bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_0310_orf1
Length=198
Score E
Sequences producing significant alignments: (Bits) Value
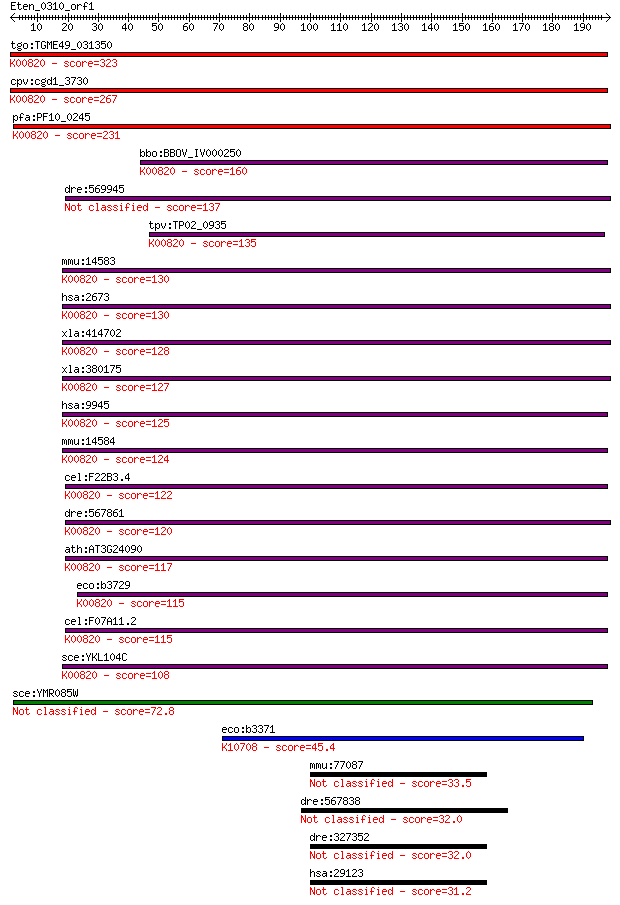
tgo:TGME49_031350 glucosamine--fructose-6-phosphate aminotrans... 323 3e-88
cpv:cgd1_3730 glucosamine-fructose-6-phosphate aminotransferas... 267 2e-71
pfa:PF10_0245 glucosamine-fructose-6-phosphate aminotransferas... 231 9e-61
bbo:BBOV_IV000250 21.m02906; glucosamine--fructose-6-phosphate... 160 4e-39
dre:569945 gfpt2; glutamine-fructose-6-phosphate transaminase ... 137 4e-32
tpv:TP02_0935 glucosamine-fructose-6-phosphate aminotransferas... 135 7e-32
mmu:14583 Gfpt1, 2810423A18Rik, AI324119, AI449986, GFA, GFAT,... 130 3e-30
hsa:2673 GFPT1, GFA, GFAT, GFAT_1, GFAT1, GFAT1m, GFPT; glutam... 130 3e-30
xla:414702 gfpt1, MGC83201, gfat; glutamine--fructose-6-phosph... 128 2e-29
xla:380175 gfpt2, MGC53126, gfpt1; glutamine-fructose-6-phosph... 127 2e-29
hsa:9945 GFPT2, FLJ10380, GFAT2; glutamine-fructose-6-phosphat... 125 1e-28
mmu:14584 Gfpt2, AI480523, GFAT2; glutamine fructose-6-phospha... 124 3e-28
cel:F22B3.4 hypothetical protein; K00820 glucosamine--fructose... 122 1e-27
dre:567861 gfpt1; glutamine-fructose-6-phosphate transaminase ... 120 3e-27
ath:AT3G24090 glutamine-fructose-6-phosphate transaminase (iso... 117 2e-26
eco:b3729 glmS, ECK3722, JW3707; L-glutamine:D-fructose-6-phos... 115 1e-25
cel:F07A11.2 hypothetical protein; K00820 glucosamine--fructos... 115 1e-25
sce:YKL104C GFA1; Gfa1p (EC:2.6.1.16); K00820 glucosamine--fru... 108 2e-23
sce:YMR085W Putative protein of unknown function; YMR085W and ... 72.8 7e-13
eco:b3371 frlB, ECK3359, JW5700, yhfN; fructoselysine-6-P-degl... 45.4 1e-04
mmu:77087 Ankrd11, 2410104C19Rik, 3010027A04Rik, 6330578C09Rik... 33.5 0.55
dre:567838 MGC172359, heatr6; zgc:172359 32.0 1.3
dre:327352 fi04c06; wu:fi04c06 32.0 1.3
hsa:29123 ANKRD11, ANCO-1, ANCO1, LZ16, T13; ankyrin repeat do... 31.2 2.4
> tgo:TGME49_031350 glucosamine--fructose-6-phosphate aminotransferase
(isomerizing), putative (EC:2.6.1.16); K00820 glucosamine--fructose-6-phosphate
aminotransferase (isomerizing)
[EC:2.6.1.16]
Length=603
Score = 323 bits (828), Expect = 3e-88, Method: Compositional matrix adjust.
Identities = 149/197 (75%), Positives = 181/197 (91%), Gaps = 0/197 (0%)
Query 1 ALVASWFWQNLRSEEYPDRCSALLDALHRLPVYAGMTLNAHGACKRIAEKLKDSQTLFVL 60
+L+A+WF QN ++ +PDRCSAL+DA+HRLPVYAGMTLN C+ IAE+LKD++TLFVL
Sbjct 406 SLIAAWFAQNQPTQAFPDRCSALMDAIHRLPVYAGMTLNCRALCQNIAERLKDAKTLFVL 465
Query 61 GKGFGYPVALEGALKIKEISYIHAEGFAGGALKHGPFALIDDKEQTPVIMIVLGDQHAAL 120
GKGFGYPVALEGALKIKE++Y+HAEGF GALKHGPFALID+KE+TPVI+++L DQHAA
Sbjct 466 GKGFGYPVALEGALKIKELAYLHAEGFPAGALKHGPFALIDEKEKTPVILVLLADQHAAS 525
Query 121 MLNAAQQVKARGAYLICLTDFPELVQDVADDYIVIPSNGPLTALLAAVPLQLLAYELSIA 180
+LNAAQQVKARGA+LIC+TD P++V+D+ADD +V+PSNGPLTALLA +PLQLLAYEL+IA
Sbjct 526 LLNAAQQVKARGAHLICVTDEPDIVKDLADDVLVVPSNGPLTALLACIPLQLLAYELAIA 585
Query 181 RGINPDKPRGLAKTVTV 197
+GINPDKPRGLAKTVTV
Sbjct 586 KGINPDKPRGLAKTVTV 602
> cpv:cgd1_3730 glucosamine-fructose-6-phosphate aminotransferase
(EC:2.6.1.16); K00820 glucosamine--fructose-6-phosphate
aminotransferase (isomerizing) [EC:2.6.1.16]
Length=696
Score = 267 bits (682), Expect = 2e-71, Method: Compositional matrix adjust.
Identities = 123/197 (62%), Positives = 159/197 (80%), Gaps = 1/197 (0%)
Query 1 ALVASWFWQNLRSEEYPDRCSALLDALHRLPVYAGMTLNAHGACKRIAEKLKDSQTLFVL 60
+L+A+WF QN R RC LL+A+HR+P+ G++L A C++IAE +KD+ ++FVL
Sbjct 500 SLIAAWFAQN-RDSVISQRCQELLEAIHRVPISVGVSLQAKDQCEQIAEMIKDNNSIFVL 558
Query 61 GKGFGYPVALEGALKIKEISYIHAEGFAGGALKHGPFALIDDKEQTPVIMIVLGDQHAAL 120
GKG+GYPVALEGALKIKEISYIH+EG++ GALKHGPFALID QTPVI+++L D++ +L
Sbjct 559 GKGYGYPVALEGALKIKEISYIHSEGYSAGALKHGPFALIDKDSQTPVILVILSDENQSL 618
Query 121 MLNAAQQVKARGAYLICLTDFPELVQDVADDYIVIPSNGPLTALLAAVPLQLLAYELSIA 180
M+N AQQVKARGA +IC+TD L +D+ + ++IPSNGPLTAL A +PLQL+AY L+I
Sbjct 619 MMNVAQQVKARGARVICITDDENLCKDIDCEKVLIPSNGPLTALNAVIPLQLIAYYLAIK 678
Query 181 RGINPDKPRGLAKTVTV 197
RGINPDKPRGLAK VTV
Sbjct 679 RGINPDKPRGLAKAVTV 695
> pfa:PF10_0245 glucosamine-fructose-6-phosphate aminotransferase,
putative; K00820 glucosamine--fructose-6-phosphate aminotransferase
(isomerizing) [EC:2.6.1.16]
Length=829
Score = 231 bits (590), Expect = 9e-61, Method: Composition-based stats.
Identities = 100/198 (50%), Positives = 150/198 (75%), Gaps = 1/198 (0%)
Query 2 LVASWFWQNLRSEEYPDRCSALLDALHRLPVYAGMTLNA-HGACKRIAEKLKDSQTLFVL 60
L+A WF+Q+ ++ + ++ ++L+++LHRLP+Y G+T+ + CK ++EK K+++++ ++
Sbjct 632 LIALWFFQHKKNNQSSNKATSLINSLHRLPLYTGVTIKSCENTCKTLSEKFKNTKSMLII 691
Query 61 GKGFGYPVALEGALKIKEISYIHAEGFAGGALKHGPFALIDDKEQTPVIMIVLGDQHAAL 120
G G YP+A EGALKIKE++YIH EGF G +LKHGP+AL+ ++ PVIM++ D
Sbjct 692 GNGLSYPIAQEGALKIKELAYIHCEGFTGASLKHGPYALLGGEDNIPVIMLLFNDNTKNA 751
Query 121 MLNAAQQVKARGAYLICLTDFPELVQDVADDYIVIPSNGPLTALLAAVPLQLLAYELSIA 180
M+N +Q+K+RGA+++CLTD LV+ ADD I+IP+NG LT LLA +PLQ+LAY S+
Sbjct 752 MINTGEQIKSRGAHIVCLTDDENLVKHFADDIILIPNNGILTPLLAVIPLQMLAYYTSVN 811
Query 181 RGINPDKPRGLAKTVTVS 198
+GINPDKPR LAKTVTVS
Sbjct 812 KGINPDKPRCLAKTVTVS 829
> bbo:BBOV_IV000250 21.m02906; glucosamine--fructose-6-phosphate
aminotransferase (EC:2.6.1.16); K00820 glucosamine--fructose-6-phosphate
aminotransferase (isomerizing) [EC:2.6.1.16]
Length=723
Score = 160 bits (404), Expect = 4e-39, Method: Compositional matrix adjust.
Identities = 79/154 (51%), Positives = 111/154 (72%), Gaps = 2/154 (1%)
Query 44 CKRIAEKLKDSQTLFVLGKGFGYPVALEGALKIKEISYIHAEGFAGGALKHGPFALIDDK 103
C+R+AE+L + +++V+G G GY +A E ALK KEI+YIHAEG A G +KHG A ID K
Sbjct 571 CQRVAERLCNVDSMYVIGSGEGYAIAQEAALKFKEITYIHAEGIASGTMKHGSLASIDPK 630
Query 104 EQTPVIMIVLGDQHAALMLNAAQQVKARGAYLICLTDFPELVQDVADDYIVIPSNGPLTA 163
+ TPVI I+ D+ + +NA +Q+KARGA++I L P + + + D++I IP G LTA
Sbjct 631 KHTPVICIMTPDE-PEVTVNATKQLKARGAFIIMLASNPAMGEGM-DEFIQIPECGMLTA 688
Query 164 LLAAVPLQLLAYELSIARGINPDKPRGLAKTVTV 197
A VP+Q++AY++++ RG NPD PRGLAKTVTV
Sbjct 689 ACAVVPVQMMAYKIAMLRGWNPDTPRGLAKTVTV 722
> dre:569945 gfpt2; glutamine-fructose-6-phosphate transaminase
2 (EC:2.6.1.16)
Length=681
Score = 137 bits (344), Expect = 4e-32, Method: Compositional matrix adjust.
Identities = 79/183 (43%), Positives = 108/183 (59%), Gaps = 6/183 (3%)
Query 19 RCSALLDALHRLPVYAGMTLNAHGACKRIAEKLKDSQTLFVLGKGFGYPVALEGALKIKE 78
R ++ L++LP L K IAE+L ++L V+G+GF Y LEGALKIKE
Sbjct 502 RRQEIISGLNQLPELIKKVLAQDDNIKTIAEELHHQRSLLVMGRGFNYATCLEGALKIKE 561
Query 79 ISYIHAEGFAGGALKHGPFALIDDKEQTPVIMIVLGDQHAALMLNAAQQVKARGAY--LI 136
I+Y+H+EG G LKHGP ALID + PVIMI++ D NA QQV AR +I
Sbjct 562 ITYMHSEGILAGELKHGPLALID--KHMPVIMIIMRDACYQKCHNALQQVTARQGRPIII 619
Query 137 CLTDFPELVQDVADDYIVIPSN-GPLTALLAAVPLQLLAYELSIARGINPDKPRGLAKTV 195
C D PE + +A I +P L +L+ +PLQL+++ L++ RG + D PR LAK+V
Sbjct 620 CCQDDPE-ISKMAYKTIELPQTVDCLQGILSVIPLQLISFHLAVLRGYDVDCPRNLAKSV 678
Query 196 TVS 198
TV
Sbjct 679 TVE 681
> tpv:TP02_0935 glucosamine-fructose-6-phosphate aminotransferase;
K00820 glucosamine--fructose-6-phosphate aminotransferase
(isomerizing) [EC:2.6.1.16]
Length=810
Score = 135 bits (341), Expect = 7e-32, Method: Composition-based stats.
Identities = 68/150 (45%), Positives = 101/150 (67%), Gaps = 2/150 (1%)
Query 47 IAEKLKDSQTLFVLGKGFGYPVALEGALKIKEISYIHAEGFAGGALKHGPFALIDDKEQT 106
+A+ L + +++LG+G G+ VALE +LK+KE++YI AEG GA+KHG +A+I +++ T
Sbjct 661 LAQWLLKEKIVYILGRGCGHAVALEASLKMKEVAYIQAEGVLSGAMKHGIYAMIKEEDTT 720
Query 107 PVIMIVLGDQHAALMLNAAQQVKARGAYLICLTDFPELVQDVADDYIVIPSNGPLTALLA 166
P I I+ + + +N+ Q+KARG Y+I +TD + V D AD I IPS G LT LA
Sbjct 721 PTISIITSEDK-EMTINSTMQIKARGGYIIVITDLEDEV-DFADVLIRIPSIGALTPALA 778
Query 167 AVPLQLLAYELSIARGINPDKPRGLAKTVT 196
VP+Q++ +++ NPD PRGLAKTVT
Sbjct 779 IVPIQIITSKIATLSKRNPDIPRGLAKTVT 808
> mmu:14583 Gfpt1, 2810423A18Rik, AI324119, AI449986, GFA, GFAT,
GFAT1, GFAT1m, Gfpt; glutamine fructose-6-phosphate transaminase
1 (EC:2.6.1.16); K00820 glucosamine--fructose-6-phosphate
aminotransferase (isomerizing) [EC:2.6.1.16]
Length=681
Score = 130 bits (326), Expect = 3e-30, Method: Compositional matrix adjust.
Identities = 73/183 (39%), Positives = 108/183 (59%), Gaps = 4/183 (2%)
Query 18 DRCSALLDALHRLPVYAGMTLNAHGACKRIAEKLKDSQTLFVLGKGFGYPVALEGALKIK 77
+R ++ L RLP L+ +++A +L +++ ++G+G+ Y LEGALKIK
Sbjct 501 ERRKEIMLGLKRLPDLIKEVLSMDDEIQKLATELYHQKSVLIMGRGYHYATCLEGALKIK 560
Query 78 EISYIHAEGFAGGALKHGPFALIDDKEQTPVIMIVLGDQHAALMLNAAQQVKARGAY--L 135
EI+Y+H+EG G LKHGP AL+D + PVIMI++ D A NA QQV AR +
Sbjct 561 EITYMHSEGILAGELKHGPLALVD--KLMPVIMIIMRDHTYAKCQNALQQVVARQGRPVV 618
Query 136 ICLTDFPELVQDVADDYIVIPSNGPLTALLAAVPLQLLAYELSIARGINPDKPRGLAKTV 195
IC + E +++ V S L +L+ +PLQLLA+ L++ RG + D PR LAK+V
Sbjct 619 ICDKEDTETIKNTKRTIKVPHSVDCLQGILSVIPLQLLAFHLAVLRGYDVDFPRNLAKSV 678
Query 196 TVS 198
TV
Sbjct 679 TVE 681
> hsa:2673 GFPT1, GFA, GFAT, GFAT_1, GFAT1, GFAT1m, GFPT; glutamine--fructose-6-phosphate
transaminase 1 (EC:2.6.1.16); K00820
glucosamine--fructose-6-phosphate aminotransferase (isomerizing)
[EC:2.6.1.16]
Length=681
Score = 130 bits (326), Expect = 3e-30, Method: Compositional matrix adjust.
Identities = 73/183 (39%), Positives = 108/183 (59%), Gaps = 4/183 (2%)
Query 18 DRCSALLDALHRLPVYAGMTLNAHGACKRIAEKLKDSQTLFVLGKGFGYPVALEGALKIK 77
+R ++ L RLP L+ +++A +L +++ ++G+G+ Y LEGALKIK
Sbjct 501 ERRKEIMLGLKRLPDLIKEVLSMDDEIQKLATELYHQKSVLIMGRGYHYATCLEGALKIK 560
Query 78 EISYIHAEGFAGGALKHGPFALIDDKEQTPVIMIVLGDQHAALMLNAAQQVKARGAY--L 135
EI+Y+H+EG G LKHGP AL+D + PVIMI++ D A NA QQV AR +
Sbjct 561 EITYMHSEGILAGELKHGPLALVD--KLMPVIMIIMRDHTYAKCQNALQQVVARQGRPVV 618
Query 136 ICLTDFPELVQDVADDYIVIPSNGPLTALLAAVPLQLLAYELSIARGINPDKPRGLAKTV 195
IC + E +++ V S L +L+ +PLQLLA+ L++ RG + D PR LAK+V
Sbjct 619 ICDKEDTETIKNTKRTIKVPHSVDCLQGILSVIPLQLLAFHLAVLRGYDVDFPRNLAKSV 678
Query 196 TVS 198
TV
Sbjct 679 TVE 681
> xla:414702 gfpt1, MGC83201, gfat; glutamine--fructose-6-phosphate
transaminase 1; K00820 glucosamine--fructose-6-phosphate
aminotransferase (isomerizing) [EC:2.6.1.16]
Length=681
Score = 128 bits (321), Expect = 2e-29, Method: Compositional matrix adjust.
Identities = 71/183 (38%), Positives = 106/183 (57%), Gaps = 4/183 (2%)
Query 18 DRCSALLDALHRLPVYAGMTLNAHGACKRIAEKLKDSQTLFVLGKGFGYPVALEGALKIK 77
+R +++ L LP L+ +++A +L +++ ++G+GF Y +EGALKIK
Sbjct 501 ERRKQIINGLKILPDNIKEVLSLDDEIQKLASELYQQKSVLIMGRGFHYATCMEGALKIK 560
Query 78 EISYIHAEGFAGGALKHGPFALIDDKEQTPVIMIVLGDQHAALMLNAAQQVKARGA--YL 135
EI+Y+H+EG G LKHGP ALID + PVIMI++ D NA QQV AR +
Sbjct 561 EITYMHSEGILAGELKHGPLALID--KLMPVIMIIMRDHSYTKCQNALQQVVARQGRPVV 618
Query 136 ICLTDFPELVQDVADDYIVIPSNGPLTALLAAVPLQLLAYELSIARGINPDKPRGLAKTV 195
IC + E + + V + L +L+ +PLQLLA+ L++ RG + D PR LAK+V
Sbjct 619 ICDKEDTETINSIKRTIKVPHTVDCLQGILSVIPLQLLAFHLAVLRGYDVDCPRNLAKSV 678
Query 196 TVS 198
TV
Sbjct 679 TVE 681
> xla:380175 gfpt2, MGC53126, gfpt1; glutamine-fructose-6-phosphate
transaminase 2 (EC:2.6.1.16); K00820 glucosamine--fructose-6-phosphate
aminotransferase (isomerizing) [EC:2.6.1.16]
Length=681
Score = 127 bits (319), Expect = 2e-29, Method: Compositional matrix adjust.
Identities = 70/183 (38%), Positives = 107/183 (58%), Gaps = 4/183 (2%)
Query 18 DRCSALLDALHRLPVYAGMTLNAHGACKRIAEKLKDSQTLFVLGKGFGYPVALEGALKIK 77
+R +++AL LP L+ +++A +L +++ ++G+G+ Y +EGALKIK
Sbjct 501 ERRKQIMNALQILPDNIKEVLSLDDEIQKLASELYQQKSVLIMGRGYHYSTCMEGALKIK 560
Query 78 EISYIHAEGFAGGALKHGPFALIDDKEQTPVIMIVLGDQHAALMLNAAQQVKARGA--YL 135
EI+Y+H+EG G LKHGP AL+D + PVIMI++ D NA QQV AR +
Sbjct 561 EITYMHSEGILAGELKHGPLALVD--KLMPVIMIIMRDHAYTKCQNALQQVVARQGRPVV 618
Query 136 ICLTDFPELVQDVADDYIVIPSNGPLTALLAAVPLQLLAYELSIARGINPDKPRGLAKTV 195
IC + E + + V + L +L+ +PLQLLA+ L++ RG + D PR LAK+V
Sbjct 619 ICDKEDTETINSIKRTIKVPHTVDCLQGVLSVIPLQLLAFHLAVLRGYDVDCPRNLAKSV 678
Query 196 TVS 198
TV
Sbjct 679 TVE 681
> hsa:9945 GFPT2, FLJ10380, GFAT2; glutamine-fructose-6-phosphate
transaminase 2 (EC:2.6.1.16); K00820 glucosamine--fructose-6-phosphate
aminotransferase (isomerizing) [EC:2.6.1.16]
Length=682
Score = 125 bits (313), Expect = 1e-28, Method: Compositional matrix adjust.
Identities = 75/183 (40%), Positives = 105/183 (57%), Gaps = 6/183 (3%)
Query 18 DRCSALLDALHRLPVYAGMTLNAHGACKRIAEKLKDSQTLFVLGKGFGYPVALEGALKIK 77
+R ++ L LP L+ +A +L ++L V+G+G+ Y LEGALKIK
Sbjct 502 NRRQEIIRGLRSLPELIKEVLSLEEKIHDLALELYTQRSLLVMGRGYNYATCLEGALKIK 561
Query 78 EISYIHAEGFAGGALKHGPFALIDDKEQTPVIMIVLGDQHAALMLNAAQQVKARGAY--L 135
EI+Y+H+EG G LKHGP ALID +Q PVIM+++ D A NA QQV AR +
Sbjct 562 EITYMHSEGILAGELKHGPLALID--KQMPVIMVIMKDPCFAKCQNALQQVTARQGRPII 619
Query 136 ICLTDFPELVQDVADDYIVIPSN-GPLTALLAAVPLQLLAYELSIARGINPDKPRGLAKT 194
+C D E A I +P L +L+ +PLQLL++ L++ RG + D PR LAK+
Sbjct 620 LCSKDDTE-SSKFAYKTIELPHTVDCLQGILSVIPLQLLSFHLAVLRGYDVDFPRNLAKS 678
Query 195 VTV 197
VTV
Sbjct 679 VTV 681
> mmu:14584 Gfpt2, AI480523, GFAT2; glutamine fructose-6-phosphate
transaminase 2 (EC:2.6.1.16); K00820 glucosamine--fructose-6-phosphate
aminotransferase (isomerizing) [EC:2.6.1.16]
Length=682
Score = 124 bits (310), Expect = 3e-28, Method: Compositional matrix adjust.
Identities = 74/183 (40%), Positives = 105/183 (57%), Gaps = 6/183 (3%)
Query 18 DRCSALLDALHRLPVYAGMTLNAHGACKRIAEKLKDSQTLFVLGKGFGYPVALEGALKIK 77
+R ++ L LP L+ +A +L ++L V+G+G+ Y LEGALKIK
Sbjct 502 NRRQEIIRGLRSLPELIKEVLSLDEKIHDLALELYTQRSLLVMGRGYNYATCLEGALKIK 561
Query 78 EISYIHAEGFAGGALKHGPFALIDDKEQTPVIMIVLGDQHAALMLNAAQQVKARGAY--L 135
EI+Y+H+EG G LKHGP AL+D +Q PVIM+++ D A NA QQV AR +
Sbjct 562 EITYMHSEGILAGELKHGPLALVD--KQMPVIMVIMKDPCFAKCQNALQQVTARQGRPII 619
Query 136 ICLTDFPELVQDVADDYIVIPSN-GPLTALLAAVPLQLLAYELSIARGINPDKPRGLAKT 194
+C D E A I +P L +L+ +PLQLL++ L++ RG + D PR LAK+
Sbjct 620 LCSKDDTE-SSKFAYKTIELPHTVDCLQGILSVIPLQLLSFHLAVLRGYDVDFPRNLAKS 678
Query 195 VTV 197
VTV
Sbjct 679 VTV 681
> cel:F22B3.4 hypothetical protein; K00820 glucosamine--fructose-6-phosphate
aminotransferase (isomerizing) [EC:2.6.1.16]
Length=710
Score = 122 bits (305), Expect = 1e-27, Method: Compositional matrix adjust.
Identities = 70/182 (38%), Positives = 103/182 (56%), Gaps = 5/182 (2%)
Query 19 RCSALLDALHRLPVYAGMTLNAHGACKRIAEKLKDSQTLFVLGKGFGYPVALEGALKIKE 78
R + ++DAL+ LP+ L+ +IAE++ ++L ++G+G + LEGALKIKE
Sbjct 530 RRAEIIDALNNLPILIRDVLDLDDQVLKIAEQIYKDKSLLIMGRGLNFATCLEGALKIKE 589
Query 79 ISYIHAEGFAGGALKHGPFALIDDKEQTPVIMIVLGDQHAALMLNAAQQVKARGAYLICL 138
+SY+H EG G LKHGP A++D E + M+V D LNA QQV AR I +
Sbjct 590 LSYMHCEGIMSGELKHGPLAMVD--EFLSICMVVCNDHVYKKSLNALQQVVARKGAPIII 647
Query 139 TDFPELVQDVA--DDYIVIPSN-GPLTALLAAVPLQLLAYELSIARGINPDKPRGLAKTV 195
D D+A + +P + +L +PLQLL+Y ++ G N D+PR LAK+V
Sbjct 648 ADSSVPESDLAGMKHVLRVPRTVDCVQNILTVIPLQLLSYHIAELNGANVDRPRNLAKSV 707
Query 196 TV 197
TV
Sbjct 708 TV 709
> dre:567861 gfpt1; glutamine-fructose-6-phosphate transaminase
1 (EC:2.6.1.16); K00820 glucosamine--fructose-6-phosphate
aminotransferase (isomerizing) [EC:2.6.1.16]
Length=682
Score = 120 bits (301), Expect = 3e-27, Method: Compositional matrix adjust.
Identities = 69/182 (37%), Positives = 102/182 (56%), Gaps = 4/182 (2%)
Query 19 RCSALLDALHRLPVYAGMTLNAHGACKRIAEKLKDSQTLFVLGKGFGYPVALEGALKIKE 78
R ++ L LP L +++A +L +++ ++G+G+ Y LEGALKIKE
Sbjct 503 RRKEIIQGLRILPDLIKEVLTLDEEIQKLAAELYPQKSVLIMGRGYHYATCLEGALKIKE 562
Query 79 ISYIHAEGFAGGALKHGPFALIDDKEQTPVIMIVLGDQHAALMLNAAQQVKARGAY--LI 136
I+Y+H+EG G LKHGP AL+D + PVIMI++ D NA QQV AR +I
Sbjct 563 ITYMHSEGILAGELKHGPLALVD--KLMPVIMIIMRDHTYTKCQNALQQVVARQGRPIVI 620
Query 137 CLTDFPELVQDVADDYIVIPSNGPLTALLAAVPLQLLAYELSIARGINPDKPRGLAKTVT 196
C + E + + V L +L+ +PLQLL++ L++ RG + D PR LAK+VT
Sbjct 621 CDKEDNETINNSKRTIKVPHCVDCLQGILSVIPLQLLSFHLAVLRGYDVDCPRNLAKSVT 680
Query 197 VS 198
V
Sbjct 681 VE 682
> ath:AT3G24090 glutamine-fructose-6-phosphate transaminase (isomerizing)/
sugar binding / transaminase (EC:2.6.1.16); K00820
glucosamine--fructose-6-phosphate aminotransferase (isomerizing)
[EC:2.6.1.16]
Length=695
Score = 117 bits (293), Expect = 2e-26, Method: Compositional matrix adjust.
Identities = 71/183 (38%), Positives = 100/183 (54%), Gaps = 6/183 (3%)
Query 19 RCSALLDALHRLPVYAGMTLNAHGACKRIAEKLKDSQTLFVLGKGFGYPVALEGALKIKE 78
R A++D L LP L K +A+ L D Q+L V G+G+ Y ALEGALK+KE
Sbjct 514 RREAIIDGLLDLPYKVKEVLKLDDEMKDLAQLLIDEQSLLVFGRGYNYATALEGALKVKE 573
Query 79 ISYIHAEGFAGGALKHGPFALIDDKEQTPVIMIVLGDQHAALMLNAAQQVKARGAYLICL 138
++ +H+EG G +KHGP AL+D E P+ +I D + + QQ+ AR LI +
Sbjct 574 VALMHSEGILAGEMKHGPLALVD--ENLPIAVIATRDACFSKQQSVIQQLHARKGRLIVM 631
Query 139 T---DFPELVQDVADDYIVIPS-NGPLTALLAAVPLQLLAYELSIARGINPDKPRGLAKT 194
D + + I +P L ++ VPLQLLAY L++ RG N D+PR LAK+
Sbjct 632 CSKGDAASVSSSGSCRAIEVPQVEDCLQPVINIVPLQLLAYHLTVLRGHNVDQPRNLAKS 691
Query 195 VTV 197
VT
Sbjct 692 VTT 694
> eco:b3729 glmS, ECK3722, JW3707; L-glutamine:D-fructose-6-phosphate
aminotransferase (EC:2.6.1.16); K00820 glucosamine--fructose-6-phosphate
aminotransferase (isomerizing) [EC:2.6.1.16]
Length=609
Score = 115 bits (288), Expect = 1e-25, Method: Compositional matrix adjust.
Identities = 72/182 (39%), Positives = 102/182 (56%), Gaps = 14/182 (7%)
Query 23 LLDALHRLPVYAGMTLNAHGACKRIAEKLKDSQTLFVLGKGFGYPVALEGALKIKEISYI 82
++ L LP L+ + +AE D LG+G YP+ALEGALK+KEISYI
Sbjct 434 IVHGLQALPSRIEQMLSQDKRIEALAEDFSDKHHALFLGRGDQYPIALEGALKLKEISYI 493
Query 83 HAEGFAGGALKHGPFALIDDKEQTPVIMIVLGDQHAALMLNAAQQVKARGAYLICLTDFP 142
HAE +A G LKHGP ALID PVI++ ++ + + ++V+ARG L D
Sbjct 494 HAEAYAAGELKHGPLALID--ADMPVIVVAPNNELLEKLKSNIEEVRARGGQLYVFAD-- 549
Query 143 ELVQD---VADDYIVIPSNGPLTALLA----AVPLQLLAYELSIARGINPDKPRGLAKTV 195
QD V+ D + I + ++A VPLQLLAY +++ +G + D+PR LAK+V
Sbjct 550 ---QDAGFVSSDNMHIIEMPHVEEVIAPIFYTVPLQLLAYHVALIKGTDVDQPRNLAKSV 606
Query 196 TV 197
TV
Sbjct 607 TV 608
> cel:F07A11.2 hypothetical protein; K00820 glucosamine--fructose-6-phosphate
aminotransferase (isomerizing) [EC:2.6.1.16]
Length=712
Score = 115 bits (287), Expect = 1e-25, Method: Compositional matrix adjust.
Identities = 69/182 (37%), Positives = 100/182 (54%), Gaps = 5/182 (2%)
Query 19 RCSALLDALHRLPVYAGMTLNAHGACKRIAEKLKDSQTLFVLGKGFGYPVALEGALKIKE 78
R ++DAL+ LP L IA+++ ++L ++G+G + LEGALKIKE
Sbjct 532 RREEIIDALNDLPELIREVLQLDEKVLDIAKQIYKEKSLLIMGRGLNFATCLEGALKIKE 591
Query 79 ISYIHAEGFAGGALKHGPFALIDDKEQTPVIMIVLGDQHAALMLNAAQQVKARGAYLICL 138
+SY+H EG G LKHGP A++D E + M+V D+ LNA QQV AR I +
Sbjct 592 LSYMHCEGIMSGELKHGPLAMVD--EFLSICMVVCNDRVYKKSLNALQQVVARKGAPIII 649
Query 139 TDFPELVQDVA--DDYIVIPSN-GPLTALLAAVPLQLLAYELSIARGINPDKPRGLAKTV 195
D D+A + +P + +L +PLQLL+Y ++ G N D+PR LAK+V
Sbjct 650 ADCTVPEGDLAGMKHILRVPKTVDCVQNILTVIPLQLLSYHIAELNGANVDRPRNLAKSV 709
Query 196 TV 197
TV
Sbjct 710 TV 711
> sce:YKL104C GFA1; Gfa1p (EC:2.6.1.16); K00820 glucosamine--fructose-6-phosphate
aminotransferase (isomerizing) [EC:2.6.1.16]
Length=717
Score = 108 bits (269), Expect = 2e-23, Method: Compositional matrix adjust.
Identities = 68/187 (36%), Positives = 106/187 (56%), Gaps = 9/187 (4%)
Query 18 DRCSALLDALHRLPVYAGMTLNAHGACKRI-AEKLKDSQTLFVLGKGFGYPVALEGALKI 76
DR ++ L +P L K++ A +LKD ++L +LG+G+ + ALEGALKI
Sbjct 532 DRRIEIIQGLKLIPGQIKQVLKLEPRIKKLCATELKDQKSLLLLGRGYQFAAALEGALKI 591
Query 77 KEISYIHAEGFAGGALKHGPFALIDDKEQTPVIMIVLGDQHAALMLNAAQQVKARGAYLI 136
KEISY+H+EG G LKHG AL+D E P+I D ++++ +QV AR + I
Sbjct 592 KEISYMHSEGVLAGELKHGVLALVD--ENLPIIAFGTRDSLFPKVVSSIEQVTARKGHPI 649
Query 137 CLTDFPELV-----QDVADDYIVIPSN-GPLTALLAAVPLQLLAYELSIARGINPDKPRG 190
+ + + V + + + +P L L+ +PLQL++Y L++ +GI+ D PR
Sbjct 650 IICNENDEVWAQKSKSIDLQTLEVPQTVDCLQGLINIIPLQLMSYWLAVNKGIDVDFPRN 709
Query 191 LAKTVTV 197
LAK+VTV
Sbjct 710 LAKSVTV 716
> sce:YMR085W Putative protein of unknown function; YMR085W and
adjacent ORF YMR084W are merged in related strains
Length=432
Score = 72.8 bits (177), Expect = 7e-13, Method: Compositional matrix adjust.
Identities = 58/198 (29%), Positives = 91/198 (45%), Gaps = 11/198 (5%)
Query 2 LVASWFWQNLRSEEYPDRCSALLDALHRLPVYAGMTLNAHGACKRIAEK-LKDSQTLFVL 60
++A W ++L S+ +R ++ AL +P L + +K LK T +L
Sbjct 233 MIALWMSEDLVSK--IERRKEIIQALTIVPSQIKEVLELEPLIIELCDKKLKQHDTFLLL 290
Query 61 GKGFGYPVALEGALKIKEISYIHAEGFAGGALKHGPFALIDDKEQTPVIMIVLGDQHAAL 120
G+G+ + ALEGA K+KEISY+H+E L H A+ D P+I D +
Sbjct 291 GRGYQFASALEGASKMKEISYVHSESILTNELGHRVLAVASD--NPPIIAFATKDAFSPK 348
Query 121 MLNAAQQVKAR--GAYLICLTDFPELVQDVADDYIV---IPSN-GPLTALLAAVPLQLLA 174
+ + Q+ R +IC QD +V +P L +L +PLQL++
Sbjct 349 IASCIDQIIERKGNPIIICNKGHKIWEQDKQKGNVVTLEVPQTVDCLQGILNVIPLQLIS 408
Query 175 YELSIARGINPDKPRGLA 192
Y L+I + I D PR A
Sbjct 409 YWLAIKKDIGVDLPRDSA 426
> eco:b3371 frlB, ECK3359, JW5700, yhfN; fructoselysine-6-P-deglycase;
K10708 fructoselysine 6-phosphate deglycase [EC:3.5.-.-]
Length=340
Score = 45.4 bits (106), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 31/119 (26%), Positives = 53/119 (44%), Gaps = 12/119 (10%)
Query 71 EGALKIKEISYIHAEGFAGGALKHGPFALIDDKEQTPVIMIVLGDQHAALMLNAAQQVKA 130
EG + + E ++ H G +HGP +++ P + ++ D+ A VK
Sbjct 226 EGIVTLMEFTWTHGCVIESGEFRHGPLEIVEPG--VPFLFLLGNDESRHTTERAINFVKQ 283
Query 131 RGAYLICLTDFPELVQDVADDYIVIPSNGPLTALLAAVPLQLLAYELSIARGINPDKPR 189
R +I + D+ E+ Q + + L L VP++ L Y LSI + NPD+ R
Sbjct 284 RTDNVIVI-DYAEISQGL---------HPWLAPFLMFVPMEWLCYYLSIYKDHNPDERR 332
> mmu:77087 Ankrd11, 2410104C19Rik, 3010027A04Rik, 6330578C09Rik,
9530048I21Rik, AA930108, Gm176, Yod; ankyrin repeat domain
11
Length=2643
Score = 33.5 bits (75), Expect = 0.55, Method: Composition-based stats.
Identities = 21/70 (30%), Positives = 36/70 (51%), Gaps = 12/70 (17%)
Query 100 IDDKEQTPVIMIVLGDQHAALMLNAAQ----QVKAR-----GAYLICLTDFPEL---VQD 147
+DDK +++ QH A LNA Q Q+KA+ G +C+ + P + D
Sbjct 2574 VDDKYDRMKTCLLMRQQHEAAALNAVQRMEWQLKAQELDPAGHKSLCVNEVPSFYVPMVD 2633
Query 148 VADDYIVIPS 157
V DD++++P+
Sbjct 2634 VNDDFVLLPA 2643
> dre:567838 MGC172359, heatr6; zgc:172359
Length=1201
Score = 32.0 bits (71), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 24/68 (35%), Positives = 36/68 (52%), Gaps = 6/68 (8%)
Query 97 FALIDDKEQTPVIMIVLGDQHAALMLNAAQQVKARGAYLICLTDFPELVQDVADDYIVIP 156
F+ + DK Q I I+LG ++ L A V+A G Y++ FP L +DV ++
Sbjct 811 FSQLPDKTQVLCITILLGLTYSENSLVKAAAVRALGVYIL----FPCLREDVM--FVADT 864
Query 157 SNGPLTAL 164
+N LTAL
Sbjct 865 ANAILTAL 872
> dre:327352 fi04c06; wu:fi04c06
Length=2649
Score = 32.0 bits (71), Expect = 1.3, Method: Composition-based stats.
Identities = 20/70 (28%), Positives = 35/70 (50%), Gaps = 12/70 (17%)
Query 100 IDDKEQTPVIMIVLGDQHAALMLNAAQ----QVKAR-----GAYLICLTDFPEL---VQD 147
+DDK +++ QH A LNA Q Q+K + G +C+ + P + D
Sbjct 2580 VDDKYDRMKTCLLMRQQHEAAALNAVQRMEWQLKVQELDPAGHKSLCVNEVPSFYVPMVD 2639
Query 148 VADDYIVIPS 157
V DD++++P+
Sbjct 2640 VNDDFVLLPA 2649
> hsa:29123 ANKRD11, ANCO-1, ANCO1, LZ16, T13; ankyrin repeat
domain 11
Length=2663
Score = 31.2 bits (69), Expect = 2.4, Method: Composition-based stats.
Identities = 20/70 (28%), Positives = 35/70 (50%), Gaps = 12/70 (17%)
Query 100 IDDKEQTPVIMIVLGDQHAALMLNAAQ----QVKAR-----GAYLICLTDFPEL---VQD 147
+DDK +++ QH A LNA Q Q+K + G +C+ + P + D
Sbjct 2594 VDDKYDRMKTCLLMRQQHEAAALNAVQRMEWQLKVQELDPAGHKSLCVNEVPSFYVPMVD 2653
Query 148 VADDYIVIPS 157
V DD++++P+
Sbjct 2654 VNDDFVLLPA 2663
Lambda K H
0.321 0.138 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5866798756
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40