bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_0267_orf2
Length=191
Score E
Sequences producing significant alignments: (Bits) Value
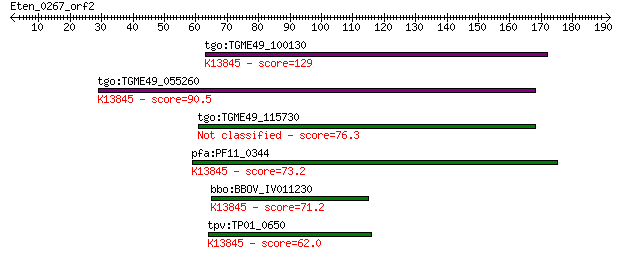
tgo:TGME49_100130 apical membrane antigen, putative ; K13845 a... 129 7e-30
tgo:TGME49_055260 apical membrane antigen 1, putative ; K13845... 90.5 3e-18
tgo:TGME49_115730 apical membrane antigen, putative 76.3 7e-14
pfa:PF11_0344 AMA1, AMA-1, Pf83, RMA-1, RMA1; apical membrane ... 73.2 5e-13
bbo:BBOV_IV011230 23.m06405; apical membrane antigen 1; K13845... 71.2 2e-12
tpv:TP01_0650 apical membrane antigen 1; K13845 apical merozoi... 62.0 1e-09
> tgo:TGME49_100130 apical membrane antigen, putative ; K13845
apical merozoite antigen 1
Length=493
Score = 129 bits (324), Expect = 7e-30, Method: Compositional matrix adjust.
Identities = 59/110 (53%), Positives = 80/110 (72%), Gaps = 5/110 (4%)
Query 63 SNPWGDSMQKFNIPYTHGSGVYVDLGNEKTVSNKKYREPAGRCPVMGKEIRLQQPTTDSS 122
+NPW ++++N+P HGSGVYVDLGN K +S KKYREP G+CP GK I+ QPTT+
Sbjct 54 ANPWAKILERYNVPLVHGSGVYVDLGNTKILSKKKYREPGGKCPNYGKYIKTYQPTTNPE 113
Query 123 IWPGNYLEKVPTKGSPQDTRPLGGGFAMWDTTPV-KISPLTLSELEALAK 171
IWP ++L+ VP +PQDT PLGGGFAM P+ +ISP++L +L+ A+
Sbjct 114 IWPNDFLKPVPYANTPQDTMPLGGGFAM----PMHQISPVSLKDLKDEAE 159
> tgo:TGME49_055260 apical membrane antigen 1, putative ; K13845
apical merozoite antigen 1
Length=569
Score = 90.5 bits (223), Expect = 3e-18, Method: Compositional matrix adjust.
Identities = 61/154 (39%), Positives = 81/154 (52%), Gaps = 17/154 (11%)
Query 29 FGLRRLCCISAVAAFC-LFGAKPSQAAASNGSQVAS-----NPWGDS------MQKFNIP 76
GL + + + A C +F + S + S SQ S NP+ + M++FN+
Sbjct 29 MGLVGVASLLVLVADCTIFASGLSSSTRSRESQTLSASTSGNPFQANVEMKTFMERFNLT 88
Query 77 YTHGSGVYVDLGNEKTVSNKKYREPAGRCPVMGKEIRLQQPTTDSSIWPGNYLEKVPT-K 135
+ H SG+YVDLG +K V YREPAG CP+ GK I LQQP D + N+LE VPT K
Sbjct 89 HHHQSGIYVDLGQDKEVDGTLYREPAGLCPIWGKHIELQQP--DRPPYRNNFLEDVPTEK 146
Query 136 GSPQDTRPLGGGFAMWDTTPV--KISPLTLSELE 167
Q PL GGF + TP +ISP + LE
Sbjct 147 EYKQSGNPLPGGFNLNFVTPSGQRISPFPMELLE 180
> tgo:TGME49_115730 apical membrane antigen, putative
Length=388
Score = 76.3 bits (186), Expect = 7e-14, Method: Compositional matrix adjust.
Identities = 46/115 (40%), Positives = 62/115 (53%), Gaps = 17/115 (14%)
Query 61 VASNPWG------DSMQKFNIPYTHGSGVYVDLGNEKTVSNKKYREPAGRCPVMGKEIRL 114
V NPW D M++FNIP HGSG++VDLG + + YRE G+CPV GK I++
Sbjct 95 VKQNPWATTTAFADFMKRFNIPQVHGSGIFVDLGRD----TEGYREVGGKCPVFGKAIQM 150
Query 115 QQPTTDSSIWPGNYLEKVPTKGSPQDTRPLGGGFA--MWDTTPVKISPLTLSELE 167
QP S+ N+L+ PT +PL GGF T+ K SP+ S L+
Sbjct 151 HQPAEYSN----NFLDDAPTSND-ASKKPLPGGFNNPQVYTSGQKFSPIDDSLLQ 200
> pfa:PF11_0344 AMA1, AMA-1, Pf83, RMA-1, RMA1; apical membrane
antigen 1, AMA1; K13845 apical merozoite antigen 1
Length=622
Score = 73.2 bits (178), Expect = 5e-13, Method: Composition-based stats.
Identities = 43/119 (36%), Positives = 62/119 (52%), Gaps = 18/119 (15%)
Query 59 SQVASNPWGDSMQKFNIPYTHGSGVYVDLGNEKTVSNKKYREPAGRCPVMGKEIRLQQPT 118
S NPW + M K++I HGSG+ VDLG + V+ +YR P+G+CPV GK I ++
Sbjct 103 SNYMGNPWTEYMAKYDIEEVHGSGIRVDLGEDAEVAGTQYRLPSGKCPVFGKGIIIENSN 162
Query 119 TD--SSIWPGN-YLEKVPTKGSPQDTRPLGGGFAMWDTTPVKISPLTLSELEALAKQQR 174
T + + GN YL+ GGFA T P+ +SP+TL E+ K +
Sbjct 163 TTFLTPVATGNQYLKD--------------GGFAFPPTEPL-MSPMTLDEMRHFYKDNK 206
> bbo:BBOV_IV011230 23.m06405; apical membrane antigen 1; K13845
apical merozoite antigen 1
Length=605
Score = 71.2 bits (173), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 30/50 (60%), Positives = 37/50 (74%), Gaps = 0/50 (0%)
Query 65 PWGDSMQKFNIPYTHGSGVYVDLGNEKTVSNKKYREPAGRCPVMGKEIRL 114
PW MQKF+IP HGSG+YVDLG ++V +K YR P G+CPV+GK I L
Sbjct 96 PWIKYMQKFDIPRNHGSGIYVDLGGYESVGSKSYRMPVGKCPVVGKIIDL 145
> tpv:TP01_0650 apical membrane antigen 1; K13845 apical merozoite
antigen 1
Length=785
Score = 62.0 bits (149), Expect = 1e-09, Method: Composition-based stats.
Identities = 29/52 (55%), Positives = 34/52 (65%), Gaps = 0/52 (0%)
Query 64 NPWGDSMQKFNIPYTHGSGVYVDLGNEKTVSNKKYREPAGRCPVMGKEIRLQ 115
N W + M KF+I HGSGVYVDLG TV YR P G+CPV+GK I L+
Sbjct 248 NKWTEFMAKFDIAKVHGSGVYVDLGESATVGIYDYRMPIGKCPVVGKAIILE 299
Lambda K H
0.312 0.128 0.382
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5493959020
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40