bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_0282_orf1
Length=233
Score E
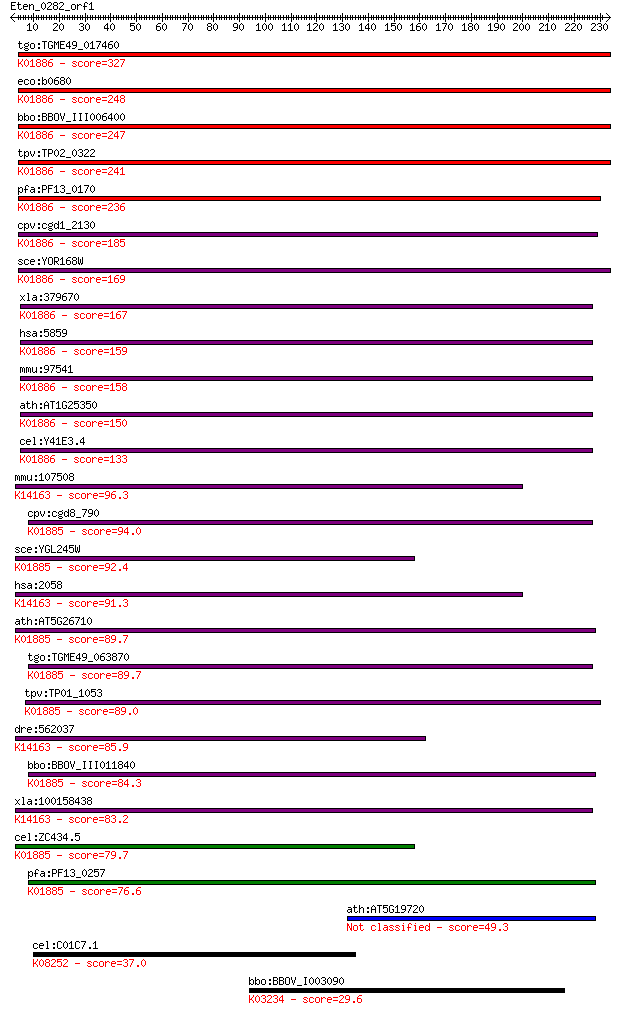
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_017460 glutaminyl-tRNA synthetase, putative (EC:6.1... 327 3e-89
eco:b0680 glnS, ECK0668, JW0666; glutamyl-tRNA synthetase (EC:... 248 2e-65
bbo:BBOV_III006400 17.m07570; glutaminyl-tRNA synthetase famil... 247 3e-65
tpv:TP02_0322 glutaminyl-tRNA synthetase; K01886 glutaminyl-tR... 241 2e-63
pfa:PF13_0170 glutaminyl-tRNA synthetase, putative (EC:6.1.1.1... 236 4e-62
cpv:cgd1_2130 glutaminyl-tRNA synthetase, of predicted bacteri... 185 1e-46
sce:YOR168W GLN4; Gln4p (EC:6.1.1.18); K01886 glutaminyl-tRNA ... 169 6e-42
xla:379670 qars, MGC69128; glutaminyl-tRNA synthetase (EC:6.1.... 167 4e-41
hsa:5859 QARS, GLNRS, PRO2195; glutaminyl-tRNA synthetase (EC:... 159 1e-38
mmu:97541 Qars, 1110018N24Rik, 1200016L19Rik, C80286, GLNRS, M... 158 1e-38
ath:AT1G25350 OVA9; OVA9 (ovule abortion 9); glutamine-tRNA li... 150 3e-36
cel:Y41E3.4 ers-1; glutamyl(E)/glutaminyl(Q) tRNA Synthetase f... 133 6e-31
mmu:107508 Eprs, 2410081F06Rik, 3010002K18Rik, C79379, Qprs; g... 96.3 9e-20
cpv:cgd8_790 glutamate--tRNA ligase ; K01885 glutamyl-tRNA syn... 94.0 4e-19
sce:YGL245W GUS1, GSN1; GluRS (EC:6.1.1.17); K01885 glutamyl-t... 92.4 1e-18
hsa:2058 EPRS, DKFZp313B047, EARS, GLUPRORS, PARS, QARS, QPRS;... 91.3 3e-18
ath:AT5G26710 glutamate-tRNA ligase, putative / glutamyl-tRNA ... 89.7 7e-18
tgo:TGME49_063870 glutamyl-tRNA synthetase, putative (EC:6.1.1... 89.7 8e-18
tpv:TP01_1053 glutamyl-tRNA synthetase; K01885 glutamyl-tRNA s... 89.0 1e-17
dre:562037 eprs, wu:fb38d08, wu:ft34d10; glutamyl-prolyl-tRNA ... 85.9 1e-16
bbo:BBOV_III011840 17.m08008; glutamyl-tRNA synthetase family ... 84.3 4e-16
xla:100158438 eprs; glutamyl-prolyl-tRNA synthetase; K14163 bi... 83.2 7e-16
cel:ZC434.5 ers-2; glutamyl(E)/glutaminyl(Q) tRNA Synthetase f... 79.7 7e-15
pfa:PF13_0257 glutamate--tRNA ligase, putative (EC:6.1.1.17); ... 76.6 6e-14
ath:AT5G19720 tRNA synthetase class I (E and Q) family protein 49.3 1e-05
cel:C01C7.1 ark-1; A Ras-regulating Kinase family member (ark-... 37.0 0.054
bbo:BBOV_I003090 19.m02240; elongation factor 2, EF-2; K03234 ... 29.6 9.8
> tgo:TGME49_017460 glutaminyl-tRNA synthetase, putative (EC:6.1.1.18);
K01886 glutaminyl-tRNA synthetase [EC:6.1.1.18]
Length=860
Score = 327 bits (837), Expect = 3e-89, Method: Compositional matrix adjust.
Identities = 155/231 (67%), Positives = 188/231 (81%), Gaps = 1/231 (0%)
Query 4 NRQLYDWFQEALGITRTRQIEFARLNVTYTVMSKRKLLALVNEKVVAGWDDPRLSTLSAL 63
+R LYDWFQE L ITRTRQIEFARLNVTYTVMSKRKLLALV EK V GWDDPRL TLS L
Sbjct 483 HRILYDWFQEKLEITRTRQIEFARLNVTYTVMSKRKLLALVTEKWVDGWDDPRLPTLSGL 542
Query 64 RRRGVPPAAIRDFCERVGVARRPSTIQIELLERCIREYLDERAPRRFAVKQPLLVKITNF 123
RRRGVPP+A+RDFC++VGVARR STI++E+LE+CIR+ L A RRFA++ P+ V ITN+
Sbjct 543 RRRGVPPSALRDFCDKVGVARRESTIKVEVLEKCIRDALHVVAHRRFAIQDPIAVTITNY 602
Query 124 GAE-EVLTIPSDPKRPAAGSRQLAFTNELYIDAEDFMENPPKEFHRLSPGKEVRLRSAYW 182
G + E +T + P+ + G+R+L F+ +LYID +DFMENPP + RL+PG EVRL+ AYW
Sbjct 603 GDKVETITAANHPEDESFGTRELHFSKKLYIDRDDFMENPPAGYRRLAPGAEVRLKHAYW 662
Query 183 IKCNEVEKDANGNITAIICSYDPQTLNCSVAPDGRKVKGTIHWLPAKYAVP 233
IKC +V KDA+G +T ++C+YDPQT NCSVAPDGRKVKG IHWL K AVP
Sbjct 663 IKCVDVVKDASGLVTELLCTYDPQTKNCSVAPDGRKVKGAIHWLSEKDAVP 713
> eco:b0680 glnS, ECK0668, JW0666; glutamyl-tRNA synthetase (EC:6.1.1.18);
K01886 glutaminyl-tRNA synthetase [EC:6.1.1.18]
Length=554
Score = 248 bits (632), Expect = 2e-65, Method: Compositional matrix adjust.
Identities = 126/232 (54%), Positives = 164/232 (70%), Gaps = 3/232 (1%)
Query 4 NRQLYDWFQEALGI-TRTRQIEFARLNVTYTVMSKRKLLALVNEKVVAGWDDPRLSTLSA 62
NR+LYDW + + I RQ EF+RLN+ YTVMSKRKL LV +K V GWDDPR+ T+S
Sbjct 237 NRRLYDWVLDNITIPVHPRQYEFSRLNLEYTVMSKRKLNLLVTDKHVEGWDDPRMPTISG 296
Query 63 LRRRGVPPAAIRDFCERVGVARRPSTIQIELLERCIREYLDERAPRRFAVKQPLLVKITN 122
LRRRG A+IR+FC+R+GV ++ +TI++ LE CIRE L+E APR AV P+ + I N
Sbjct 297 LRRRGYTAASIREFCKRIGVTKQDNTIEMASLESCIREDLNENAPRAMAVIDPVKLVIEN 356
Query 123 FGAE-EVLTIPSDPKRPAAGSRQLAFTNELYIDAEDFMENPPKEFHRLSPGKEVRLRSAY 181
+ E E++T+P+ P +P GSRQ+ F+ E++ID DF E K++ RL GKEVRLR+AY
Sbjct 357 YQGEGEMVTMPNHPNKPEMGSRQVPFSGEIWIDRADFREEANKQYKRLVLGKEVRLRNAY 416
Query 182 WIKCNEVEKDANGNITAIICSYDPQTLNCSVAPDGRKVKGTIHWLPAKYAVP 233
IK VEKDA GNIT I C+YD TL+ A DGRKVKG IHW+ A +A+P
Sbjct 417 VIKAERVEKDAEGNITTIFCTYDADTLSKDPA-DGRKVKGVIHWVSAAHALP 467
> bbo:BBOV_III006400 17.m07570; glutaminyl-tRNA synthetase family
protein (EC:6.1.1.18); K01886 glutaminyl-tRNA synthetase
[EC:6.1.1.18]
Length=596
Score = 247 bits (630), Expect = 3e-65, Method: Compositional matrix adjust.
Identities = 128/232 (55%), Positives = 155/232 (66%), Gaps = 8/232 (3%)
Query 4 NRQLYDWFQEALGITRTRQIEFARLNVTYTVMSKRKLLALVNEKVVAGWDDPRLSTLSAL 63
+R LYDWFQE L I TRQIE+ARLN+TY VM+KR L+ LV +V GWDDPR+ T+S L
Sbjct 256 HRPLYDWFQEKLEIPPTRQIEYARLNLTYCVMNKRLLIQLVKRGLVRGWDDPRMPTISGL 315
Query 64 RRRGVPPAAIRDFCERVGVARRPSTIQIELLERCIREYLDERAPRRFAVKQPLLVKITNF 123
RRRG P AAIR FC +VGVA+R + IQIELLE C+RE ++E A R FAV PLLV+I N
Sbjct 316 RRRGCPAAAIRRFCRKVGVAKRENLIQIELLEDCMREVMNENAVRLFAVIDPLLVEIENL 375
Query 124 --GAEEVLTIPSDPKRPAAGSRQLAFTNELYIDAEDFMENPPKEFHRLSPGKEVRLRSAY 181
EEV+ + A G+R+L F Y+D +DFME P FHRL G EVRLR Y
Sbjct 376 PEDFEEVVEVDG-----AEGNRRLTFGKRFYVDRKDFMEEPSTGFHRLFVGNEVRLRHCY 430
Query 182 WIKCNEVEKDANGNITAIICSYDPQTLNCSVAPDGRKVKGTIHWLPAKYAVP 233
WIKC K+ N + +IC+YDP T + PDGRKVK TIHWL K + P
Sbjct 431 WIKCINFVKN-NDKVEKLICTYDPSTKGGAAPPDGRKVKATIHWLNEKDSKP 481
> tpv:TP02_0322 glutaminyl-tRNA synthetase; K01886 glutaminyl-tRNA
synthetase [EC:6.1.1.18]
Length=619
Score = 241 bits (614), Expect = 2e-63, Method: Compositional matrix adjust.
Identities = 123/234 (52%), Positives = 159/234 (67%), Gaps = 10/234 (4%)
Query 4 NRQLYDWFQEALGITRTRQIEFARLNVTYTVMSKRKLLALVNEKVVAGWDDPRLSTLSAL 63
+R LYDWFQE LGI RTRQIEFARLN++Y VMSKR LL LVN K+V+GWDDPR+ T+S L
Sbjct 274 HRPLYDWFQEKLGILRTRQIEFARLNLSYCVMSKRLLLHLVNTKIVSGWDDPRMPTISGL 333
Query 64 RRRGVPPAAIRDFCERVGVARRPSTIQIELLERCIREYLDERAPRRFAVKQPLLVKITNF 123
RRRG P +IR+FC ++GVA+R + IQIELLE CIRE L++ + R FAV PLLV+I N
Sbjct 334 RRRGCTPESIRNFCNKIGVAKRENMIQIELLENCIREDLNKVSTRFFAVLDPLLVEIENL 393
Query 124 G---AEEVLTIPSDPKRPAAGSRQLAFTNELYIDAEDFMENPPKEFHRLSPGKEVRLRSA 180
E+V ++P +R L F+ +Y+D +DF E+P +F+RL G EVRL
Sbjct 394 DDTFTEKVEVPCNNPNY----TRTLTFSKRIYVDRKDFSESPSSDFYRLYQGNEVRLFYT 449
Query 181 YWIKCNEVEKDANGNIT-AIICSYDPQTLNCSVAPDGRKVKGTIHWLPAKYAVP 233
YWIKC +V K NG++ ++C YDP+T + RKVK TIHWL + P
Sbjct 450 YWIKCTKVVK--NGDVVEKLVCEYDPKTKGGKLEDKSRKVKATIHWLGSNDWFP 501
> pfa:PF13_0170 glutaminyl-tRNA synthetase, putative (EC:6.1.1.18);
K01886 glutaminyl-tRNA synthetase [EC:6.1.1.18]
Length=918
Score = 236 bits (603), Expect = 4e-62, Method: Composition-based stats.
Identities = 124/253 (49%), Positives = 158/253 (62%), Gaps = 27/253 (10%)
Query 4 NRQLYDWFQEALGITRTRQIEFARLNVTYTVMSKRKLLALVNEKVVAGWDDPRLSTLSAL 63
+R LY+WFQE L I +TRQIEFARLNVTY VMSKRKLL LVNEK V WDDPR+ T+S +
Sbjct 491 HRPLYEWFQEKLHIFKTRQIEFARLNVTYMVMSKRKLLTLVNEKYVKDWDDPRMPTISGM 550
Query 64 RRRGVPPAAIRDFCERVGVARRPSTIQIELLERCIREYLDERAPRRFAVKQPLLVKITNF 123
RRRG P AI+DFC +VG+A+R + I I+LLE C+RE +D++A R FA+ +PL V ITN+
Sbjct 551 RRRGYSPDAIKDFCNKVGIAKRENMIPIDLLEFCVREDMDKKAIRLFAILKPLKVIITNY 610
Query 124 GA----EEV--LTIPSDPKRPAAGSRQLAFTNELYIDAEDFMENPPKEFHRLSPGKEVRL 177
E + L + PK G R L F E++ID +DF E P F RL+P + VRL
Sbjct 611 NDAPDYEHIHNLVASNHPKMEQMGFRNLKFEKEIFIDHDDFQEIPQDNFFRLAPNRTVRL 670
Query 178 RSAYWIKCNEVEKDANGNITAIICSYDPQTLN--CSVAP-------------------DG 216
R A+ I CNEV KD G + + C+YDP + + C+ D
Sbjct 671 RYAFCITCNEVIKDDQGKVIELRCTYDPSSKSGLCTNQKKDNVNRQGGDGDNKQGGGGDN 730
Query 217 RKVKGTIHWLPAK 229
+KVK TIHWL AK
Sbjct 731 KKVKATIHWLCAK 743
> cpv:cgd1_2130 glutaminyl-tRNA synthetase, of predicted bacterial
origin (EC:6.1.1.18); K01886 glutaminyl-tRNA synthetase
[EC:6.1.1.18]
Length=640
Score = 185 bits (470), Expect = 1e-46, Method: Compositional matrix adjust.
Identities = 109/244 (44%), Positives = 140/244 (57%), Gaps = 23/244 (9%)
Query 4 NRQLYDWFQEALGITRTRQIEFARLNVTYTVMSKRKLLALVNEKVVAGWDDPRLSTLSAL 63
+R LYDW Q L IT+TRQIEFARLN+TY V+SKRKLL LVNEK VAGWDDPR+ TLSA+
Sbjct 248 HRPLYDWLQSELEITKTRQIEFARLNMTYLVLSKRKLLLLVNEKHVAGWDDPRMPTLSAV 307
Query 64 RRRGVPPAAIRDFCERVGVARRPSTIQIELLERCIREYLDERAPRRFAVKQPLLVKITN- 122
RR+G PP ++ DFC+++G ++R I I+LLE C++E L RRFA QPL V I N
Sbjct 308 RRKGYPPQSLWDFCDKIGASKRDQMIHIQLLEDCVKENLHSICQRRFACIQPLKVVIINW 367
Query 123 ---FGAEEV-LTIPSDP-KRPAAGSRQLAFTNELYIDAEDFM---------ENPPKEFHR 168
F E V +T+ + P G R L T L+I+ DF + PP +F R
Sbjct 368 DECFQKEAVEITVKNHPDDNIDMGERVLYLTRNLWIEKSDFWFDENQEIDSQQPPGDFKR 427
Query 169 LSPGKEVRLRSAYWIKCNEVEKDANGN----ITAIICSYDPQTLNCSVAPDGRKVKGTIH 224
L G VRL+ +Y I C D + + II S D N + + +K IH
Sbjct 428 LFIGGSVRLKYSYAITCKNAIIDTDTKEVKEVHCIIHSSD----NENDSNVNKKKLAAIH 483
Query 225 WLPA 228
WL
Sbjct 484 WLST 487
> sce:YOR168W GLN4; Gln4p (EC:6.1.1.18); K01886 glutaminyl-tRNA
synthetase [EC:6.1.1.18]
Length=809
Score = 169 bits (429), Expect = 6e-42, Method: Compositional matrix adjust.
Identities = 98/236 (41%), Positives = 135/236 (57%), Gaps = 9/236 (3%)
Query 4 NRQLYDWFQEALGITRTRQIEFARLNVTYTVMSKRKLLALVNEKVVAGWDDPRLSTLSAL 63
+R+ Y+W + + + R Q E+ RLN+T TV+SKRK+ LV+EK V GWDDPRL TL A+
Sbjct 465 SRESYEWLCDQVHVFRPAQREYGRLNITGTVLSKRKIAQLVDEKFVRGWDDPRLFTLEAI 524
Query 64 RRRGVPPAAIRDFCERVGVARRPSTIQIELLERCIREYLDERAPRRFAVKQPLLVKITNF 123
RRRGVPP AI F +GV + IQ+ E +R+YL++ PR V P+ V + N
Sbjct 525 RRRGVPPGAILSFINTLGVTTSTTNIQVVRFESAVRKYLEDTTPRLMFVLDPVEVVVDNL 584
Query 124 G--AEEVLTIPSDPKRPAAGSRQLAFTNELYIDAEDFMEN-PPKEFHRLSPGKEVRL-RS 179
EE+ TIP P P G R + FTN+ YI+ DF EN KEF RL+P + V L +
Sbjct 585 SDDYEELATIPYRPGTPEFGERTVPFTNKFYIERSDFSENVDDKEFFRLTPNQPVGLIKV 644
Query 180 AYWIKCNEVEKDANGNITAIICSYDPQTLNCSVAPDGRKVKGTIHWLP--AKYAVP 233
++ + +EKD G I I +YD + S +K K I W+P +KY P
Sbjct 645 SHTVSFKSLEKDEAGKIIRIHVNYDNKVEEGSKP---KKPKTYIQWVPISSKYNSP 697
> xla:379670 qars, MGC69128; glutaminyl-tRNA synthetase (EC:6.1.1.18);
K01886 glutaminyl-tRNA synthetase [EC:6.1.1.18]
Length=772
Score = 167 bits (422), Expect = 4e-41, Method: Compositional matrix adjust.
Identities = 93/225 (41%), Positives = 129/225 (57%), Gaps = 10/225 (4%)
Query 5 RQLYDWFQEALGITRTRQIEFARLNVTYTVMSKRKLLALVNEKVVAGWDDPRLSTLSALR 64
R Y W L + Q E+ RLN+ YTV+SKRK++ LV V WDDPRL TL+ALR
Sbjct 461 RSSYFWLCNTLDVYCPVQWEYGRLNLHYTVVSKRKIIKLVETGAVRDWDDPRLFTLTALR 520
Query 65 RRGVPPAAIRDFCERVGVARRPSTIQIELLERCIREYLDERAPRRFAVKQPLLVKITNFG 124
RRG PP AI +FC RVGV +T++ LLE C+R+ L+E APR AV +PL V ITN
Sbjct 521 RRGFPPEAINNFCARVGVTVAQTTMEPHLLESCVRDVLNETAPRIMAVLEPLKVTITNLP 580
Query 125 AEEVL--TIPSDPKRPAAGSRQLAFTNELYIDAEDFMENPPKEFHRLSPGKEVRLRSA-Y 181
AE+ + ++P+ P + G + F++ +YI+ DF E K + RL+P + V LR A Y
Sbjct 581 AEKAIDVSVPNFPADESKGFHVVPFSSTIYIEQSDFREVMEKGYKRLTPDQPVGLRHAGY 640
Query 182 WIKCNEVEKDANGNITAIICSYDPQTLNCSVAPDGRKVKGTIHWL 226
I + KD +GN+ + + C+ K K IHW+
Sbjct 641 VISLQNIVKDQSGNVIEL-------EVTCTKTDVAEKPKAFIHWV 678
> hsa:5859 QARS, GLNRS, PRO2195; glutaminyl-tRNA synthetase (EC:6.1.1.18);
K01886 glutaminyl-tRNA synthetase [EC:6.1.1.18]
Length=775
Score = 159 bits (401), Expect = 1e-38, Method: Compositional matrix adjust.
Identities = 95/225 (42%), Positives = 126/225 (56%), Gaps = 10/225 (4%)
Query 5 RQLYDWFQEALGITRTRQIEFARLNVTYTVMSKRKLLALVNEKVVAGWDDPRLSTLSALR 64
R Y W AL + Q E+ RLN+ Y V+SKRK+L LV V WDDPRL TL+ALR
Sbjct 464 RSSYFWLCNALDVYCPVQWEYGRLNLHYAVVSKRKILQLVATGAVRDWDDPRLFTLTALR 523
Query 65 RRGVPPAAIRDFCERVGVARRPSTIQIELLERCIREYLDERAPRRFAVKQPLLVKITNFG 124
RRG PP AI +FC RVGV +T++ LLE C+R+ L++ APR AV + L V ITNF
Sbjct 524 RRGFPPEAINNFCARVGVTVAQTTMEPHLLEACVRDVLNDTAPRAMAVLESLRVIITNFP 583
Query 125 AEEVLTI--PSDPKRPAAGSRQLAFTNELYIDAEDFMENPPKEFHRLSPGKEVRLR-SAY 181
A + L I P+ P G Q+ F ++I+ DF E P F RL+ G+ V LR + Y
Sbjct 584 AAKSLDIQVPNFPADETKGFHQVPFAPIVFIERTDFKEEPEPGFKRLAWGQPVGLRHTGY 643
Query 182 WIKCNEVEKDANGNITAIICSYDPQTLNCSVAPDGRKVKGTIHWL 226
I+ V K +G + ++ + C A G K K IHW+
Sbjct 644 VIELQHVVKGPSGCVESL-------EVTCRRADAGEKPKAFIHWV 681
> mmu:97541 Qars, 1110018N24Rik, 1200016L19Rik, C80286, GLNRS,
MGC105238; glutaminyl-tRNA synthetase (EC:6.1.1.18); K01886
glutaminyl-tRNA synthetase [EC:6.1.1.18]
Length=606
Score = 158 bits (400), Expect = 1e-38, Method: Compositional matrix adjust.
Identities = 92/225 (40%), Positives = 127/225 (56%), Gaps = 10/225 (4%)
Query 5 RQLYDWFQEALGITRTRQIEFARLNVTYTVMSKRKLLALVNEKVVAGWDDPRLSTLSALR 64
R Y W AL + Q E+ RLN+ Y V+SKRK+L LV V WDDPRL TL+ALR
Sbjct 295 RSSYFWLCNALKVYCPVQWEYGRLNLHYAVVSKRKILQLVAAGAVRDWDDPRLFTLTALR 354
Query 65 RRGVPPAAIRDFCERVGVARRPSTIQIELLERCIREYLDERAPRRFAVKQPLLVKITNFG 124
RRG PP AI +FC RVGV +T++ LLE C+R+ L++ APR AV +PL V ITNF
Sbjct 355 RRGFPPEAINNFCARVGVTVAQTTMEPHLLEACVRDVLNDAAPRAMAVLEPLQVVITNFP 414
Query 125 AEEVLTI--PSDPKRPAAGSRQLAFTNELYIDAEDFMENPPKEFHRLSPGKEVRLR-SAY 181
A + L I P+ P G Q+ F + ++I+ DF E + RL+ G+ V LR + Y
Sbjct 415 APKPLDIRVPNFPADETKGFHQVPFASTVFIERSDFKEESEPGYKRLASGQPVGLRHTGY 474
Query 182 WIKCNEVEKDANGNITAIICSYDPQTLNCSVAPDGRKVKGTIHWL 226
I+ + + ++G + + + C A G K K IHW+
Sbjct 475 VIELQNIVRGSSGCVERL-------EVTCRRADAGEKPKAFIHWV 512
> ath:AT1G25350 OVA9; OVA9 (ovule abortion 9); glutamine-tRNA
ligase (EC:6.1.1.18); K01886 glutaminyl-tRNA synthetase [EC:6.1.1.18]
Length=795
Score = 150 bits (380), Expect = 3e-36, Method: Compositional matrix adjust.
Identities = 92/229 (40%), Positives = 123/229 (53%), Gaps = 15/229 (6%)
Query 5 RQLYDWFQEALGITRTRQIEFARLNVTYTVMSKRKLLALVNEKVVAGWDDPRLSTLSALR 64
R Y W +L + E++RLNVT TVMSKRKL +V K V GWDDPRL TLS LR
Sbjct 476 RASYYWLLHSLSLYMPYVWEYSRLNVTNTVMSKRKLNYIVTNKYVDGWDDPRLLTLSGLR 535
Query 65 RRGVPPAAIRDFCERVGVARRP-STIQIELLERCIREYLDERAPRRFAVKQPLLVKITNF 123
RRGV AI F +G+ R S I + LE IRE L++ APR V PL V ITN
Sbjct 536 RRGVTSTAINAFVRGIGITRSDGSMIHVSRLEHHIREELNKTAPRTMVVLNPLKVVITNL 595
Query 124 GAEEVLTI-----PSDPKRPAAGSRQLAFTNELYIDAEDFMENPPKEFHRLSPGKEVRLR 178
+++++ + P + ++ F+ +YID DF K+++ L+PGK V LR
Sbjct 596 ESDKLIELDAKRWPDAQNDDPSAFYKVPFSRVVYIDQSDFRMKDSKDYYGLAPGKSVLLR 655
Query 179 SAYWIKC-NEVEKDANGNITAIICSYDPQTLNCSVAPDGRKVKGTIHWL 226
A+ IKC N V D N + I YDP+ + K KG +HW+
Sbjct 656 YAFPIKCTNVVFADDNETVREIHAEYDPEKKS--------KPKGVLHWV 696
> cel:Y41E3.4 ers-1; glutamyl(E)/glutaminyl(Q) tRNA Synthetase
family member (ers-1); K01886 glutaminyl-tRNA synthetase [EC:6.1.1.18]
Length=786
Score = 133 bits (334), Expect = 6e-31, Method: Compositional matrix adjust.
Identities = 83/230 (36%), Positives = 123/230 (53%), Gaps = 17/230 (7%)
Query 5 RQLYDWFQEALGITRTRQIEFARLNVTYTVMSKRKLLALVNEKVVAGWDDPRLSTLSALR 64
R Y W AL I Q E+ RLNV YTV+SKRK+L L+ K V WDDPRL TL+ALR
Sbjct 470 RSSYYWLCNALDIYCPVQWEYGRLNVNYTVVSKRKILKLITTKTVNDWDDPRLFTLTALR 529
Query 65 RRGVPPAAIRDFCERVGVARRPSTIQIELLERCIREYLDERAPRRFAVKQPLLVKITNF- 123
RRG+P AI F ++G+ I +L+ +R+YL+ APR AV + L + I NF
Sbjct 530 RRGIPSEAINRFVAKLGLTMSQMVIDPHVLDATVRDYLNIHAPRTMAVLEGLKLTIENFS 589
Query 124 -----GAEEVLTIPSDPKRPAAGSRQLAFTNELYIDAEDFM-ENPPKEFHRLSPGKEVRL 177
+ +V PSDP P S ++ E++I+ D+ ++ K F RL+P + V L
Sbjct 590 ELNLPSSVDVPDFPSDPTDPRKHS--VSVDREIFIEKSDYKPDDSDKSFRRLTPKQAVGL 647
Query 178 RS-AYWIKCNEVEKDANGNITAIICSYDPQTLNCSVAPDGRKVKGTIHWL 226
+ ++ + KDA G++T ++ + + + K K IHW+
Sbjct 648 KHIGLVLRFVKEVKDAEGHVTEVVVKAEKLS-------EKDKPKAFIHWV 690
> mmu:107508 Eprs, 2410081F06Rik, 3010002K18Rik, C79379, Qprs;
glutamyl-prolyl-tRNA synthetase (EC:6.1.1.15 6.1.1.17); K14163
bifunctional glutamyl/prolyl-tRNA synthetase [EC:6.1.1.17
6.1.1.15]
Length=1512
Score = 96.3 bits (238), Expect = 9e-20, Method: Compositional matrix adjust.
Identities = 61/198 (30%), Positives = 103/198 (52%), Gaps = 10/198 (5%)
Query 3 RNRQLYDWFQEALGITRTRQIEFARLNVTYTVMSKRKLLALVNEKVVAGWDDPRLSTLSA 62
R+ Q Y W EALGI + E++RLN+ TV+SKRKL VNE +V GWDDPR T+
Sbjct 402 RDEQFY-WIIEALGIRKPYIWEYSRLNLNNTVLSKRKLTWFVNEGLVDGWDDPRFPTVRG 460
Query 63 LRRRGVPPAAIRDFCERVGVARRPSTIQIELLERCIREYLDERAPRRFAVKQPLLVKITN 122
+ RRG+ ++ F G +R ++ + + ++ +D APR A+ + +V +
Sbjct 461 VLRRGMTVEGLKQFIAAQGSSRSVVNMEWDKIWAFNKKVIDPVAPRYVALLKKEVVPVNV 520
Query 123 FGA-EEVLTIPSDPKRPAAGSRQLAFTNELYIDAEDFMENPPKEFHRLSPGKEVRLRSAY 181
A EE+ + PK P G + + ++ +++I+ D S G+ V +
Sbjct 521 LDAQEEMKEVARHPKNPDVGLKPVWYSPKVFIEGAD--------AETFSEGEMVTFINWG 572
Query 182 WIKCNEVEKDANGNITAI 199
I ++ K+A+G IT++
Sbjct 573 NINITKIHKNADGKITSL 590
> cpv:cgd8_790 glutamate--tRNA ligase ; K01885 glutamyl-tRNA synthetase
[EC:6.1.1.17]
Length=555
Score = 94.0 bits (232), Expect = 4e-19, Method: Compositional matrix adjust.
Identities = 66/224 (29%), Positives = 103/224 (45%), Gaps = 23/224 (10%)
Query 8 YDWFQEALGITRTRQIEFARLNVTYTVMSKRKLLALVNEKVVAGWDDPRLSTLSALRRRG 67
Y+W ALG+ EF+RLN TV+SKRKL +VN +V GWDDPR T+ + RRG
Sbjct 238 YNWVINALGLRPVEIYEFSRLNFVNTVLSKRKLTWIVNNGLVEGWDDPRFPTVRGIVRRG 297
Query 68 VPPAAIRDFCERVGVARRPSTIQIELLERCIREYLDERAPRRFAVKQPLLVKITNFGAEE 127
+ P+A+ F G ++ + ++ + L ++ +D PR FAV Q + + EE
Sbjct 298 LSPSALLQFVTEQGPSKNSNLMEWDKLWTINKQLMDPVVPRFFAVGQDAVALNLSEAPEE 357
Query 128 VLTIPSD--PKRPAAGSRQLAFTNELYIDAEDFMENPPKEFHRLSPGKEVRLR---SAYW 182
L D K P G Q+ + ID +D + + G+E+ L +A
Sbjct 358 PLINQRDLHQKNPDLGKGQIVMYKSILIDRDDATQ--------ILSGEEITLMKWGNAII 409
Query 183 IKCNEVEKDANGNITAIICSYDPQTLNCSVAPDGRKVKGTIHWL 226
++++K G + ++ D R K IHWL
Sbjct 410 QDISQIDKQPTGIFEGKL----------NLEGDFRSTKKKIHWL 443
> sce:YGL245W GUS1, GSN1; GluRS (EC:6.1.1.17); K01885 glutamyl-tRNA
synthetase [EC:6.1.1.17]
Length=708
Score = 92.4 bits (228), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 53/160 (33%), Positives = 88/160 (55%), Gaps = 8/160 (5%)
Query 3 RNRQLYDWFQEALGITRTRQIEFARLNVTYTVMSKRKLLALVNEKVVAGWDDPRLSTLSA 62
RN Q YDW +AL + + +FAR+N T++SKRKL +V++ +V WDDPR T+
Sbjct 407 RNAQ-YDWMLQALRLRKVHIWDFARINFVRTLLSKRKLQWMVDKDLVGNWDDPRFPTVRG 465
Query 63 LRRRGVPPAAIRDFCERVGVARRPSTIQIELLERCIREYLDERAPRRFAVKQPLLVKITN 122
+RRRG+ +R+F G +R ++ L+ ++ +D APR A+ P VKI
Sbjct 466 VRRRGMTVEGLRNFVLSQGPSRNVINLEWNLIWAFNKKVIDPIAPRHTAIVNP--VKIHL 523
Query 123 FGAE-----EVLTIPSDPKRPAAGSRQLAFTNELYIDAED 157
G+E ++ P K PA G +++ + ++ +D +D
Sbjct 524 EGSEAPQEPKIEMKPKHKKNPAVGEKKVIYYKDIVVDKDD 563
> hsa:2058 EPRS, DKFZp313B047, EARS, GLUPRORS, PARS, QARS, QPRS;
glutamyl-prolyl-tRNA synthetase (EC:6.1.1.15 6.1.1.17); K14163
bifunctional glutamyl/prolyl-tRNA synthetase [EC:6.1.1.17
6.1.1.15]
Length=1512
Score = 91.3 bits (225), Expect = 3e-18, Method: Compositional matrix adjust.
Identities = 58/198 (29%), Positives = 102/198 (51%), Gaps = 10/198 (5%)
Query 3 RNRQLYDWFQEALGITRTRQIEFARLNVTYTVMSKRKLLALVNEKVVAGWDDPRLSTLSA 62
R+ Q Y W EALGI + E++RLN+ TV+SKRKL VNE +V GWDDPR T+
Sbjct 402 RDEQFY-WIIEALGIRKPYIWEYSRLNLNNTVLSKRKLTWFVNEGLVDGWDDPRFPTVRG 460
Query 63 LRRRGVPPAAIRDFCERVGVARRPSTIQIELLERCIREYLDERAPRRFAVKQPLLVKITN 122
+ RRG+ ++ F G +R ++ + + ++ +D APR A+ + ++ +
Sbjct 461 VLRRGMTVEGLKQFIAAQGSSRSVVNMEWDKIWAFNKKVIDPVAPRYVALLKKEVIPVNV 520
Query 123 FGA-EEVLTIPSDPKRPAAGSRQLAFTNELYIDAEDFMENPPKEFHRLSPGKEVRLRSAY 181
A EE+ + PK P G + + ++ +++I+ D S G+ V +
Sbjct 521 PEAQEEMKEVAKHPKNPEVGLKPVWYSPKVFIEGAD--------AETFSEGEMVTFINWG 572
Query 182 WIKCNEVEKDANGNITAI 199
+ ++ K+A+G I ++
Sbjct 573 NLNITKIHKNADGKIISL 590
> ath:AT5G26710 glutamate-tRNA ligase, putative / glutamyl-tRNA
synthetase, putatuve / GluRS, putative (EC:6.1.1.17); K01885
glutamyl-tRNA synthetase [EC:6.1.1.17]
Length=719
Score = 89.7 bits (221), Expect = 7e-18, Method: Compositional matrix adjust.
Identities = 69/229 (30%), Positives = 112/229 (48%), Gaps = 21/229 (9%)
Query 3 RNRQLYDWFQEALGITRTRQIEFARLNVTYTVMSKRKLLALVNEKVVAGWDDPRLSTLSA 62
RN Q + ++ +G+ + + EF+RLN+ +T++SKRKLL V +V GWDDPR T+
Sbjct 418 RNAQYFKVLED-MGLRQVQLYEFSRLNLVFTLLSKRKLLWFVQTGLVDGWDDPRFPTVQG 476
Query 63 LRRRGVPPAAIRDFCERVGVARRPSTIQIELLERCIREYLDERAPRRFAV--KQPLLVKI 120
+ RRG+ A+ F G ++ + ++ + L + +D PR AV ++ +L +
Sbjct 477 IVRRGLKIEALIQFILEQGASKNLNLMEWDKLWSINKRIIDPVCPRHTAVVAERRVLFTL 536
Query 121 TNFGAEE--VLTIPSDPKRPAAGSRQLAFTNELYIDAEDFMENPPKEFHRLSPGKEVRLR 178
T+ G +E V IP K AG + FT ++++ D +S G+EV L
Sbjct 537 TD-GPDEPFVRMIPKHKKFEGAGEKATTFTKSIWLEEAD--------ASAISVGEEVTLM 587
Query 179 SAYWIKCNEVEKDANGNITAIICSYDPQTLNCSVAPDGRKVKGTIHWLP 227
E+ KD G +TA+ LN + K+K T WLP
Sbjct 588 DWGNAIVKEITKDEEGRVTAL-----SGVLNLQGSVKTTKLKLT--WLP 629
> tgo:TGME49_063870 glutamyl-tRNA synthetase, putative (EC:6.1.1.18);
K01885 glutamyl-tRNA synthetase [EC:6.1.1.17]
Length=786
Score = 89.7 bits (221), Expect = 8e-18, Method: Compositional matrix adjust.
Identities = 73/229 (31%), Positives = 113/229 (49%), Gaps = 33/229 (14%)
Query 8 YDWFQEALGITRTRQIEFARLNVTYTVMSKRKLLALVNEKVVAGWDDPRLSTLSALRRRG 67
Y W Q+A G+ EF+RL T++SKRKL V+ +V GWDDPR+ T+ +RRRG
Sbjct 452 YQWVQQAAGLPPVHIYEFSRLCFVKTLLSKRKLKQFVDSGLVEGWDDPRMPTVRGIRRRG 511
Query 68 VPPAAIRDFCERVGVARRPSTIQIELLERCIREYLDERAPRRFAVKQ---PLLVKITNFG 124
+ A+ +F G ++ + ++ + L ++ +D PR AV + P+ +K G
Sbjct 512 LQVEALLEFILEQGPSKAGNLMEWDKLWTKNKQIIDPIVPRFMAVGKDAVPVCIK----G 567
Query 125 AEEVLTIPSDPKRPAA-----GSRQLAFTNELYIDAEDFMENPPKEFHRLSPGKEVRLRS 179
A E T+ S +R A G L N+++ID +D G+EV L
Sbjct 568 APE--TVESKKRRMHAKNESLGEADLLLFNKVFIDRDDAA--------LCEDGEEVTL-- 615
Query 180 AYWIKC--NEVEKDANGNITAIICSYDPQTLNCSVAPDGRKVKGTIHWL 226
+W C ++V K A+G I+ I TL+ + D RK K +HWL
Sbjct 616 MHWGNCIFDKVVKTASGEISEI-----QATLH--LEGDFRKTKKKLHWL 657
> tpv:TP01_1053 glutamyl-tRNA synthetase; K01885 glutamyl-tRNA
synthetase [EC:6.1.1.17]
Length=530
Score = 89.0 bits (219), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 65/224 (29%), Positives = 105/224 (46%), Gaps = 25/224 (11%)
Query 7 LYDWFQEALGITRTRQIEFARLNVTYTVMSKRKLLALVNEKVVAGWDDPRLSTLSALRRR 66
LY+W E + EF+R+N T +SKRKL V K+V GWDDPR+ T+ + RR
Sbjct 267 LYNWVLEKCELRHVEIYEFSRMNFVRTTLSKRKLRWFVENKLVTGWDDPRMPTVKGILRR 326
Query 67 GVPPAAIRDFCERVGVARRPSTIQIELLERCIREYLDERAPRRFAV-KQPLLVKITNFGA 125
G+ A+ +F G ++ + ++ + L ++ +D +PR AV +++K+ N+
Sbjct 327 GLSVKALFEFILDQGPSKSVNLMEWDKLWAKNKQIIDPESPRYTAVVSDHVVLKVVNYEE 386
Query 126 EEVLTIPSDPKRPAAGSRQLAFTNELYIDAEDFMENPPKEFHRLSPGKEVRLRSAYWIKC 185
T P PK P L F +++ I+ +DF + + +EV L W
Sbjct 387 PATKTRPLHPKNPDLREIDLVFGDQVMIERDDF--------NLIEKSEEVTLMK--W--- 433
Query 186 NEVEKDANGNITAIICSYDPQTLNCSVAPDGRKVKGTIHWLPAK 229
GN A + + Q L ++ D + K IHWLP K
Sbjct 434 --------GN--AFVDKANLQ-LKLNLQGDFKLTKKKIHWLPLK 466
> dre:562037 eprs, wu:fb38d08, wu:ft34d10; glutamyl-prolyl-tRNA
synthetase; K14163 bifunctional glutamyl/prolyl-tRNA synthetase
[EC:6.1.1.17 6.1.1.15]
Length=1511
Score = 85.9 bits (211), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 53/163 (32%), Positives = 87/163 (53%), Gaps = 5/163 (3%)
Query 3 RNRQLYDWFQEALGITRTRQIEFARLNVTYTVMSKRKLLALVNEKVVAGWDDPRLSTLSA 62
R+ Q Y W EALG+ + E+ARLN+ TV+SKRKL V++ V GWDDPR T+
Sbjct 401 RDEQFY-WVIEALGLRKPYIWEYARLNLNNTVLSKRKLTWFVDKGYVDGWDDPRFPTVRG 459
Query 63 LRRRGVPPAAIRDFCERVGVARRPSTIQIELLERCIREYLDERAPRRFAVKQPLLVKITN 122
+ RRG+ ++ F G +R ++ + + ++ +D APR A+ ++ +
Sbjct 460 VLRRGMTVDGLKQFIAAQGGSRSVVNMEWDKIWAFNKKVIDPVAPRFTALLSSQVIPVCV 519
Query 123 FGAEEVLT-IPSDPKRPAAGSRQLAFTNELYI---DAEDFMEN 161
A+E + +P PK G +Q+ + +++I DAE F E
Sbjct 520 SEAKETMKEVPKHPKNADVGLKQVWYGPKVFIEGADAETFTEG 562
> bbo:BBOV_III011840 17.m08008; glutamyl-tRNA synthetase family
protein (EC:6.1.1.18); K01885 glutamyl-tRNA synthetase [EC:6.1.1.17]
Length=730
Score = 84.3 bits (207), Expect = 4e-16, Method: Compositional matrix adjust.
Identities = 69/225 (30%), Positives = 103/225 (45%), Gaps = 31/225 (13%)
Query 8 YDWFQEALGITRTRQIEFARLNVTYTVMSKRKLLALVNEKVVAGWDDPRLSTLSALRRRG 67
Y WF + + EF+RLN TV+SKRKL V+E +V GWDDPR+ T+ + RRG
Sbjct 437 YQWFIQQCNLRPVEIYEFSRLNFVKTVLSKRKLQWFVDEGIVTGWDDPRMPTVRGIMRRG 496
Query 68 VPPAAIRDFCERVGVARRPSTIQIELLERCIREYLDERAPRRFAVK-QPLLVKITNFGAE 126
+ A+ DF G ++ + ++ + L ++ +D PR AV+ + +K+ NF
Sbjct 497 LTKKALFDFILEQGPSKAVNLMEWDKLWAKNKQIIDPIVPRYAAVEIDAVELKLDNFSQV 556
Query 127 EVLTIPS----DPKRPAAGSRQLAFTNELYIDAEDFMENPPKEFHRLSPGKEVRLRSAYW 182
E+ PS PK P G + FT + +D D E + G+EV L W
Sbjct 557 EL--PPSKRMLHPKDPNMGECDIWFTPTVLLDRVDADE--------IVDGEEVTLMR--W 604
Query 183 IKCNEVEKDANGNITAIICSYDPQTLNCSVAPDGRKVKGTIHWLP 227
GN + S T + D +K K +HWLP
Sbjct 605 -----------GN---VFVSKPAFTGQINPGGDFKKTKKKLHWLP 635
> xla:100158438 eprs; glutamyl-prolyl-tRNA synthetase; K14163
bifunctional glutamyl/prolyl-tRNA synthetase [EC:6.1.1.17 6.1.1.15]
Length=1499
Score = 83.2 bits (204), Expect = 7e-16, Method: Compositional matrix adjust.
Identities = 63/227 (27%), Positives = 110/227 (48%), Gaps = 21/227 (9%)
Query 3 RNRQLYDWFQEALGITRTRQIEFARLNVTYTVMSKRKLLALVNEKVVAGWDDPRLSTLSA 62
R+ Q Y W +ALGI + E++RLN+ TV+SKR+L VNE +V GWDDPR T+
Sbjct 393 RDEQFY-WIIDALGIRKPYIWEYSRLNLNNTVLSKRRLTWFVNEGLVNGWDDPRFPTVRG 451
Query 63 LRRRGVPPAAIRDFCERVGVARRPSTIQIELLERCIREYLDERAPRRFAVKQPLLVKITN 122
+ RRG+ ++ F G +R ++ + + ++ +D APR A+ + +V +
Sbjct 452 VLRRGMTVEGLKQFIAAQGSSRSVVNMEWDKIWAFNKKVIDPVAPRYTALLRNQVVPVNI 511
Query 123 FGAEE-VLTIPSDPKRPAAGSRQLAFTNELYIDAEDFMENPPKEFHRLSPGKEVRLRSAY 181
A+E +L + PK G + + + ++ I+ D LS G+ V +
Sbjct 512 PDAQEGMLEVAKHPKNTEVGLKPVWYGPKVLIEGAD--------AETLSEGETVTFINWG 563
Query 182 WIKCNEVEKDANGNITAIICSYDPQTLNCSVAPDGRKVKGT--IHWL 226
+ ++ +D +G I Q+L+ + + + K T I WL
Sbjct 564 NLIITKIHRDQSGKI---------QSLDAKLNLENKDYKKTTKITWL 601
> cel:ZC434.5 ers-2; glutamyl(E)/glutaminyl(Q) tRNA Synthetase
family member (ers-2); K01885 glutamyl-tRNA synthetase [EC:6.1.1.17]
Length=1149
Score = 79.7 bits (195), Expect = 7e-15, Method: Composition-based stats.
Identities = 52/158 (32%), Positives = 87/158 (55%), Gaps = 4/158 (2%)
Query 3 RNRQLYDWFQEALGITRTRQIEFARLNVTYTVMSKRKLLALVNEKVVAGWDDPRLSTLSA 62
R+ Q Y + +ALG+ R E+ARLN+T TVMSKRKL V+E V GWDDPRL T+
Sbjct 398 RDDQYY-FICDALGLRRPHIWEYARLNMTNTVMSKRKLTWFVDEGHVEGWDDPRLPTVRG 456
Query 63 LRRRGVPPAAIRDFCERVGVARRPSTIQIELLERCIREYLDERAPRRFAV--KQPLL-VK 119
+ RRG+ ++ F G +R ++ + + ++ +D APR A+ PL+ ++
Sbjct 457 VMRRGLTVEGLKQFIVAQGGSRSVVMMEWDKIWAFNKKVIDPVAPRYTALDSTSPLVSIE 516
Query 120 ITNFGAEEVLTIPSDPKRPAAGSRQLAFTNELYIDAED 157
+T+ +++ + PK GS+ + +L ++ D
Sbjct 517 LTDSISDDTSNVSLHPKNAEIGSKDVHKGKKLLLEQVD 554
> pfa:PF13_0257 glutamate--tRNA ligase, putative (EC:6.1.1.17);
K01885 glutamyl-tRNA synthetase [EC:6.1.1.17]
Length=863
Score = 76.6 bits (187), Expect = 6e-14, Method: Composition-based stats.
Identities = 66/227 (29%), Positives = 104/227 (45%), Gaps = 27/227 (11%)
Query 8 YDWFQEALGITRTRQIEFARLNVTYTVMSKRKLLALVNEKVVAGWDDPRLSTLSALRRRG 67
Y+WF L + + EF+RL TVMSKRKL V VV W DPR T+ + RRG
Sbjct 520 YNWFISTLNLRKVYIYEFSRLAFVKTVMSKRKLKWFVENNVVDSWVDPRFPTIKGILRRG 579
Query 68 VPPAAIRDFCERVGVARRPSTIQIELLERCIREYLDERAPRRFAVKQ--PLLVKITNFGA 125
+ A+ F G ++ + +Q + L ++ +D PR AV + +L+ +T+
Sbjct 580 LTKEALFQFILEQGPSKAGNLMQWDKLWSINKQIIDPIIPRYAAVDKNSSILLILTDL-T 638
Query 126 EEVLTIPSD--PKRPAAGSRQLAFTNELYIDAED---FMENPPKEFHRLSPGKEVRLRSA 180
++V+ D K + G+ + + N+ I+ ED +EN +E+ L
Sbjct 639 DQVIQKERDLHMKNKSLGTCNMYYNNKYLIELEDAQTLLEN-----------EEITLIKL 687
Query 181 YWIKCNEVEKDANGNITAIICSYDPQTLNCSVAPDGRKVKGTIHWLP 227
I +EK+ NG I I L+ + D + K IHWLP
Sbjct 688 GNIIIKNIEKE-NGKIKQI------NALS-NFHGDFKTTKKKIHWLP 726
> ath:AT5G19720 tRNA synthetase class I (E and Q) family protein
Length=170
Score = 49.3 bits (116), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 29/97 (29%), Positives = 44/97 (45%), Gaps = 9/97 (9%)
Query 132 PSDPKRPAAGSRQLAFTNELYIDAEDFMENPPKEFHRLSPGKEVRLRSAYWIKC-NEVEK 190
P P + ++ + +Y++ DF K ++ L+PGK V LR + IKC N V
Sbjct 9 PDAPNDDPSAFYKVPLSRVIYMEQSDFQNEDSKNYYGLAPGKSVLLRYTFPIKCTNVVFA 68
Query 191 DANGNITAIICSYDPQTLNCSVAPDGRKVKGTIHWLP 227
D + I YDP+ K KG +HW+P
Sbjct 69 DDTKTVCEIYVEYDPEK--------KIKPKGVLHWVP 97
> cel:C01C7.1 ark-1; A Ras-regulating Kinase family member (ark-1);
K08252 receptor protein-tyrosine kinase [EC:2.7.10.1]
Length=1043
Score = 37.0 bits (84), Expect = 0.054, Method: Compositional matrix adjust.
Identities = 39/129 (30%), Positives = 53/129 (41%), Gaps = 21/129 (16%)
Query 10 WFQEALGITRTRQIEFARLNVTYTVMSKRKLLALVNEKVVAGWDDPRLSTLSALR----R 65
+ QEA +TR R RL Y V+ K + LV+E G S L L R
Sbjct 160 FLQEAAIMTRMRHEHVVRL---YGVVLDTKKIMLVSELATCG------SLLECLHKPALR 210
Query 66 RGVPPAAIRDFCERVGVARRPSTIQIELLERCIREYLDERAPRRFAVKQPLLVKITNFGA 125
P + D+ E++ + + L+R I L A R V P LVKI++FG
Sbjct 211 DSFPVHVLCDYAEQIAMG-----MSYLELQRLIHRDL---AARNVLVFSPKLVKISDFGL 262
Query 126 EEVLTIPSD 134
L I D
Sbjct 263 SRSLGIGED 271
> bbo:BBOV_I003090 19.m02240; elongation factor 2, EF-2; K03234
elongation factor 2
Length=833
Score = 29.6 bits (65), Expect = 9.8, Method: Compositional matrix adjust.
Identities = 32/124 (25%), Positives = 56/124 (45%), Gaps = 15/124 (12%)
Query 94 LERCIREYLDERAPRRFAVKQPLLVKITNFGAEEVLTIPSDPKRPAAGSRQLAFTNELYI 153
+E C+++ DE A F V P++ GAE +T S K P +R L++
Sbjct 529 VEICLKDLRDEYAQIDFIVSDPVVSYRETVGAESSITCLS--KSPNKHNR-------LFM 579
Query 154 DAEDFMENPPK--EFHRLSPGKEVRLRSAYWIKCNEVEKDANGNITAIICSYDPQTLNCS 211
AE F E + E ++++ + R R+ + N+ E D N + I + P+T +
Sbjct 580 KAEPFAEGLSEAIEENKITSRDDARERAN--VLANDFEWDKNAALK--IWCFGPETTGPN 635
Query 212 VAPD 215
+ D
Sbjct 636 ILVD 639
Lambda K H
0.321 0.137 0.415
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 7953524176
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40