSHRPRAASCRATA 3320
Lambda K H
0.314 0.127 0.367
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4157683456
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40