bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_0218_orf2
Length=327
Score E
Sequences producing significant alignments: (Bits) Value
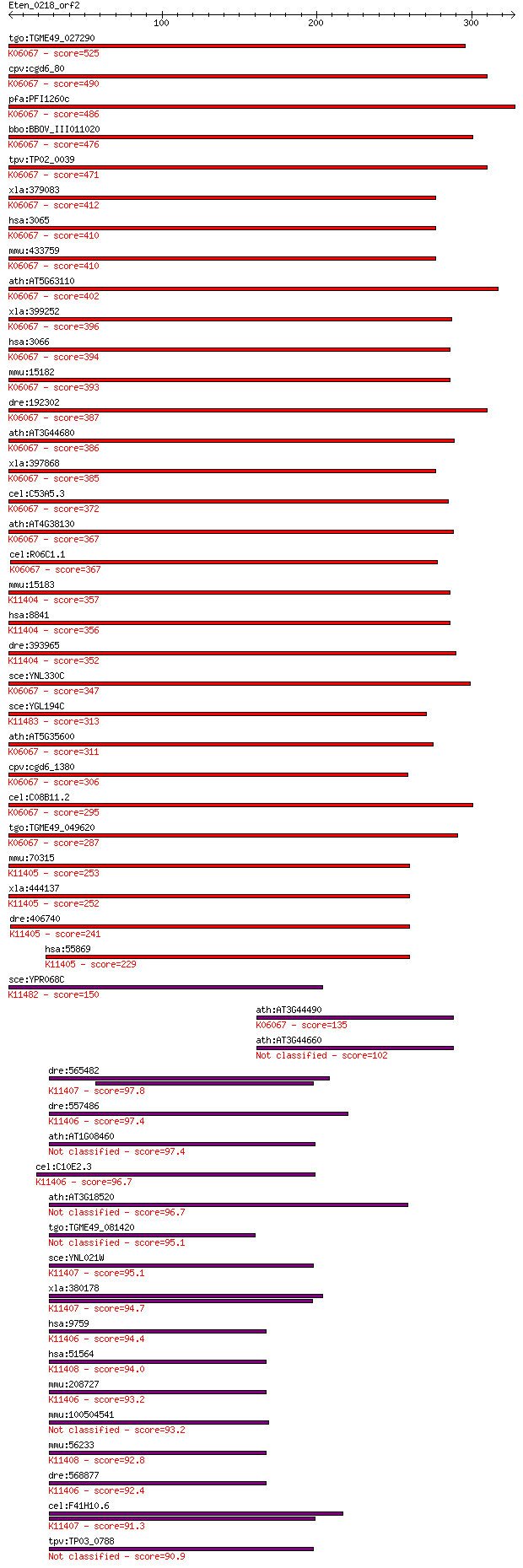
tgo:TGME49_027290 histone deacetylase, putative ; K06067 histo... 525 9e-149
cpv:cgd6_80 RPD3/HD1 histone deacetylase ; K06067 histone deac... 490 3e-138
pfa:PFI1260c PfHDAC1; histone deacetylase; K06067 histone deac... 486 6e-137
bbo:BBOV_III011020 17.m07949; histone deacetylase; K06067 hist... 476 4e-134
tpv:TP02_0039 histone deacetylase; K06067 histone deacetylase ... 471 1e-132
xla:379083 hdac1-b, HD1, MGC53583, RPD3, gon-10, hdac1, hdac1b... 412 8e-115
hsa:3065 HDAC1, DKFZp686H12203, GON-10, HD1, RPD3, RPD3L1; his... 410 2e-114
mmu:433759 Hdac1, HD1, Hdac1-ps, MGC102534, MGC118085, MommeD5... 410 3e-114
ath:AT5G63110 HDA6; HDA6 (HISTONE DEACETYLASE 6); histone deac... 402 1e-111
xla:399252 hdac2, MGC81850; histone deacetylase 2 (EC:3.5.1.98... 396 5e-110
hsa:3066 HDAC2, HD2, RPD3, YAF1; histone deacetylase 2 (EC:3.5... 394 2e-109
mmu:15182 Hdac2, D10Wsu179e, YAF1, Yy1bp, mRPD3; histone deace... 393 4e-109
dre:192302 hdac1, MGC101582, chunp6919, hdac-1, mp:zf637-2-001... 387 2e-107
ath:AT3G44680 HDA9; HDA9 (HISTONE DEACETYLASE 9); histone deac... 386 7e-107
xla:397868 hdac1-a, HD1, HDM, MGC83956, ab21, gon-10, hdac1a, ... 385 1e-106
cel:C53A5.3 hda-1; Histone DeAcetylase family member (hda-1); ... 372 1e-102
ath:AT4G38130 HD1; HD1 (HISTONE DEACETYLASE 1); basal transcri... 367 3e-101
cel:R06C1.1 hda-3; Histone DeAcetylase family member (hda-3); ... 367 3e-101
mmu:15183 Hdac3, AW537363; histone deacetylase 3 (EC:3.5.1.98)... 357 3e-98
hsa:8841 HDAC3, HD3, RPD3, RPD3-2; histone deacetylase 3 (EC:3... 356 6e-98
dre:393965 hdac3, MGC55927, zgc:55927; histone deacetylase 3 (... 352 8e-97
sce:YNL330C RPD3, MOF6, REC3, SDI2, SDS6; Histone deacetylase;... 347 5e-95
sce:YGL194C HOS2, RTL1; Histone deacetylase required for gene ... 313 6e-85
ath:AT5G35600 HDA7; HDA7 (histone deacetylase7); histone deace... 311 2e-84
cpv:cgd6_1380 histone deacetylase ; K06067 histone deacetylase... 306 7e-83
cel:C08B11.2 hda-2; Histone DeAcetylase family member (hda-2);... 295 2e-79
tgo:TGME49_049620 histone deacetylase, putative ; K06067 histo... 287 5e-77
mmu:70315 Hdac8, 2610007D20Rik; histone deacetylase 8 (EC:3.5.... 253 9e-67
xla:444137 hdac8, MGC80565; histone deacetylase 8 (EC:3.5.1.98... 252 1e-66
dre:406740 hdac8, wu:fd19a02, zgc:66196; histone deacetylase 8... 241 4e-63
hsa:55869 HDAC8, HD8, HDACL1, RPD3; histone deacetylase 8 (EC:... 229 8e-60
sce:YPR068C HOS1; Hos1p; K11482 histone deacetylase HOS1 [EC:3... 150 7e-36
ath:AT3G44490 hda17; hda17 (histone deacetylase 17); histone d... 135 2e-31
ath:AT3G44660 hda10; hda10 (histone deacetylase 10); histone d... 102 2e-21
dre:565482 hdac6, wu:fc31d02; histone deacetylase 6; K11407 hi... 97.8 5e-20
dre:557486 histone deacetylase 5-like; K11406 histone deacetyl... 97.4 7e-20
ath:AT1G08460 HDA08; HDA08; histone deacetylase 97.4 7e-20
cel:C10E2.3 hda-4; Histone DeAcetylase family member (hda-4); ... 96.7 1e-19
ath:AT3G18520 HDA15; HDA15; histone deacetylase 96.7 1e-19
tgo:TGME49_081420 histone deacetylase (EC:4.1.1.70 2.7.1.151) 95.1 3e-19
sce:YNL021W HDA1; Hda1p; K11407 histone deacetylase 6/10 [EC:3... 95.1 3e-19
xla:380178 hdac6, MGC53140, hd6, jm21; histone deacetylase 6 (... 94.7 5e-19
hsa:9759 HDAC4, AHO3, BDMR, HA6116, HD4, HDAC-A, HDACA, KIAA02... 94.4 5e-19
hsa:51564 HDAC7, DKFZp586J0917, FLJ99588, HD7A, HDAC7A; histon... 94.0 8e-19
mmu:208727 Hdac4, 4932408F19Rik, AI047285; histone deacetylase... 93.2 1e-18
mmu:100504541 histone deacetylase 9-like 93.2 1e-18
mmu:56233 Hdac7, 5830434K02Rik, HD7, HD7a, Hdac7a, mFLJ00062; ... 92.8 2e-18
dre:568877 hdac4, KIAA0288, wu:fa96d08, wu:fc56f08, zgc:152701... 92.4 2e-18
cel:F41H10.6 hypothetical protein; K11407 histone deacetylase ... 91.3 4e-18
tpv:TP03_0788 hypothetical protein 90.9 6e-18
> tgo:TGME49_027290 histone deacetylase, putative ; K06067 histone
deacetylase 1/2 [EC:3.5.1.98]
Length=451
Score = 525 bits (1352), Expect = 9e-149, Method: Compositional matrix adjust.
Identities = 241/296 (81%), Positives = 273/296 (92%), Gaps = 1/296 (0%)
Query 1 AGASIDAAKKLNYQQADLCINWSGGLHHAKRAEASGFCYVNDIVLGILELLKYHPRVMYI 60
AGASIDAAKKLN+ QAD+C+NWSGGLHHAKR+EASGFCY+NDIVLGILELLKYH RVMYI
Sbjct 115 AGASIDAAKKLNHHQADICVNWSGGLHHAKRSEASGFCYINDIVLGILELLKYHARVMYI 174
Query 61 DIDVHHGDGVEEAFYVSHRVLTVSFHKFGDFFPGTGDITDVGAAQGKYFSVNVPLNDGMD 120
DID+HHGDGVEEAFYVSHRV+TVSFHKFGDFFPGTGD+TDVGA+QGKY++VNVPLNDGMD
Sbjct 175 DIDIHHGDGVEEAFYVSHRVMTVSFHKFGDFFPGTGDVTDVGASQGKYYAVNVPLNDGMD 234
Query 121 DESFIGLFKPIIAKCVEVYRPGAVVLQCGADSLSGDRLGKFNLTVKGHAACVQFVKSFNL 180
D+SF+ LFKP+I KCV+VYRPGA+VLQCGADSL+GDRLGKFNLT+KGHAACV FVKS ++
Sbjct 235 DDSFVALFKPVITKCVDVYRPGAIVLQCGADSLTGDRLGKFNLTIKGHAACVAFVKSLDI 294
Query 181 PLLLLGGGGYTIRNVSRCWVYETAVALDRHRELSPQVPLNDYYDYYAPDFQLHLPPSPVP 240
PLL+LGGGGYTIRNV+RCW YET V LDRHRE+SP VPLNDYYDYYAPDFQLHL PS +P
Sbjct 295 PLLVLGGGGYTIRNVARCWAYETGVVLDRHREMSPHVPLNDYYDYYAPDFQLHLTPSSIP 354
Query 241 NFNSPDHLQRLKARVLQNVALLEHAPGVQFAYVPPDCFGED-EDEDEQQQLLVEWE 295
N NSP+HL+++K RVL N++ LEHAPGVQFAYVPPD FGED +DEDE Q V+ E
Sbjct 355 NSNSPEHLEKIKTRVLSNLSYLEHAPGVQFAYVPPDFFGEDNDDEDEFMQNQVDNE 410
> cpv:cgd6_80 RPD3/HD1 histone deacetylase ; K06067 histone deacetylase
1/2 [EC:3.5.1.98]
Length=460
Score = 490 bits (1262), Expect = 3e-138, Method: Compositional matrix adjust.
Identities = 224/315 (71%), Positives = 268/315 (85%), Gaps = 6/315 (1%)
Query 1 AGASIDAAKKLNYQQADLCINWSGGLHHAKRAEASGFCYVNDIVLGILELLKYHPRVMYI 60
AG SID A KLN +Q+D+CINWSGGLHHAKR+EASGFCY+NDIVLGILELLKYH RVMYI
Sbjct 126 AGGSIDGAYKLNNEQSDICINWSGGLHHAKRSEASGFCYINDIVLGILELLKYHARVMYI 185
Query 61 DIDVHHGDGVEEAFYVSHRVLTVSFHKFGDFFPGTGDITDVGAAQGKYFSVNVPLNDGMD 120
DIDVHHGDGVEEAFY+SHRVLTVSFHKFG+FFPGTGDITD+G AQGKY+SVNVPLNDG+D
Sbjct 186 DIDVHHGDGVEEAFYLSHRVLTVSFHKFGEFFPGTGDITDIGVAQGKYYSVNVPLNDGID 245
Query 121 DESFIGLFKPIIAKCVEVYRPGAVVLQCGADSLSGDRLGKFNLTVKGHAACVQFVKSFNL 180
D+SF+ LFKPII+KC+EVYRPGA+VLQCGADS+ GDRLG+FNL++KGHA CV+F K FN+
Sbjct 246 DDSFLSLFKPIISKCIEVYRPGAIVLQCGADSVRGDRLGRFNLSIKGHAECVEFCKKFNI 305
Query 181 PLLLLGGGGYTIRNVSRCWVYETAVALDRHRELSPQVPLNDYYDYYAPDFQLHLPPSPVP 240
PLL+LGGGGYTIRNV+R W YETA LDR +S +PLNDYYDY+APDF+LH+PP +P
Sbjct 306 PLLILGGGGYTIRNVARTWAYETATILDRTDLISDNIPLNDYYDYFAPDFKLHIPPLNLP 365
Query 241 NFNSPDHLQRLKARVLQNVALLEHAPGVQFAYVPPDCFGEDEDEDEQQ------QLLVEW 294
N NSP+HL+++KA+V+ N+ LEHAPGV+FAYVP D F + ++Q + L W
Sbjct 366 NMNSPEHLEKIKAKVIDNLRYLEHAPGVEFAYVPSDFFDREASNLQKQEDEEREEELSSW 425
Query 295 EGGGLAAGSSSSSSS 309
+GGG AAGS+ S +
Sbjct 426 QGGGRAAGSTESQGN 440
> pfa:PFI1260c PfHDAC1; histone deacetylase; K06067 histone deacetylase
1/2 [EC:3.5.1.98]
Length=449
Score = 486 bits (1250), Expect = 6e-137, Method: Compositional matrix adjust.
Identities = 222/329 (67%), Positives = 270/329 (82%), Gaps = 2/329 (0%)
Query 1 AGASIDAAKKLNYQQADLCINWSGGLHHAKRAEASGFCYVNDIVLGILELLKYHPRVMYI 60
AGASID A KLN+ AD+C+NWSGGLHHAK +EASGFCY+NDIVLGILELLKYH RVMYI
Sbjct 112 AGASIDGASKLNHHCADICVNWSGGLHHAKMSEASGFCYINDIVLGILELLKYHARVMYI 171
Query 61 DIDVHHGDGVEEAFYVSHRVLTVSFHKFGDFFPGTGDITDVGAAQGKYFSVNVPLNDGMD 120
DIDVHHGDGVEEAFYV+HRV+TVSFHKFGD+FPGTGDITDVG GKY+SVNVPLNDGM
Sbjct 172 DIDVHHGDGVEEAFYVTHRVMTVSFHKFGDYFPGTGDITDVGVNHGKYYSVNVPLNDGMT 231
Query 121 DESFIGLFKPIIAKCVEVYRPGAVVLQCGADSLSGDRLGKFNLTVKGHAACVQFVKSFNL 180
D++F+ LFK +I KCV+ YRPGA+++QCGADSL+GDRLG+FNLT+KGHA CV+ V+S+N+
Sbjct 232 DDAFVDLFKVVIDKCVQTYRPGAIIIQCGADSLTGDRLGRFNLTIKGHARCVEHVRSYNI 291
Query 181 PLLLLGGGGYTIRNVSRCWVYETAVALDRHRELSPQVPLNDYYDYYAPDFQLHLPPSPVP 240
PLL+LGGGGYTIRNVSRCW YET V L++H E+ Q+ LNDYYDYYAPDFQLHL PS +P
Sbjct 292 PLLVLGGGGYTIRNVSRCWAYETGVVLNKHHEMPDQISLNDYYDYYAPDFQLHLQPSNIP 351
Query 241 NFNSPDHLQRLKARVLQNVALLEHAPGVQFAYVPPDCFGEDEDE--DEQQQLLVEWEGGG 298
N+NSP+HL R+K ++ +N+ +EHAPGVQF+YVPPD F D D+ D+ Q L + GGG
Sbjct 352 NYNSPEHLSRIKMKIAENLRHIEHAPGVQFSYVPPDFFNSDIDDESDKNQYELKDDSGGG 411
Query 299 LAAGSSSSSSSSSSCRSCWSACESFVQLS 327
A G+ + S++ + + F LS
Sbjct 412 RAPGTRAKEHSTTHHLRRKNYDDDFFDLS 440
> bbo:BBOV_III011020 17.m07949; histone deacetylase; K06067 histone
deacetylase 1/2 [EC:3.5.1.98]
Length=449
Score = 476 bits (1226), Expect = 4e-134, Method: Compositional matrix adjust.
Identities = 217/300 (72%), Positives = 259/300 (86%), Gaps = 0/300 (0%)
Query 1 AGASIDAAKKLNYQQADLCINWSGGLHHAKRAEASGFCYVNDIVLGILELLKYHPRVMYI 60
+GASID A +LN QQAD+ INWSGGLHHAKR+EASGFCY+NDIVL ILELLKYH RVMYI
Sbjct 110 SGASIDGAHRLNNQQADISINWSGGLHHAKRSEASGFCYLNDIVLAILELLKYHARVMYI 169
Query 61 DIDVHHGDGVEEAFYVSHRVLTVSFHKFGDFFPGTGDITDVGAAQGKYFSVNVPLNDGMD 120
DIDVHHGDGVEEAFYV+HRV+TVSFHKFG+FFPGTGD+TDVG A GKY+SVNVPLNDGMD
Sbjct 170 DIDVHHGDGVEEAFYVTHRVMTVSFHKFGNFFPGTGDVTDVGVASGKYYSVNVPLNDGMD 229
Query 121 DESFIGLFKPIIAKCVEVYRPGAVVLQCGADSLSGDRLGKFNLTVKGHAACVQFVKSFNL 180
DESF+ +F+ ++ KCVEVY PGA+VLQCGADSL+GDRLG+FNLT KGHA CV F +S N+
Sbjct 230 DESFVDMFRTVVGKCVEVYEPGAIVLQCGADSLTGDRLGRFNLTNKGHAGCVAFCRSLNI 289
Query 181 PLLLLGGGGYTIRNVSRCWVYETAVALDRHRELSPQVPLNDYYDYYAPDFQLHLPPSPVP 240
PLL+LGGGGYTIRNV+RCW YET V LD+H E++ Q+ LN+YYDYYAPDF LHL P+ +P
Sbjct 290 PLLVLGGGGYTIRNVARCWAYETGVVLDKHNEMAEQISLNEYYDYYAPDFNLHLQPTNMP 349
Query 241 NFNSPDHLQRLKARVLQNVALLEHAPGVQFAYVPPDCFGEDEDEDEQQQLLVEWEGGGLA 300
N+N+ +HL R+K ++++N+ +E APGVQFA+VP D F D+DEDE QL V EGGG+A
Sbjct 350 NYNTSEHLDRIKMKIIENLRHVERAPGVQFAHVPNDFFQYDDDEDEAAQLEVFDEGGGVA 409
> tpv:TP02_0039 histone deacetylase; K06067 histone deacetylase
1/2 [EC:3.5.1.98]
Length=447
Score = 471 bits (1213), Expect = 1e-132, Method: Compositional matrix adjust.
Identities = 218/309 (70%), Positives = 265/309 (85%), Gaps = 0/309 (0%)
Query 1 AGASIDAAKKLNYQQADLCINWSGGLHHAKRAEASGFCYVNDIVLGILELLKYHPRVMYI 60
+GASIDAA +LN QQAD+C+NWSGGLHHAKR+EASGFCY+NDIVLGILELLKYH RVMYI
Sbjct 110 SGASIDAAHRLNNQQADICVNWSGGLHHAKRSEASGFCYLNDIVLGILELLKYHARVMYI 169
Query 61 DIDVHHGDGVEEAFYVSHRVLTVSFHKFGDFFPGTGDITDVGAAQGKYFSVNVPLNDGMD 120
DIDVHHGDGVEEAFYV+HRV+T+SFHKFG+FFPGTGD+TDVG + GKY+SVNVPLNDG+D
Sbjct 170 DIDVHHGDGVEEAFYVTHRVMTISFHKFGNFFPGTGDVTDVGVSSGKYYSVNVPLNDGID 229
Query 121 DESFIGLFKPIIAKCVEVYRPGAVVLQCGADSLSGDRLGKFNLTVKGHAACVQFVKSFNL 180
DESF+ LFK ++ KCVEVY PGA+VLQCGADSL+GDRLG+FNLT+KGHAACVQ+V+S N+
Sbjct 230 DESFVDLFKVVVGKCVEVYCPGAIVLQCGADSLTGDRLGRFNLTIKGHAACVQYVRSLNI 289
Query 181 PLLLLGGGGYTIRNVSRCWVYETAVALDRHRELSPQVPLNDYYDYYAPDFQLHLPPSPVP 240
PLL+LGGGGYTIRNV+RCW YET V L++H ++S Q+ LNDYYDYYAPDFQLHL PS +
Sbjct 290 PLLVLGGGGYTIRNVARCWAYETGVILNKHTDMSNQISLNDYYDYYAPDFQLHLTPSQMT 349
Query 241 NFNSPDHLQRLKARVLQNVALLEHAPGVQFAYVPPDCFGEDEDEDEQQQLLVEWEGGGLA 300
N+N+ +HL ++K ++L N+ +E +PGVQFA+VPPD D+D D+ Q V EGGG
Sbjct 350 NYNTKEHLDKIKVKILDNLRYVEKSPGVQFAHVPPDFLTRDDDVDDDLQKQVFDEGGGRT 409
Query 301 AGSSSSSSS 309
+ S+ S
Sbjct 410 SLSTRKRIS 418
> xla:379083 hdac1-b, HD1, MGC53583, RPD3, gon-10, hdac1, hdac1b,
rpd3l1; histone deacetylase 1 (EC:3.5.1.98); K06067 histone
deacetylase 1/2 [EC:3.5.1.98]
Length=480
Score = 412 bits (1059), Expect = 8e-115, Method: Compositional matrix adjust.
Identities = 178/276 (64%), Positives = 233/276 (84%), Gaps = 2/276 (0%)
Query 1 AGASIDAAKKLNYQQADLCINWSGGLHHAKRAEASGFCYVNDIVLGILELLKYHPRVMYI 60
G S+ +A KLN QQ D+ +NWSGGLHHAK++EASGFCYVNDIVL ILELLKYH RV+YI
Sbjct 114 TGGSVASAVKLNKQQTDISVNWSGGLHHAKKSEASGFCYVNDIVLAILELLKYHQRVVYI 173
Query 61 DIDVHHGDGVEEAFYVSHRVLTVSFHKFGDFFPGTGDITDVGAAQGKYFSVNVPLNDGMD 120
DID+HHGDGVEEAFY + RV++VSFHK+G++FPGTGD+ D+GA +GKY++VN PL DG+D
Sbjct 174 DIDIHHGDGVEEAFYTTDRVMSVSFHKYGEYFPGTGDLRDIGAGKGKYYAVNYPLRDGID 233
Query 121 DESFIGLFKPIIAKCVEVYRPGAVVLQCGADSLSGDRLGKFNLTVKGHAACVQFVKSFNL 180
DES+ +FKP++ K +E+++P AVVLQCGADSLSGDRLG FNLT+KGHA CV+F+K+FNL
Sbjct 234 DESYEAIFKPVMTKVMEMFQPSAVVLQCGADSLSGDRLGCFNLTIKGHAKCVEFIKTFNL 293
Query 181 PLLLLGGGGYTIRNVSRCWVYETAVALDRHRELSPQVPLNDYYDYYAPDFQLHLPPSPVP 240
P+L+LGGGGYTIRNV+RCW YETAVALD E+ ++P NDY++Y+ PDF+LH+ PS +
Sbjct 294 PMLMLGGGGYTIRNVARCWTYETAVALD--SEIPNELPYNDYFEYFGPDFKLHISPSNMT 351
Query 241 NFNSPDHLQRLKARVLQNVALLEHAPGVQFAYVPPD 276
N N+ ++L+++K R+ +N+ +L HAPGVQ +P D
Sbjct 352 NQNTNEYLEKIKQRLFENLRMLPHAPGVQMQAIPED 387
> hsa:3065 HDAC1, DKFZp686H12203, GON-10, HD1, RPD3, RPD3L1; histone
deacetylase 1 (EC:3.5.1.98); K06067 histone deacetylase
1/2 [EC:3.5.1.98]
Length=482
Score = 410 bits (1055), Expect = 2e-114, Method: Compositional matrix adjust.
Identities = 179/276 (64%), Positives = 234/276 (84%), Gaps = 2/276 (0%)
Query 1 AGASIDAAKKLNYQQADLCINWSGGLHHAKRAEASGFCYVNDIVLGILELLKYHPRVMYI 60
G S+ +A KLN QQ D+ +NW+GGLHHAK++EASGFCYVNDIVL ILELLKYH RV+YI
Sbjct 114 TGGSVASAVKLNKQQTDIAVNWAGGLHHAKKSEASGFCYVNDIVLAILELLKYHQRVLYI 173
Query 61 DIDVHHGDGVEEAFYVSHRVLTVSFHKFGDFFPGTGDITDVGAAQGKYFSVNVPLNDGMD 120
DID+HHGDGVEEAFY + RV+TVSFHK+G++FPGTGD+ D+GA +GKY++VN PL DG+D
Sbjct 174 DIDIHHGDGVEEAFYTTDRVMTVSFHKYGEYFPGTGDLRDIGAGKGKYYAVNYPLRDGID 233
Query 121 DESFIGLFKPIIAKCVEVYRPGAVVLQCGADSLSGDRLGKFNLTVKGHAACVQFVKSFNL 180
DES+ +FKP+++K +E+++P AVVLQCG+DSLSGDRLG FNLT+KGHA CV+FVKSFNL
Sbjct 234 DESYEAIFKPVMSKVMEMFQPSAVVLQCGSDSLSGDRLGCFNLTIKGHAKCVEFVKSFNL 293
Query 181 PLLLLGGGGYTIRNVSRCWVYETAVALDRHRELSPQVPLNDYYDYYAPDFQLHLPPSPVP 240
P+L+LGGGGYTIRNV+RCW YETAVALD E+ ++P NDY++Y+ PDF+LH+ PS +
Sbjct 294 PMLMLGGGGYTIRNVARCWTYETAVALD--TEIPNELPYNDYFEYFGPDFKLHISPSNMT 351
Query 241 NFNSPDHLQRLKARVLQNVALLEHAPGVQFAYVPPD 276
N N+ ++L+++K R+ +N+ +L HAPGVQ +P D
Sbjct 352 NQNTNEYLEKIKQRLFENLRMLPHAPGVQMQAIPED 387
> mmu:433759 Hdac1, HD1, Hdac1-ps, MGC102534, MGC118085, MommeD5,
RPD3; histone deacetylase 1 (EC:3.5.1.98); K06067 histone
deacetylase 1/2 [EC:3.5.1.98]
Length=482
Score = 410 bits (1055), Expect = 3e-114, Method: Compositional matrix adjust.
Identities = 179/276 (64%), Positives = 234/276 (84%), Gaps = 2/276 (0%)
Query 1 AGASIDAAKKLNYQQADLCINWSGGLHHAKRAEASGFCYVNDIVLGILELLKYHPRVMYI 60
G S+ +A KLN QQ D+ +NW+GGLHHAK++EASGFCYVNDIVL ILELLKYH RV+YI
Sbjct 114 TGGSVASAVKLNKQQTDIAVNWAGGLHHAKKSEASGFCYVNDIVLAILELLKYHQRVLYI 173
Query 61 DIDVHHGDGVEEAFYVSHRVLTVSFHKFGDFFPGTGDITDVGAAQGKYFSVNVPLNDGMD 120
DID+HHGDGVEEAFY + RV+TVSFHK+G++FPGTGD+ D+GA +GKY++VN PL DG+D
Sbjct 174 DIDIHHGDGVEEAFYTTDRVMTVSFHKYGEYFPGTGDLRDIGAGKGKYYAVNYPLRDGID 233
Query 121 DESFIGLFKPIIAKCVEVYRPGAVVLQCGADSLSGDRLGKFNLTVKGHAACVQFVKSFNL 180
DES+ +FKP+++K +E+++P AVVLQCG+DSLSGDRLG FNLT+KGHA CV+FVKSFNL
Sbjct 234 DESYEAIFKPVMSKVMEMFQPSAVVLQCGSDSLSGDRLGCFNLTIKGHAKCVEFVKSFNL 293
Query 181 PLLLLGGGGYTIRNVSRCWVYETAVALDRHRELSPQVPLNDYYDYYAPDFQLHLPPSPVP 240
P+L+LGGGGYTIRNV+RCW YETAVALD E+ ++P NDY++Y+ PDF+LH+ PS +
Sbjct 294 PMLMLGGGGYTIRNVARCWTYETAVALD--TEIPNELPYNDYFEYFGPDFKLHISPSNMT 351
Query 241 NFNSPDHLQRLKARVLQNVALLEHAPGVQFAYVPPD 276
N N+ ++L+++K R+ +N+ +L HAPGVQ +P D
Sbjct 352 NQNTNEYLEKIKQRLFENLRMLPHAPGVQMQAIPED 387
> ath:AT5G63110 HDA6; HDA6 (HISTONE DEACETYLASE 6); histone deacetylase;
K06067 histone deacetylase 1/2 [EC:3.5.1.98]
Length=471
Score = 402 bits (1032), Expect = 1e-111, Method: Compositional matrix adjust.
Identities = 194/318 (61%), Positives = 240/318 (75%), Gaps = 4/318 (1%)
Query 1 AGASIDAAKKLNYQQADLCINWSGGLHHAKRAEASGFCYVNDIVLGILELLKYHPRVMYI 60
AG SI AA KLN Q AD+ INW GGLHHAK++EASGFCYVNDIVLGILELLK RV+YI
Sbjct 126 AGGSIGAAVKLNRQDADIAINWGGGLHHAKKSEASGFCYVNDIVLGILELLKMFKRVLYI 185
Query 61 DIDVHHGDGVEEAFYVSHRVLTVSFHKFGDFFPGTGDITDVGAAQGKYFSVNVPLNDGMD 120
DIDVHHGDGVEEAFY + RV+TVSFHKFGDFFPGTG I DVGA +GKY+++NVPLNDGMD
Sbjct 186 DIDVHHGDGVEEAFYTTDRVMTVSFHKFGDFFPGTGHIRDVGAEKGKYYALNVPLNDGMD 245
Query 121 DESFIGLFKPIIAKCVEVYRPGAVVLQCGADSLSGDRLGKFNLTVKGHAACVQFVKSFNL 180
DESF LF+P+I K +EVY+P AVVLQCGADSLSGDRLG FNL+VKGHA C++F++S+N+
Sbjct 246 DESFRSLFRPLIQKVMEVYQPEAVVLQCGADSLSGDRLGCFNLSVKGHADCLRFLRSYNV 305
Query 181 PLLLLGGGGYTIRNVSRCWVYETAVALDRHRELSPQVPLNDYYDYYAPDFQLHLPPSPVP 240
PL++LGGGGYTIRNV+RCW YETAVA+ E ++P N+Y++Y+ PD+ LH+ PSP+
Sbjct 306 PLMVLGGGGYTIRNVARCWCYETAVAVG--VEPDNKLPYNEYFEYFGPDYTLHVDPSPME 363
Query 241 NFNSPDHLQRLKARVLQNVALLEHAPGVQFAYVPP--DCFGEDEDEDEQQQLLVEWEGGG 298
N N+P ++R++ +L+ ++ L HAP VQF + PP E ED+ E + W G
Sbjct 364 NLNTPKDMERIRNTLLEQLSGLIHAPSVQFQHTPPVNRVLDEPEDDMETRPKPRIWSGTA 423
Query 299 LAAGSSSSSSSSSSCRSC 316
S SC
Sbjct 424 TYESDSDDDDKPLHGYSC 441
> xla:399252 hdac2, MGC81850; histone deacetylase 2 (EC:3.5.1.98);
K06067 histone deacetylase 1/2 [EC:3.5.1.98]
Length=488
Score = 396 bits (1018), Expect = 5e-110, Method: Compositional matrix adjust.
Identities = 186/286 (65%), Positives = 239/286 (83%), Gaps = 2/286 (0%)
Query 1 AGASIDAAKKLNYQQADLCINWSGGLHHAKRAEASGFCYVNDIVLGILELLKYHPRVMYI 60
G S+ A KLN QQ D+ +NW+GGLHHAK++EASGFCYVNDIVLGILELLKYH RV+YI
Sbjct 115 TGGSVAGAVKLNRQQTDMAVNWAGGLHHAKKSEASGFCYVNDIVLGILELLKYHQRVLYI 174
Query 61 DIDVHHGDGVEEAFYVSHRVLTVSFHKFGDFFPGTGDITDVGAAQGKYFSVNVPLNDGMD 120
DID+HHGDGVEEAFY + RV+TVSFHK+G++FPGTGD+ D+GA +GKY++VN P+ DG+D
Sbjct 175 DIDIHHGDGVEEAFYTTDRVMTVSFHKYGEYFPGTGDLRDIGAGKGKYYAVNFPMRDGID 234
Query 121 DESFIGLFKPIIAKCVEVYRPGAVVLQCGADSLSGDRLGKFNLTVKGHAACVQFVKSFNL 180
DES+ +FKPII+K +E+Y+P AVVLQCGADSLSGDRLG FNLTVKGHA CV+ VK+FNL
Sbjct 235 DESYGQIFKPIISKVMEMYQPSAVVLQCGADSLSGDRLGCFNLTVKGHAKCVEVVKTFNL 294
Query 181 PLLLLGGGGYTIRNVSRCWVYETAVALDRHRELSPQVPLNDYYDYYAPDFQLHLPPSPVP 240
PLL+LGGGGYTIRNV+RCW YETAVALD E+ ++P NDY++Y+ PDF+LH+ PS +
Sbjct 295 PLLMLGGGGYTIRNVARCWTYETAVALD--CEIPNELPYNDYFEYFGPDFKLHISPSNMT 352
Query 241 NFNSPDHLQRLKARVLQNVALLEHAPGVQFAYVPPDCFGEDEDEDE 286
N N+P++++++K R+ +N+ +L HAPGVQ +P D ED ++E
Sbjct 353 NQNTPEYMEKIKQRLFENLRMLPHAPGVQMQAIPEDAVQEDSGDEE 398
> hsa:3066 HDAC2, HD2, RPD3, YAF1; histone deacetylase 2 (EC:3.5.1.98);
K06067 histone deacetylase 1/2 [EC:3.5.1.98]
Length=488
Score = 394 bits (1012), Expect = 2e-109, Method: Compositional matrix adjust.
Identities = 187/286 (65%), Positives = 238/286 (83%), Gaps = 3/286 (1%)
Query 1 AGASIDAAKKLNYQQADLCINWSGGLHHAKRAEASGFCYVNDIVLGILELLKYHPRVMYI 60
G S+ A KLN QQ D+ +NW+GGLHHAK++EASGFCYVNDIVL ILELLKYH RV+YI
Sbjct 115 TGGSVAGAVKLNRQQTDMAVNWAGGLHHAKKSEASGFCYVNDIVLAILELLKYHQRVLYI 174
Query 61 DIDVHHGDGVEEAFYVSHRVLTVSFHKFGDFFPGTGDITDVGAAQGKYFSVNVPLNDGMD 120
DID+HHGDGVEEAFY + RV+TVSFHK+G++FPGTGD+ D+GA +GKY++VN P+ DG+D
Sbjct 175 DIDIHHGDGVEEAFYTTDRVMTVSFHKYGEYFPGTGDLRDIGAGKGKYYAVNFPMRDGID 234
Query 121 DESFIGLFKPIIAKCVEVYRPGAVVLQCGADSLSGDRLGKFNLTVKGHAACVQFVKSFNL 180
DES+ +FKPII+K +E+Y+P AVVLQCGADSLSGDRLG FNLTVKGHA CV+ VK+FNL
Sbjct 235 DESYGQIFKPIISKVMEMYQPSAVVLQCGADSLSGDRLGCFNLTVKGHAKCVEVVKTFNL 294
Query 181 PLLLLGGGGYTIRNVSRCWVYETAVALDRHRELSPQVPLNDYYDYYAPDFQLHLPPSPVP 240
PLL+LGGGGYTIRNV+RCW YETAVALD E+ ++P NDY++Y+ PDF+LH+ PS +
Sbjct 295 PLLMLGGGGYTIRNVARCWTYETAVALD--CEIPNELPYNDYFEYFGPDFKLHISPSNMT 352
Query 241 NFNSPDHLQRLKARVLQNVALLEHAPGVQFAYVPPDCFGEDE-DED 285
N N+P++++++K R+ +N+ +L HAPGVQ +P D ED DED
Sbjct 353 NQNTPEYMEKIKQRLFENLRMLPHAPGVQMQAIPEDAVHEDSGDED 398
> mmu:15182 Hdac2, D10Wsu179e, YAF1, Yy1bp, mRPD3; histone deacetylase
2 (EC:3.5.1.98); K06067 histone deacetylase 1/2 [EC:3.5.1.98]
Length=488
Score = 393 bits (1010), Expect = 4e-109, Method: Compositional matrix adjust.
Identities = 187/286 (65%), Positives = 238/286 (83%), Gaps = 3/286 (1%)
Query 1 AGASIDAAKKLNYQQADLCINWSGGLHHAKRAEASGFCYVNDIVLGILELLKYHPRVMYI 60
G S+ A KLN QQ D+ +NW+GGLHHAK++EASGFCYVNDIVL ILELLKYH RV+YI
Sbjct 115 TGGSVAGAVKLNRQQTDMAVNWAGGLHHAKKSEASGFCYVNDIVLAILELLKYHQRVLYI 174
Query 61 DIDVHHGDGVEEAFYVSHRVLTVSFHKFGDFFPGTGDITDVGAAQGKYFSVNVPLNDGMD 120
DID+HHGDGVEEAFY + RV+TVSFHK+G++FPGTGD+ D+GA +GKY++VN P+ DG+D
Sbjct 175 DIDIHHGDGVEEAFYTTDRVMTVSFHKYGEYFPGTGDLRDIGAGKGKYYAVNFPMRDGID 234
Query 121 DESFIGLFKPIIAKCVEVYRPGAVVLQCGADSLSGDRLGKFNLTVKGHAACVQFVKSFNL 180
DES+ +FKPII+K +E+Y+P AVVLQCGADSLSGDRLG FNLTVKGHA CV+ VK+FNL
Sbjct 235 DESYGQIFKPIISKVMEMYQPSAVVLQCGADSLSGDRLGCFNLTVKGHAKCVEVVKTFNL 294
Query 181 PLLLLGGGGYTIRNVSRCWVYETAVALDRHRELSPQVPLNDYYDYYAPDFQLHLPPSPVP 240
PLL+LGGGGYTIRNV+RCW YETAVALD E+ ++P NDY++Y+ PDF+LH+ PS +
Sbjct 295 PLLMLGGGGYTIRNVARCWTYETAVALD--CEIPNELPYNDYFEYFGPDFKLHISPSNMT 352
Query 241 NFNSPDHLQRLKARVLQNVALLEHAPGVQFAYVPPDCFGEDE-DED 285
N N+P++++++K R+ +N+ +L HAPGVQ +P D ED DED
Sbjct 353 NQNTPEYMEKIKQRLFENLRMLPHAPGVQMQAIPEDAVHEDSGDED 398
> dre:192302 hdac1, MGC101582, chunp6919, hdac-1, mp:zf637-2-001987,
wu:fb19h11, wu:fi06f03, zgc:101582, zgc:65818; histone
deacetylase 1 (EC:3.5.1.98); K06067 histone deacetylase 1/2
[EC:3.5.1.98]
Length=480
Score = 387 bits (995), Expect = 2e-107, Method: Compositional matrix adjust.
Identities = 186/312 (59%), Positives = 245/312 (78%), Gaps = 5/312 (1%)
Query 1 AGASIDAAKKLNYQQADLCINWSGGLHHAKRAEASGFCYVNDIVLGILELLKYHPRVMYI 60
G S+ A KLN QQ D+ INW+GGLHHAK++EASGFCYVNDIVL ILELLKYH RV+YI
Sbjct 115 TGGSVAGAVKLNKQQTDIAINWAGGLHHAKKSEASGFCYVNDIVLAILELLKYHQRVLYI 174
Query 61 DIDVHHGDGVEEAFYVSHRVLTVSFHKFGDFFPGTGDITDVGAAQGKYFSVNVPLNDGMD 120
DID+HHGDGVEEAFY + RV+TVSFHK+G++FPGTGD+ D+GA +GKY++VN PL DG+D
Sbjct 175 DIDIHHGDGVEEAFYTTDRVMTVSFHKYGEYFPGTGDLRDIGAGKGKYYAVNYPLRDGID 234
Query 121 DESFIGLFKPIIAKCVEVYRPGAVVLQCGADSLSGDRLGKFNLTVKGHAACVQFVKSFNL 180
DES+ +FKPI++K +E+Y+P AVVLQCGADSLSGDRLG FNLT+KGHA CV+++KSFNL
Sbjct 235 DESYEAIFKPIMSKVMEMYQPSAVVLQCGADSLSGDRLGCFNLTIKGHAKCVEYMKSFNL 294
Query 181 PLLLLGGGGYTIRNVSRCWVYETAVALDRHRELSPQVPLNDYYDYYAPDFQLHLPPSPVP 240
PLL+LGGGGYTI+NV+RCW +ETAVALD + ++P NDY++Y+ PDF+LH+ P +
Sbjct 295 PLLMLGGGGYTIKNVARCWTFETAVALD--STIPNELPYNDYFEYFGPDFKLHISPFNMT 352
Query 241 NFNSPDHLQRLKARVLQNVALLEHAPGVQFAYVPPDCFGE---DEDEDEQQQLLVEWEGG 297
N N+ D+L+++K R+ +N+ +L HAPGVQ +P D E DE++D +++ +
Sbjct 353 NQNTNDYLEKIKQRLFENLRMLPHAPGVQMQAIPEDAVQEDSGDEEDDPDKRISIRAHDK 412
Query 298 GLAAGSSSSSSS 309
+A S S
Sbjct 413 RIACDEEFSDSE 424
> ath:AT3G44680 HDA9; HDA9 (HISTONE DEACETYLASE 9); histone deacetylase;
K06067 histone deacetylase 1/2 [EC:3.5.1.98]
Length=426
Score = 386 bits (991), Expect = 7e-107, Method: Compositional matrix adjust.
Identities = 177/289 (61%), Positives = 228/289 (78%), Gaps = 3/289 (1%)
Query 1 AGASIDAAKKLNYQQADLCINWSGGLHHAKRAEASGFCYVNDIVLGILELLKYHPRVMYI 60
AG +IDAA++LN + D+ INW+GGLHHAK+ +ASGFCY+ND+VLGILELLK+HPRV+YI
Sbjct 110 AGGTIDAARRLNNKLCDIAINWAGGLHHAKKCDASGFCYINDLVLGILELLKHHPRVLYI 169
Query 61 DIDVHHGDGVEEAFYVSHRVLTVSFHKFGD-FFPGTGDITDVGAAQGKYFSVNVPLNDGM 119
DIDVHHGDGVEEAFY + RV+TVSFHKFGD FFPGTGD+ ++G +GK++++NVPL DG+
Sbjct 170 DIDVHHGDGVEEAFYFTDRVMTVSFHKFGDKFFPGTGDVKEIGEREGKFYAINVPLKDGI 229
Query 120 DDESFIGLFKPIIAKCVEVYRPGAVVLQCGADSLSGDRLGKFNLTVKGHAACVQFVKSFN 179
DD SF LF+ II+K VE+Y+PGA+VLQCGADSL+ DRLG FNL++ GHA CV+FVK FN
Sbjct 230 DDSSFNRLFRTIISKVVEIYQPGAIVLQCGADSLARDRLGCFNLSIDGHAECVKFVKKFN 289
Query 180 LPLLLLGGGGYTIRNVSRCWVYETAVALDRHRELSPQVPLNDYYDYYAPDFQLHLPPSPV 239
LPLL+ GGGGYT NV+RCW ET + LD EL ++P NDY Y+APDF L +P +
Sbjct 290 LPLLVTGGGGYTKENVARCWTVETGILLD--TELPNEIPENDYIKYFAPDFSLKIPGGHI 347
Query 240 PNFNSPDHLQRLKARVLQNVALLEHAPGVQFAYVPPDCFGEDEDEDEQQ 288
N N+ ++ +K ++L+N+ ++HAP VQ VPPD + D DEDEQ
Sbjct 348 ENLNTKSYISSIKVQILENLRYIQHAPSVQMQEVPPDFYIPDFDEDEQN 396
> xla:397868 hdac1-a, HD1, HDM, MGC83956, ab21, gon-10, hdac1a,
rpd3, rpd3l1; histone deacetylase 1 (EC:3.5.1.98); K06067
histone deacetylase 1/2 [EC:3.5.1.98]
Length=480
Score = 385 bits (990), Expect = 1e-106, Method: Compositional matrix adjust.
Identities = 180/276 (65%), Positives = 233/276 (84%), Gaps = 2/276 (0%)
Query 1 AGASIDAAKKLNYQQADLCINWSGGLHHAKRAEASGFCYVNDIVLGILELLKYHPRVMYI 60
AG S+ +A KLN QQ D+ +NWSGGLHHAK++EASGFCYVNDIVL ILELLKYH RV+YI
Sbjct 114 AGGSVASAVKLNKQQTDISVNWSGGLHHAKKSEASGFCYVNDIVLAILELLKYHQRVVYI 173
Query 61 DIDVHHGDGVEEAFYVSHRVLTVSFHKFGDFFPGTGDITDVGAAQGKYFSVNVPLNDGMD 120
DID+HHGDGVEEAFY + RV+TVSFHK+G++FPGTGD+ D+GA +GKY++VN L DG+D
Sbjct 174 DIDIHHGDGVEEAFYTTDRVMTVSFHKYGEYFPGTGDLRDIGAGKGKYYAVNYALRDGID 233
Query 121 DESFIGLFKPIIAKCVEVYRPGAVVLQCGADSLSGDRLGKFNLTVKGHAACVQFVKSFNL 180
DES+ +FKP+++K +E+++P AVVLQCGADSLSGDRLG FNLT+KGHA CV+F+K+FNL
Sbjct 234 DESYEAIFKPVMSKVMEMFQPSAVVLQCGADSLSGDRLGCFNLTIKGHAKCVEFIKTFNL 293
Query 181 PLLLLGGGGYTIRNVSRCWVYETAVALDRHRELSPQVPLNDYYDYYAPDFQLHLPPSPVP 240
PLL+LGGGGYTIRNV+RCW YETAVALD E+ ++P NDY++Y+ PDF+LH+ PS +
Sbjct 294 PLLMLGGGGYTIRNVARCWTYETAVALD--SEIPNELPYNDYFEYFGPDFKLHISPSNMT 351
Query 241 NFNSPDHLQRLKARVLQNVALLEHAPGVQFAYVPPD 276
N N+ ++L+++K R+ +N+ +L HAPGVQ V D
Sbjct 352 NQNTNEYLEKIKQRLFENLRMLPHAPGVQMQAVAED 387
> cel:C53A5.3 hda-1; Histone DeAcetylase family member (hda-1);
K06067 histone deacetylase 1/2 [EC:3.5.1.98]
Length=461
Score = 372 bits (955), Expect = 1e-102, Method: Compositional matrix adjust.
Identities = 168/284 (59%), Positives = 219/284 (77%), Gaps = 2/284 (0%)
Query 1 AGASIDAAKKLNYQQADLCINWSGGLHHAKRAEASGFCYVNDIVLGILELLKYHPRVMYI 60
+G S+ AA KLN Q+ D+ INW GGLHHAK++EASGFCY NDIVLGILELLKYH RV+Y+
Sbjct 118 SGGSLAAATKLNKQKVDIAINWMGGLHHAKKSEASGFCYTNDIVLGILELLKYHKRVLYV 177
Query 61 DIDVHHGDGVEEAFYVSHRVLTVSFHKFGDFFPGTGDITDVGAAQGKYFSVNVPLNDGMD 120
DIDVHHGDGVEEAFY + RV+TVSFHK+GDFFPGTGD+ D+GA +GK +SVNVPL DG+
Sbjct 178 DIDVHHGDGVEEAFYTTDRVMTVSFHKYGDFFPGTGDLKDIGAGKGKLYSVNVPLRDGIT 237
Query 121 DESFIGLFKPIIAKCVEVYRPGAVVLQCGADSLSGDRLGKFNLTVKGHAACVQFVKSFNL 180
D S+ +FKPI+ K +E + P AVVLQCGADSL+GDRLG FNLT+KGH C +F +S+N+
Sbjct 238 DVSYQSIFKPIMTKVMERFDPCAVVLQCGADSLNGDRLGPFNLTLKGHGECARFFRSYNV 297
Query 181 PLLLLGGGGYTIRNVSRCWVYETAVALDRHRELSPQVPLNDYYDYYAPDFQLHLPPSPVP 240
PL+++GGGGYT RNV+RCW YET++A+D +E+ ++P NDY++Y+ P+++LH+ S
Sbjct 298 PLMMVGGGGYTPRNVARCWTYETSIAVD--KEVPNELPYNDYFEYFGPNYRLHIESSNAA 355
Query 241 NFNSPDHLQRLKARVLQNVALLEHAPGVQFAYVPPDCFGEDEDE 284
N NS D L +L+ V+ N+ L P VQ +P D D+
Sbjct 356 NENSSDMLAKLQTDVIANLEQLTFVPSVQMRPIPEDALSALNDD 399
> ath:AT4G38130 HD1; HD1 (HISTONE DEACETYLASE 1); basal transcription
repressor/ histone deacetylase/ protein binding; K06067
histone deacetylase 1/2 [EC:3.5.1.98]
Length=469
Score = 367 bits (943), Expect = 3e-101, Method: Compositional matrix adjust.
Identities = 180/287 (62%), Positives = 229/287 (79%), Gaps = 2/287 (0%)
Query 1 AGASIDAAKKLNYQQADLCINWSGGLHHAKRAEASGFCYVNDIVLGILELLKYHPRVMYI 60
AG S+ + KLN+ D+ INW+GGLHHAK+ EASGFCYVNDIVL ILELLK H RV+Y+
Sbjct 122 AGGSVGGSVKLNHGLCDIAINWAGGLHHAKKCEASGFCYVNDIVLAILELLKQHERVLYV 181
Query 61 DIDVHHGDGVEEAFYVSHRVLTVSFHKFGDFFPGTGDITDVGAAQGKYFSVNVPLNDGMD 120
DID+HHGDGVEEAFY + RV+TVSFHKFGD+FPGTG I D+G GKY+S+NVPL+DG+D
Sbjct 182 DIDIHHGDGVEEAFYATDRVMTVSFHKFGDYFPGTGHIQDIGYGSGKYYSLNVPLDDGID 241
Query 121 DESFIGLFKPIIAKCVEVYRPGAVVLQCGADSLSGDRLGKFNLTVKGHAACVQFVKSFNL 180
DES+ LFKPI+ K +E++RPGAVVLQCGADSLSGDRLG FNL++KGHA CV+F++SFN+
Sbjct 242 DESYHLLFKPIMGKVMEIFRPGAVVLQCGADSLSGDRLGCFNLSIKGHAECVKFMRSFNV 301
Query 181 PLLLLGGGGYTIRNVSRCWVYETAVALDRHRELSPQVPLNDYYDYYAPDFQLHLPPSPVP 240
PLLLLGGGGYTIRNV+RCW YET VAL E+ ++P ++YY+Y+ PD+ LH+ PS +
Sbjct 302 PLLLLGGGGYTIRNVARCWCYETGVALG--VEVEDKMPEHEYYEYFGPDYTLHVAPSNME 359
Query 241 NFNSPDHLQRLKARVLQNVALLEHAPGVQFAYVPPDCFGEDEDEDEQ 287
N NS L+ ++ +L N++ L+HAP V F PPD + DED++
Sbjct 360 NKNSRQMLEEIRNDLLHNLSKLQHAPSVPFQERPPDTETPEVDEDQE 406
> cel:R06C1.1 hda-3; Histone DeAcetylase family member (hda-3);
K06067 histone deacetylase 1/2 [EC:3.5.1.98]
Length=465
Score = 367 bits (942), Expect = 3e-101, Method: Compositional matrix adjust.
Identities = 165/277 (59%), Positives = 221/277 (79%), Gaps = 3/277 (1%)
Query 2 GASIDAAKKLNYQQADLCINWSGGLHHAKRAEASGFCYVNDIVLGILELLKYHPRVMYID 61
G S+ AA +LN Q++++ INW GGLHHAK++EASGFCY NDIVL ILELLK+H RV+YID
Sbjct 114 GGSLAAAARLNRQESEIAINWMGGLHHAKKSEASGFCYSNDIVLAILELLKHHKRVLYID 173
Query 62 IDVHHGDGVEEAFYVSHRVLTVSFHKFGDFFPGTGDITDVGAAQGKYFSVNVPLNDGMDD 121
IDVHHGDGVEEAFY + RV+TVSFHK G++FPGTGD+ DVGA GKY+++NVPL DG+DD
Sbjct 174 IDVHHGDGVEEAFYTTDRVMTVSFHKHGEYFPGTGDLKDVGAGSGKYYALNVPLRDGVDD 233
Query 122 ESFIGLFKPIIAKCVEVYRPGAVVLQCGADSLSGDRLGKFNLTVKGHAACVQFVKSFNLP 181
++ +F+ I+ + + ++P AVVLQCGADSL+GDRLG FNLT GH CV+++KSFN+P
Sbjct 234 VTYERIFRTIMGEVMARFQPEAVVLQCGADSLAGDRLGVFNLTTYGHGKCVEYMKSFNVP 293
Query 182 LLLLGGGGYTIRNVSRCWVYETAVALDRHRELSPQVPLNDYYDYYAPDFQLHLPP-SPVP 240
LLL+GGGGYTIRNVSRCW+YETA+AL+ +E+S +PL+DY+DY+ PD++LH+ P + +
Sbjct 294 LLLVGGGGYTIRNVSRCWLYETAIALN--QEVSDDLPLHDYFDYFIPDYKLHIKPLAALS 351
Query 241 NFNSPDHLQRLKARVLQNVALLEHAPGVQFAYVPPDC 277
NFN+P+ + + +L+N+ L H P VQ + C
Sbjct 352 NFNTPEFIDQTIVALLENLKQLPHVPSVQMQSISTSC 388
> mmu:15183 Hdac3, AW537363; histone deacetylase 3 (EC:3.5.1.98);
K11404 histone deacetylase 3 [EC:3.5.1.98]
Length=428
Score = 357 bits (916), Expect = 3e-98, Method: Compositional matrix adjust.
Identities = 163/287 (56%), Positives = 225/287 (78%), Gaps = 4/287 (1%)
Query 1 AGASIDAAKKLNYQQADLCINWSGGLHHAKRAEASGFCYVNDIVLGILELLKYHPRVMYI 60
GAS+ A +LN + D+ INW+GGLHHAK+ EASGFCYVNDIV+GILELLKYHPRV+YI
Sbjct 108 TGASLQGATQLNNKICDIAINWAGGLHHAKKFEASGFCYVNDIVIGILELLKYHPRVLYI 167
Query 61 DIDVHHGDGVEEAFYVSHRVLTVSFHKFGD-FFPGTGDITDVGAAQGKYFSVNVPLNDGM 119
DID+HHGDGV+EAFY++ RV+TVSFHK+G+ FFPGTGD+ +VGA G+Y+ +NVPL DG+
Sbjct 168 DIDIHHGDGVQEAFYLTDRVMTVSFHKYGNYFFPGTGDMYEVGAESGRYYCLNVPLRDGI 227
Query 120 DDESFIGLFKPIIAKCVEVYRPGAVVLQCGADSLSGDRLGKFNLTVKGHAACVQFVKSFN 179
DD+S+ LF+P+I++ V+ Y+P +VLQCGADSL DRLG FNL+++GH CV++VKSFN
Sbjct 228 DDQSYKHLFQPVISQVVDFYQPTCIVLQCGADSLGCDRLGCFNLSIRGHGECVEYVKSFN 287
Query 180 LPLLLLGGGGYTIRNVSRCWVYETAVALDRHRELSPQVPLNDYYDYYAPDFQLHLPPSP- 238
+PLL+LGGGGYT+RNV+RCW YET++ ++ +S ++P ++Y++Y+APDF LH S
Sbjct 288 IPLLVLGGGGYTVRNVARCWTYETSLLVE--EAISEELPYSEYFEYFAPDFTLHPDVSTR 345
Query 239 VPNFNSPDHLQRLKARVLQNVALLEHAPGVQFAYVPPDCFGEDEDED 285
+ N NS +L +++ + +N+ +L HAP VQ VP D D ++
Sbjct 346 IENQNSRQYLDQIRQTIFENLKMLNHAPSVQIHDVPADLLTYDRTDE 392
> hsa:8841 HDAC3, HD3, RPD3, RPD3-2; histone deacetylase 3 (EC:3.5.1.98);
K11404 histone deacetylase 3 [EC:3.5.1.98]
Length=428
Score = 356 bits (914), Expect = 6e-98, Method: Compositional matrix adjust.
Identities = 163/287 (56%), Positives = 224/287 (78%), Gaps = 4/287 (1%)
Query 1 AGASIDAAKKLNYQQADLCINWSGGLHHAKRAEASGFCYVNDIVLGILELLKYHPRVMYI 60
GAS+ A +LN + D+ INW+GGLHHAK+ EASGFCYVNDIV+GILELLKYHPRV+YI
Sbjct 108 TGASLQGATQLNNKICDIAINWAGGLHHAKKFEASGFCYVNDIVIGILELLKYHPRVLYI 167
Query 61 DIDVHHGDGVEEAFYVSHRVLTVSFHKFGD-FFPGTGDITDVGAAQGKYFSVNVPLNDGM 119
DID+HHGDGV+EAFY++ RV+TVSFHK+G+ FFPGTGD+ +VGA G+Y+ +NVPL DG+
Sbjct 168 DIDIHHGDGVQEAFYLTDRVMTVSFHKYGNYFFPGTGDMYEVGAESGRYYCLNVPLRDGI 227
Query 120 DDESFIGLFKPIIAKCVEVYRPGAVVLQCGADSLSGDRLGKFNLTVKGHAACVQFVKSFN 179
DD+S+ LF+P+I + V+ Y+P +VLQCGADSL DRLG FNL+++GH CV++VKSFN
Sbjct 228 DDQSYKHLFQPVINQVVDFYQPTCIVLQCGADSLGCDRLGCFNLSIRGHGECVEYVKSFN 287
Query 180 LPLLLLGGGGYTIRNVSRCWVYETAVALDRHRELSPQVPLNDYYDYYAPDFQLHLPPSP- 238
+PLL+LGGGGYT+RNV+RCW YET++ ++ +S ++P ++Y++Y+APDF LH S
Sbjct 288 IPLLVLGGGGYTVRNVARCWTYETSLLVE--EAISEELPYSEYFEYFAPDFTLHPDVSTR 345
Query 239 VPNFNSPDHLQRLKARVLQNVALLEHAPGVQFAYVPPDCFGEDEDED 285
+ N NS +L +++ + +N+ +L HAP VQ VP D D ++
Sbjct 346 IENQNSRQYLDQIRQTIFENLKMLNHAPSVQIHDVPADLLTYDRTDE 392
> dre:393965 hdac3, MGC55927, zgc:55927; histone deacetylase 3
(EC:3.5.1.98); K11404 histone deacetylase 3 [EC:3.5.1.98]
Length=428
Score = 352 bits (904), Expect = 8e-97, Method: Compositional matrix adjust.
Identities = 163/292 (55%), Positives = 226/292 (77%), Gaps = 5/292 (1%)
Query 1 AGASIDAAKKLNYQQADLCINWSGGLHHAKRAEASGFCYVNDIVLGILELLKYHPRVMYI 60
GAS+ A +LN++ D+ INW+GGLHHAK+ EASGFCYVNDIV+ ILELLKYHPRV+YI
Sbjct 108 TGASLQGATQLNHKICDIAINWAGGLHHAKKFEASGFCYVNDIVISILELLKYHPRVLYI 167
Query 61 DIDVHHGDGVEEAFYVSHRVLTVSFHKFGD-FFPGTGDITDVGAAQGKYFSVNVPLNDGM 119
DID+HHGDGV+EAFY++ RV+TVSFHK+G+ FFPGTGD+ +VGA G+Y+ +NVPL DG+
Sbjct 168 DIDIHHGDGVQEAFYLTDRVMTVSFHKYGNYFFPGTGDMYEVGAESGRYYCLNVPLRDGI 227
Query 120 DDESFIGLFKPIIAKCVEVYRPGAVVLQCGADSLSGDRLGKFNLTVKGHAACVQFVKSFN 179
DD+S+ LF+P+I + V+ Y+P +VLQCGADSL DRLG FNL+++GH CV+FVK F
Sbjct 228 DDQSYRQLFQPVIKQVVDFYQPTCIVLQCGADSLGCDRLGCFNLSIRGHGECVEFVKGFK 287
Query 180 LPLLLLGGGGYTIRNVSRCWVYETAVALDRHRELSPQVPLNDYYDYYAPDFQLHLPPSP- 238
+PLL+LGGGGYT+RNV+RCW +ET++ ++ +S ++P ++Y++Y+APDF LH S
Sbjct 288 IPLLVLGGGGYTVRNVARCWTFETSLLVE--ESISDELPYSEYFEYFAPDFTLHPDVSTR 345
Query 239 VPNFNSPDHLQRLKARVLQNVALLEHAPGVQFAYVPPDCFG-EDEDEDEQQQ 289
+ N NS +L++++ V +N+ +L HAP VQ VP D E DE + ++
Sbjct 346 IENQNSRQYLEQIRQTVFENLKMLNHAPSVQIRDVPSDLLSYERPDEADPEE 397
> sce:YNL330C RPD3, MOF6, REC3, SDI2, SDS6; Histone deacetylase;
regulates transcription and silencing; plays a role in regulating
Ty1 transposition; K06067 histone deacetylase 1/2 [EC:3.5.1.98]
Length=433
Score = 347 bits (889), Expect = 5e-95, Method: Compositional matrix adjust.
Identities = 158/298 (53%), Positives = 222/298 (74%), Gaps = 4/298 (1%)
Query 1 AGASIDAAKKLNYQQADLCINWSGGLHHAKRAEASGFCYVNDIVLGILELLKYHPRVMYI 60
G S++ A +LN + D+ +N++GGLHHAK++EASGFCY+NDIVLGI+ELL+YHPRV+YI
Sbjct 124 GGGSMEGAARLNRGKCDVAVNYAGGLHHAKKSEASGFCYLNDIVLGIIELLRYHPRVLYI 183
Query 61 DIDVHHGDGVEEAFYVSHRVLTVSFHKFGDFFPGTGDITDVGAAQGKYFSVNVPLNDGMD 120
DIDVHHGDGVEEAFY + RV+T SFHK+G+FFPGTG++ D+G GK ++VNVPL DG+D
Sbjct 184 DIDVHHGDGVEEAFYTTDRVMTCSFHKYGEFFPGTGELRDIGVGAGKNYAVNVPLRDGID 243
Query 121 DESFIGLFKPIIAKCVEVYRPGAVVLQCGADSLSGDRLGKFNLTVKGHAACVQFVKSFNL 180
D ++ +F+P+I K +E Y+P AVVLQCG DSLSGDRLG FNL+++GHA CV +VKSF +
Sbjct 244 DATYRSVFEPVIKKIMEWYQPSAVVLQCGGDSLSGDRLGCFNLSMEGHANCVNYVKSFGI 303
Query 181 PLLLLGGGGYTIRNVSRCWVYETAVALDRHRELSPQVPLNDYYDYYAPDFQLHLPPSPVP 240
P++++GGGGYT+RNV+R W +ET L + L +P N+YY+YY PD++L + PS +
Sbjct 304 PMMVVGGGGYTMRNVARTWCFET--GLLNNVVLDKDLPYNEYYEYYGPDYKLSVRPSNMF 361
Query 241 NFNSPDHLQRLKARVLQNVALLEHAPGVQFAYVPPDCFGEDEDEDEQQQLLVEWEGGG 298
N N+P++L ++ + N+ ++AP VQ + P D ED + E+ + GG
Sbjct 362 NVNTPEYLDKVMTNIFANLENTKYAPSVQLNHTPRD--AEDLGDVEEDSAEAKDTKGG 417
> sce:YGL194C HOS2, RTL1; Histone deacetylase required for gene
activation via specific deacetylation of lysines in H3 and
H4 histone tails; subunit of the Set3 complex, a meiotic-specific
repressor of sporulation specific genes that contains
deacetylase activity; K11483 histone deacetylase HOS2 [EC:3.5.1.98]
Length=452
Score = 313 bits (802), Expect = 6e-85, Method: Compositional matrix adjust.
Identities = 148/274 (54%), Positives = 198/274 (72%), Gaps = 7/274 (2%)
Query 1 AGASIDAAKKLNYQQADLCINWSGGLHHAKRAEASGFCYVNDIVLGILELLKYHPRVMYI 60
GAS+DA +KL Q+D+ INWSGGLHHAK+ SGFCYVNDIVL IL LL+YHPR++YI
Sbjct 132 TGASLDATRKLINNQSDIAINWSGGLHHAKKNSPSGFCYVNDIVLSILNLLRYHPRILYI 191
Query 61 DIDVHHGDGVEEAFYVSHRVLTVSFHKF-GDFFPGTGDITDVGAAQGKYFSVNVPLNDGM 119
DID+HHGDGV+EAFY + RV T+SFHK+ G+FFPGTGD+T++G +GK+F++NVPL DG+
Sbjct 192 DIDLHHGDGVQEAFYTTDRVFTLSFHKYNGEFFPGTGDLTEIGCDKGKHFALNVPLEDGI 251
Query 120 DDESFIGLFKPIIAKCVEVYRPGAVVLQCGADSLSGDRLGKFNLTVKGHAACVQFVKSFN 179
DD+S+I LFK I+ + ++P +V QCGADSL DRLG FNL +K H CV+FVKSF
Sbjct 252 DDDSYINLFKSIVDPLIMTFKPTLIVQQCGADSLGHDRLGCFNLNIKAHGECVKFVKSFG 311
Query 180 LPLLLLGGGGYTIRNVSRCWVYETAVALD--RHRELSPQVPLNDYYDYYAPDFQLH-LPP 236
LP+L++GGGGYT RNVSR W YET + D ++ +P + D + PD+ L+ +
Sbjct 312 LPMLVVGGGGYTPRNVSRLWTYETGILNDVLLPEDIPEDIP---FRDSFGPDYSLYPMLD 368
Query 237 SPVPNFNSPDHLQRLKARVLQNVALLEHAPGVQF 270
N NS L+ ++ R L+N+ L+ AP V+
Sbjct 369 DLYENKNSKKLLEDIRIRCLENIRYLQGAPSVRM 402
> ath:AT5G35600 HDA7; HDA7 (histone deacetylase7); histone deacetylase;
K06067 histone deacetylase 1/2 [EC:3.5.1.98]
Length=409
Score = 311 bits (798), Expect = 2e-84, Method: Compositional matrix adjust.
Identities = 148/274 (54%), Positives = 198/274 (72%), Gaps = 6/274 (2%)
Query 1 AGASIDAAKKLNYQQADLCINWSGGLHHAKRAEASGFCYVNDIVLGILELLKYHPRVMYI 60
AG SI AA KLN Q+AD+ INW+GG+HH K+ +ASGF YVND+VL ILELLK RV+YI
Sbjct 121 AGGSISAAAKLNRQEADIAINWAGGMHHVKKDKASGFGYVNDVVLAILELLKSFKRVLYI 180
Query 61 DIDVHHGDGVEEAFYVSHRVLTVSFHKFGDFFPGTGDITDVGAAQGKYFSVNVPLNDGMD 120
+I HGD VEEAF + RV+TVSFHK GD TGDI+D G +G+Y+S+N PL DG+D
Sbjct 181 EIGFPHGDEVEEAFKDTDRVMTVSFHKVGD----TGDISDYGEGKGQYYSLNAPLKDGLD 236
Query 121 DESFIGLFKPIIAKCVEVYRPGAVVLQCGADSLSGDRLGKFNLTVKGHAACVQFVKSFNL 180
D S GLF P+I + +E+Y P +VLQCGADSL+GD G FNL++KGH C+Q+V+SFN+
Sbjct 237 DFSLRGLFIPVIHRAMEIYEPEVIVLQCGADSLAGDPFGTFNLSIKGHGDCLQYVRSFNV 296
Query 181 PLLLLGGGGYTIRNVSRCWVYETAVALDRHRELSPQVPLNDYYDYYAPDFQLHLPPSPVP 240
PL++LGGGGYT+ NV+RCW YETA+A+ +L +P NDY Y+ PD++LH+ P+
Sbjct 297 PLMILGGGGYTLPNVARCWCYETAIAVG--EQLDNDLPGNDYMKYFRPDYKLHILPTNRQ 354
Query 241 NFNSPDHLQRLKARVLQNVALLEHAPGVQFAYVP 274
N N+ + ++ +L ++L+ HAP V F P
Sbjct 355 NLNTRLDIITMRETLLAQLSLVMHAPSVPFQDTP 388
> cpv:cgd6_1380 histone deacetylase ; K06067 histone deacetylase
1/2 [EC:3.5.1.98]
Length=440
Score = 306 bits (784), Expect = 7e-83, Method: Compositional matrix adjust.
Identities = 146/259 (56%), Positives = 188/259 (72%), Gaps = 3/259 (1%)
Query 1 AGASIDAAKKLNYQQADLCINWSGGLHHAKRAEASGFCYVNDIVLGILELLKYHPRVMYI 60
AG S+ A+ +N INW+GGLHH K+ EASGFCYVND VLG LE LKY RV Y+
Sbjct 118 AGGSLGGAQSVNELGYQYAINWAGGLHHGKKHEASGFCYVNDCVLGALEFLKYQHRVCYV 177
Query 61 DIDVHHGDGVEEAFYVSHRVLTVSFHKFGDFFPGTGDITDVGAAQGKYFSVNVPLNDGMD 120
DID+HHGDGVEEAFY S R + VSFHK+GD+FPGTG + DVG +G +SVNVPL DG+D
Sbjct 178 DIDIHHGDGVEEAFYTSPRCMCVSFHKYGDYFPGTGALNDVGVEEGLGYSVNVPLKDGVD 237
Query 121 DESFIGLFKPIIAKCVEVYRPGAVVLQCGADSLSGDRLGKFNLTVKGHAACVQFVKSFNL 180
D +FI LF ++ +E YRPGA+VLQCGADSLSGDRLG FNL++KGH V F+K FN+
Sbjct 238 DATFIDLFTKVMTLVMENYRPGAIVLQCGADSLSGDRLGCFNLSLKGHGHAVSFLKKFNV 297
Query 181 PLLLLGGGGYTIRNVSRCWVYETAVALDRHRELSPQVP-LNDYYDYYAPDFQLHLPPSPV 239
PLL+LGGGGYT+RNV +CW YET++ +D + + Q+P +++Y YY PDF L + S +
Sbjct 298 PLLILGGGGYTLRNVPKCWTYETSLIVDTY--IDEQLPNSSNFYGYYGPDFSLAVRTSNM 355
Query 240 PNFNSPDHLQRLKARVLQN 258
N NS + + ++ +N
Sbjct 356 ENLNSRQDCEEIYRKISEN 374
> cel:C08B11.2 hda-2; Histone DeAcetylase family member (hda-2);
K06067 histone deacetylase 1/2 [EC:3.5.1.98]
Length=507
Score = 295 bits (754), Expect = 2e-79, Method: Compositional matrix adjust.
Identities = 150/310 (48%), Positives = 204/310 (65%), Gaps = 15/310 (4%)
Query 1 AGASIDAAKKLNYQQADLCINWSGGLHHAKRAEASGFCYVNDIVLGILELLKYHPRVMYI 60
AG S++ A++LN++ D+ INW GGLHHAK++EASGFCYVNDIVLGILELLKYH RV+YI
Sbjct 135 AGGSVEGARRLNHKMNDIVINWPGGLHHAKKSEASGFCYVNDIVLGILELLKYHKRVLYI 194
Query 61 DIDVHHGDGVEEAFYVSHRVLTVSFHKFGDFFPGTGDITDVGAAQGKYFSVNVPLNDGMD 120
DID+HHGDGV+EAF S RV+TVSFH+FG +FPG+G I D G GKYF++NVPL +
Sbjct 195 DIDIHHGDGVQEAFNNSDRVMTVSFHRFGQYFPGSGSIMDKGVGPGKYFAINVPLMAAIR 254
Query 121 DESFIGLFKPIIAKCVEVYRPGAVVLQCGADSLSGDRLGKFNLTVKGHAACVQFVKSFNL 180
DE ++ LF+ +I+ E + P A+VLQCG+DSL DRLG+F L+ HA V++VKS
Sbjct 255 DEPYLKLFESVISGVEENFNPEAIVLQCGSDSLCEDRLGQFALSFNAHARAVKYVKSLGK 314
Query 181 PLLLLGGGGYTIRNVSRCWVYETAVALDRHRELSPQVPLNDYYD-YYAPDFQLHLPPSPV 239
PL++LGGGGYT+RNV+RCW ET V L + ++P Y Y+ P L P+ V
Sbjct 315 PLMVLGGGGYTLRNVARCWALETGVILGLR--MDDEIPGTSLYSHYFTPRL---LRPNLV 369
Query 240 PNF---NSPDHLQRLKARVLQNVALLEHAPGVQFAYVPPDCFGEDEDEDEQQQLL----- 291
P NS +L ++ L + ++ AP VQ + E E +E ++L
Sbjct 370 PKMNDANSAAYLASIEKETLACLRMIRGAPSVQMQNIVGIRLDEIEQIEENERLQKSSKS 429
Query 292 -VEWEGGGLA 300
+E+E G ++
Sbjct 430 SIEYEVGKVS 439
> tgo:TGME49_049620 histone deacetylase, putative ; K06067 histone
deacetylase 1/2 [EC:3.5.1.98]
Length=734
Score = 287 bits (734), Expect = 5e-77, Method: Compositional matrix adjust.
Identities = 144/291 (49%), Positives = 196/291 (67%), Gaps = 6/291 (2%)
Query 1 AGASIDAAKKLNYQQADLCINWSGGLHHAKRAEASGFCYVNDIVLGILELLKYHPRVMYI 60
+G SI+ A+++ + D INW+GGLHH K+ EASGFCY+ND VL LE L++ RV+Y+
Sbjct 412 SGGSIEGARRVVRGEYDWAINWAGGLHHGKKHEASGFCYINDCVLAALEFLRFKHRVLYV 471
Query 61 DIDVHHGDGVEEAFYVSHRVLTVSFHKFGDFFPGTGDITDVGAAQGKYFSVNVPLNDGMD 120
D+D+HHGDGVEEAFY S RVL SFHK+GD+FPGTG + DVG +G +SVNVPL++G+D
Sbjct 472 DVDIHHGDGVEEAFYTSPRVLCCSFHKYGDYFPGTGALDDVGIDEGLGYSVNVPLSEGVD 531
Query 121 DESFIGLFKPIIAKCVEVYRPGAVVLQCGADSLSGDRLGKFNLTVKGHAACVQFVKSFNL 180
D + LF+ ++ +E YRP AVVLQCGADSLSGDRLG FNL++ GH+ V++ S ++
Sbjct 532 DAMYTRLFRQVMDMIMESYRPEAVVLQCGADSLSGDRLGCFNLSIDGHSEAVRYFCSKDV 591
Query 181 PLLLLGGGGYTIRNVSRCWVYETAVALDRHRELSPQVP-LNDYYDYYAPDFQLHLPPSPV 239
P L LGGGGYT+RNV RCW ET L L+P +P ++Y YY PD LH+ S +
Sbjct 592 PALFLGGGGYTLRNVPRCWATETGYVLG--LSLNPNIPEESEYVGYYGPDSTLHVRTSNM 649
Query 240 PNFNSPDHLQRLKARVLQNVALLEHAPGVQFAYVPPDCFGEDEDEDEQQQL 290
N N ++Q + R+ Q L +H V A + + F +D D+ Q+
Sbjct 650 ENRNEDSYVQHVIERISQT--LKDHVRPVG-AQISANYFLDDSVMDKSDQI 697
> mmu:70315 Hdac8, 2610007D20Rik; histone deacetylase 8 (EC:3.5.1.98);
K11405 histone deacetylase 8 [EC:3.5.1.98]
Length=377
Score = 253 bits (645), Expect = 9e-67, Method: Compositional matrix adjust.
Identities = 118/260 (45%), Positives = 176/260 (67%), Gaps = 3/260 (1%)
Query 1 AGASIDAAKKLNYQQADLCINWSGGLHHAKRAEASGFCYVNDIVLGILELLKYHPRVMYI 60
G +I AA+ L + + INWSGG HHAK+ EASGFCY+ND VLGIL L + R++Y+
Sbjct 116 GGGTITAAQCLIDGKCKVAINWSGGWHHAKKDEASGFCYLNDAVLGILRLRRKFDRILYV 175
Query 61 DIDVHHGDGVEEAFYVSHRVLTVSFHKFG-DFFPGTGDITDVGAAQGKYFSVNVPLNDGM 119
D+D+HHGDGVE+AF + +V+TVS HKF FFPGTGD++DVG +G+Y+SVNVP+ DG+
Sbjct 176 DLDLHHGDGVEDAFSFTSKVMTVSLHKFSPGFFPGTGDMSDVGLGKGRYYSVNVPIQDGI 235
Query 120 DDESFIGLFKPIIAKCVEVYRPGAVVLQCGADSLSGDRLGKFNLTVKGHAACVQFVKSFN 179
DE + + + ++ + + + P AVVLQ GAD+++GD + FN+T G C+++V +
Sbjct 236 QDEKYYHICESVLKEVYQAFNPKAVVLQLGADTIAGDPMCSFNMTPVGIGKCLKYVLQWQ 295
Query 180 LPLLLLGGGGYTIRNVSRCWVYETAVALDRHRELSPQVPLNDYYDYYAPDFQLHLPPSPV 239
L L+LGGGGY + N +RCW Y T V L + LS ++P ++++ Y PD+ L + PS
Sbjct 296 LATLILGGGGYNLANTARCWTYLTGVILG--KTLSSEIPDHEFFTAYGPDYVLEITPSCR 353
Query 240 PNFNSPDHLQRLKARVLQNV 259
P+ N P +Q++ + N+
Sbjct 354 PDRNEPHRIQQILNYIKGNL 373
> xla:444137 hdac8, MGC80565; histone deacetylase 8 (EC:3.5.1.98);
K11405 histone deacetylase 8 [EC:3.5.1.98]
Length=325
Score = 252 bits (644), Expect = 1e-66, Method: Compositional matrix adjust.
Identities = 115/260 (44%), Positives = 178/260 (68%), Gaps = 3/260 (1%)
Query 1 AGASIDAAKKLNYQQADLCINWSGGLHHAKRAEASGFCYVNDIVLGILELLKYHPRVMYI 60
GA++ AA++L + + +NW GG HHAK+ EASGFCY+ND VLGIL+L + RV+Y+
Sbjct 64 GGATLTAAEQLIEGKTRIAVNWPGGWHHAKKDEASGFCYLNDAVLGILKLREKFDRVLYV 123
Query 61 DIDVHHGDGVEEAFYVSHRVLTVSFHKFG-DFFPGTGDITDVGAAQGKYFSVNVPLNDGM 119
D+D+HHGDGVE+AF + +V+TVS HKF FFPGTGD++D+G +G+Y+S+NVPL DG+
Sbjct 124 DMDLHHGDGVEDAFSFTSKVMTVSLHKFSPGFFPGTGDVSDIGLGKGRYYSINVPLQDGI 183
Query 120 DDESFIGLFKPIIAKCVEVYRPGAVVLQCGADSLSGDRLGKFNLTVKGHAACVQFVKSFN 179
D+ + + + ++ + + P AVVLQ GAD+++GD + FN+T +G C+++V +
Sbjct 184 QDDKYYQICEGVLKEVFTTFNPEAVVLQLGADTIAGDPMCSFNMTPEGIGKCLKYVLQWQ 243
Query 180 LPLLLLGGGGYTIRNVSRCWVYETAVALDRHRELSPQVPLNDYYDYYAPDFQLHLPPSPV 239
LP L+LGGGGY + N +RCW Y TA+ + R LS ++P ++++ Y PD+ L + PS
Sbjct 244 LPTLILGGGGYHLPNTARCWTYLTALIVG--RTLSSEIPDHEFFTEYGPDYVLEITPSCR 301
Query 240 PNFNSPDHLQRLKARVLQNV 259
P+ N +Q + + N+
Sbjct 302 PDRNDTQKVQEILQSIKGNL 321
> dre:406740 hdac8, wu:fd19a02, zgc:66196; histone deacetylase
8 (EC:3.5.1.98); K11405 histone deacetylase 8 [EC:3.5.1.98]
Length=378
Score = 241 bits (614), Expect = 4e-63, Method: Compositional matrix adjust.
Identities = 118/259 (45%), Positives = 177/259 (68%), Gaps = 3/259 (1%)
Query 2 GASIDAAKKLNYQQADLCINWSGGLHHAKRAEASGFCYVNDIVLGILELLKYHPRVMYID 61
GA++ AA+ L + D+ INW+GG HHAK+ EASG CYVND VLGIL+L + + RV+Y+D
Sbjct 118 GATLTAAQNLLDGKCDVAINWAGGWHHAKKDEASGSCYVNDAVLGILKLREKYDRVLYVD 177
Query 62 IDVHHGDGVEEAFYVSHRVLTVSFHKFG-DFFPGTGDITDVGAAQGKYFSVNVPLNDGMD 120
+D+HHGDGVE+AF + +V+TVS HKF FFPGTGD+TD G +G++++VNVP DG+
Sbjct 178 VDLHHGDGVEDAFSFTSKVMTVSLHKFSPGFFPGTGDVTDTGLGKGRWYAVNVPFEDGVR 237
Query 121 DESFIGLFKPIIAKCVEVYRPGAVVLQCGADSLSGDRLGKFNLTVKGHAACVQFVKSFNL 180
D+ + F ++ + ++ P AVV+Q GAD+++GD + FN+T G A C+ ++ + L
Sbjct 238 DDRYCQTFTSVMQEVKALFNPEAVVMQLGADTMAGDPMCSFNMTPVGVAKCLTYILGWEL 297
Query 181 PLLLLGGGGYTIRNVSRCWVYETAVALDRHRELSPQVPLNDYYDYYAPDFQLHLPPSPVP 240
P LLLGGGGY + N +RCW Y T L + LS ++P ++++ Y PD+ L + PS P
Sbjct 298 PTLLLGGGGYNLANTARCWTYLTGTVLG--QTLSSEIPDHEFFTEYGPDYSLEISPSCRP 355
Query 241 NFNSPDHLQRLKARVLQNV 259
+ N HL+R+ + + N+
Sbjct 356 DRNESQHLERVISTIKGNL 374
> hsa:55869 HDAC8, HD8, HDACL1, RPD3; histone deacetylase 8 (EC:3.5.1.98);
K11405 histone deacetylase 8 [EC:3.5.1.98]
Length=286
Score = 229 bits (585), Expect = 8e-60, Method: Compositional matrix adjust.
Identities = 105/236 (44%), Positives = 159/236 (67%), Gaps = 3/236 (1%)
Query 25 GLHHAKRAEASGFCYVNDIVLGILELLKYHPRVMYIDIDVHHGDGVEEAFYVSHRVLTVS 84
LH R EASGFCY+ND VLGIL L + R++Y+D+D+HHGDGVE+AF + +V+TVS
Sbjct 49 ALHKQMRDEASGFCYLNDAVLGILRLRRKFERILYVDLDLHHGDGVEDAFSFTSKVMTVS 108
Query 85 FHKFG-DFFPGTGDITDVGAAQGKYFSVNVPLNDGMDDESFIGLFKPIIAKCVEVYRPGA 143
HKF FFPGTGD++DVG +G+Y+SVNVP+ DG+ DE + + + ++ + + + P A
Sbjct 109 LHKFSPGFFPGTGDVSDVGLGKGRYYSVNVPIQDGIQDEKYYQICESVLKEVYQAFNPKA 168
Query 144 VVLQCGADSLSGDRLGKFNLTVKGHAACVQFVKSFNLPLLLLGGGGYTIRNVSRCWVYET 203
VVLQ GAD+++GD + FN+T G C++++ + L L+LGGGGY + N +RCW Y T
Sbjct 169 VVLQLGADTIAGDPMCSFNMTPVGIGKCLKYILQWQLATLILGGGGYNLANTARCWTYLT 228
Query 204 AVALDRHRELSPQVPLNDYYDYYAPDFQLHLPPSPVPNFNSPDHLQRLKARVLQNV 259
V L + LS ++P ++++ Y PD+ L + PS P+ N P +Q++ + N+
Sbjct 229 GVILG--KTLSSEIPDHEFFTAYGPDYVLEITPSCRPDRNEPHRIQQILNYIKGNL 282
> sce:YPR068C HOS1; Hos1p; K11482 histone deacetylase HOS1 [EC:3.5.1.98]
Length=470
Score = 150 bits (379), Expect = 7e-36, Method: Compositional matrix adjust.
Identities = 81/207 (39%), Positives = 122/207 (58%), Gaps = 7/207 (3%)
Query 1 AGASIDAAKKLNYQQADLCINWSGGLHHAKRAEASGFCYVNDIVLGILELLKYH-PRVMY 59
GA+++ L+ + + INW GG HHA + ASGFCY+ND+VL I L K ++ Y
Sbjct 184 TGATLNLLDHLSPTERLIGINWDGGRHHAFKQRASGFCYINDVVLLIQRLRKAKLNKITY 243
Query 60 IDIDVHHGDGVEEAFYVSHRVLTVSFHKFG-DFFPGTGDITDVGAAQGKYFSVNVPLNDG 118
+D D+HHGDGVE+AF S ++ T+S H + FFPGTG ++D + VN+PL G
Sbjct 244 VDFDLHHGDGVEKAFQYSKQIQTISVHLYEPGFFPGTGSLSDSRKDKN---VVNIPLKHG 300
Query 119 MDDESFIGLFKPIIAKCVEVYRPGAVVLQCGADSLSGDRLGKFNLTVKGHAA-CVQFVKS 177
DD + I+ +E + P A++++CG D L GDR ++ LT++G + + +KS
Sbjct 301 CDDNYLELIASKIVNPLIERHEPEALIIECGGDGLLGDRFNEWQLTIRGLSRIIINIMKS 360
Query 178 F-NLPLLLLGGGGYTIRNVSRCWVYET 203
+ + LLGGGGY +SR + Y T
Sbjct 361 YPRAHIFLLGGGGYNDLLMSRFYTYLT 387
> ath:AT3G44490 hda17; hda17 (histone deacetylase 17); histone
deacetylase; K06067 histone deacetylase 1/2 [EC:3.5.1.98]
Length=158
Score = 135 bits (341), Expect = 2e-31, Method: Compositional matrix adjust.
Identities = 62/127 (48%), Positives = 84/127 (66%), Gaps = 2/127 (1%)
Query 161 FNLTVKGHAACVQFVKSFNLPLLLLGGGGYTIRNVSRCWVYETAVALDRHRELSPQVPLN 220
F++ GHA CV+FVK FNLPLL+ GGGGYT NV+RCW ET + LD EL ++ N
Sbjct 3 FSMLFTGHAECVKFVKKFNLPLLVTGGGGYTKENVARCWTVETGILLDT--ELPNEISEN 60
Query 221 DYYDYYAPDFQLHLPPSPVPNFNSPDHLQRLKARVLQNVALLEHAPGVQFAYVPPDCFGE 280
DY Y+APDF L +P + N N+ ++ +K ++L+N+ ++HAP VQ VPPD +
Sbjct 61 DYIKYFAPDFSLKIPGGHIENLNTKSYISSIKVQILENLRYIQHAPSVQMQEVPPDFYIP 120
Query 281 DEDEDEQ 287
D DEDEQ
Sbjct 121 DFDEDEQ 127
> ath:AT3G44660 hda10; hda10 (histone deacetylase 10); histone
deacetylase
Length=142
Score = 102 bits (254), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 51/127 (40%), Positives = 71/127 (55%), Gaps = 18/127 (14%)
Query 161 FNLTVKGHAACVQFVKSFNLPLLLLGGGGYTIRNVSRCWVYETAVALDRHRELSPQVPLN 220
F++ GHA C GGYT NV+RCW ET + LD EL ++P N
Sbjct 3 FSMLFTGHAEC----------------GGYTKENVARCWTVETGILLDT--ELPNEIPEN 44
Query 221 DYYDYYAPDFQLHLPPSPVPNFNSPDHLQRLKARVLQNVALLEHAPGVQFAYVPPDCFGE 280
DY Y+APDF L +P + N N+ ++ +K ++L+N+ ++HAP VQ VPPD +
Sbjct 45 DYIKYFAPDFSLKIPGGHIENLNTKSYISSIKVQILENLRYIQHAPSVQMQEVPPDFYIP 104
Query 281 DEDEDEQ 287
D DEDEQ
Sbjct 105 DFDEDEQ 111
> dre:565482 hdac6, wu:fc31d02; histone deacetylase 6; K11407
histone deacetylase 6/10 [EC:3.5.1.98]
Length=858
Score = 97.8 bits (242), Expect = 5e-20, Method: Compositional matrix adjust.
Identities = 59/190 (31%), Positives = 97/190 (51%), Gaps = 9/190 (4%)
Query 27 HHAKRAEASGFCYVNDIVLGIL---ELLKYHPRVMYIDIDVHHGDGVEEAFYVSHRVLTV 83
HHA++ A GFC+ N L + + RV+ +D DVHHG+G + F VL +
Sbjct 350 HHAEKDTACGFCFFNTAALTARYAQSITRESLRVLIVDWDVHHGNGTQHIFEEDDSVLYI 409
Query 84 SFHKF--GDFFPGTGDIT--DVGAAQGKYFSVNVPLNDG-MDDESFIGLFKPIIAKCVEV 138
S H++ G FFP + D VG +G+ ++VN+P N G M D ++ F ++
Sbjct 410 SLHRYEDGAFFPNSEDANYDKVGLGKGRGYNVNIPWNGGKMGDPEYMAAFHHLVMPIARE 469
Query 139 YRPGAVVLQCGADSLSGDRLGKFNLTVKGHAACVQFVKSFNL-PLLLLGGGGYTIRNVSR 197
+ P V++ G D+ GD LG F +T +G+A + S +L++ GGY + ++S
Sbjct 470 FAPELVLVSAGFDAARGDPLGGFQVTPEGYAHLTHQLMSLAAGRVLIILEGGYNLTSISE 529
Query 198 CWVYETAVAL 207
T++ L
Sbjct 530 SMSMCTSMLL 539
Score = 64.7 bits (156), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 40/147 (27%), Positives = 75/147 (51%), Gaps = 6/147 (4%)
Query 57 VMYIDIDVHHGDGVEEAFYVSHRVLTVSFHKF--GDFFP--GTGDITDVGAAQGKYFSVN 112
V+ +D DVHHG G++ F VL S H++ G F+P D + VG+ G+ +++N
Sbjct 1 VLIVDWDVHHGQGIQYIFEEDPSVLYFSVHRYEDGSFWPHLKESDSSSVGSGAGQGYNIN 60
Query 113 VPLND-GMDDESFIGLFKPIIAKCVEVYRPGAVVLQCGADSLSGDRLGKFNLTVKGHAAC 171
+P N GM+ +I F+ ++ ++P V++ G D++ GD G ++ + +
Sbjct 61 LPWNKVGMESGDYITAFQQLLLPVAYEFQPQLVLVAAGFDAVIGDPKGGMQVSPECFSIL 120
Query 172 VQFVKSFNLPLLLLG-GGGYTIRNVSR 197
+K L+L GGY +++ +
Sbjct 121 THMLKGVAQGRLVLALEGGYNLQSTAE 147
> dre:557486 histone deacetylase 5-like; K11406 histone deacetylase
4/5 [EC:3.5.1.98]
Length=1115
Score = 97.4 bits (241), Expect = 7e-20, Method: Composition-based stats.
Identities = 65/206 (31%), Positives = 110/206 (53%), Gaps = 18/206 (8%)
Query 27 HHAKRAEASGFCYVNDIVLGILELLKYH---PRVMYIDIDVHHGDGVEEAFYVSHRVLTV 83
HHA+ + A GFC+ N + + +LL+ +++ ID D+HHG+G ++AFY VL +
Sbjct 828 HHAEESTAMGFCFFNSVAI-TAKLLQQKLGVGKILIIDWDIHHGNGTQQAFYNDPNVLYI 886
Query 84 SFHKF--GDFFPGTGDITDVGAAQGKYFSVNVPLNDG----MDDESFIGLFKPIIAKCVE 137
S H++ G+FFPG+G +VG G+ F+VN+ G M D ++ F+ ++
Sbjct 887 SLHRYDDGNFFPGSGAPEEVGVGPGEGFNVNIAWTGGVEPPMGDVEYLTAFRTVVMPIAN 946
Query 138 VYRPGAVVLQCGADSLSGDR--LGKFNLTVK--GHAACVQFVKSFNLPLLLLGGGGYTIR 193
+ P V++ G D++ G + LG +N+T K GH Q +K ++L GG+ +
Sbjct 947 EFSPDVVLVSAGFDAVEGHQSPLGGYNVTAKCFGHLT-KQLMKLAGGRVVLALEGGHDLT 1005
Query 194 NVSRCWVYETAVALDRHRELSPQVPL 219
+ C E+ V EL+P +PL
Sbjct 1006 AI--CDASESCVEALLGDELNP-LPL 1028
> ath:AT1G08460 HDA08; HDA08; histone deacetylase
Length=377
Score = 97.4 bits (241), Expect = 7e-20, Method: Compositional matrix adjust.
Identities = 60/183 (32%), Positives = 94/183 (51%), Gaps = 11/183 (6%)
Query 27 HHAKRAEASGFCYVNDIVLGILELLKYHP--RVMYIDIDVHHGDGVEEAFYVSHRVLTVS 84
HH++ +A G+C++N+ L + L RV IDIDVH+G+G E FY S +VLTVS
Sbjct 144 HHSQPTQADGYCFLNNAALAVKLALNSGSCSRVAVIDIDVHYGNGTAEGFYTSDKVLTVS 203
Query 85 FH----KFGDFFPGTGDITDVGAAQGKYFSVNVPLNDGMDDESFIGLFKPIIAKCVEVYR 140
H +G P G I ++G G +++NVPL +G D + ++ V +
Sbjct 204 LHMNHGSWGSSHPQKGSIDELGEDVGLGYNLNVPLPNGTGDRGYEYAMNELVVPAVRRFG 263
Query 141 PGAVVLQCGADSLSGDRLGKFNLTVKGHAACVQFVKSF-----NLPLLLLGGGGYTIRNV 195
P VVL G DS + D G+ +LT+ G+ Q ++ + LL++ GGY +
Sbjct 264 PDMVVLVVGQDSSAFDPNGRQSLTMNGYRRIGQIMRGVAEEHSHGRLLMVQEGGYHVTYA 323
Query 196 SRC 198
+ C
Sbjct 324 AYC 326
> cel:C10E2.3 hda-4; Histone DeAcetylase family member (hda-4);
K11406 histone deacetylase 4/5 [EC:3.5.1.98]
Length=869
Score = 96.7 bits (239), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 57/190 (30%), Positives = 102/190 (53%), Gaps = 12/190 (6%)
Query 19 CINWSGGLHHAKRAEASGFCYVNDIVLGILELLKYHP----RVMYIDIDVHHGDGVEEAF 74
CI G HHA+ +A GFC+ N++ + + L +P ++ ID DVHHG+G + +F
Sbjct 601 CIRPPG--HHAEHEQAMGFCFFNNVAVAVKVLQTKYPAQCAKIAIIDWDVHHGNGTQLSF 658
Query 75 YVSHRVLTVSFHKF--GDFFPGTGDITDVGAAQGKYFSVNVPLN-DGMDDESFIGLFKPI 131
VL +S H+ G+FFPGTG +T+VG K +VNVP + D M D ++ ++ +
Sbjct 659 ENDPNVLYMSLHRHDKGNFFPGTGSVTEVGKNDAKGLTVNVPFSGDVMRDPEYLAAWRTV 718
Query 132 IAKCVEVYRPGAVVLQCGADSLSG--DRLGKFNLTVKGHAACVQFVKSFNLPLLLLG-GG 188
I + + P +++ G D+ G + LG + +T + + + ++ ++L G
Sbjct 719 IEPVMASFCPDFIIVSAGFDACHGHPNALGGYEVTPEMFGYMTKSLLNYASGKVVLALEG 778
Query 189 GYTIRNVSRC 198
GY ++++S
Sbjct 779 GYDLKSISEA 788
> ath:AT3G18520 HDA15; HDA15; histone deacetylase
Length=552
Score = 96.7 bits (239), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 73/244 (29%), Positives = 122/244 (50%), Gaps = 16/244 (6%)
Query 27 HHAKRAEASGFC-YVNDIVLGILELLKYHPRVMYIDIDVHHGDGVEEAFYVSHRVLTVSF 85
HHA A GFC + N V ++ +V+ +D DVHHG+G +E F + VL +S
Sbjct 276 HHAGVRHAMGFCLHNNAAVAALVAQAAGAKKVLIVDWDVHHGNGTQEIFEQNKSVLYISL 335
Query 86 HKF--GDFFPGTGDITDVGAAQGKYFSVNVPLN-DGMDDESFIGLFKPIIAKCVEVYRPG 142
H+ G+F+PGTG +VG+ G+ + VNVP + G+ D+ +I F+ ++ + P
Sbjct 336 HRHEGGNFYPGTGAADEVGSNGGEGYCVNVPWSCGGVGDKDYIFAFQHVVLPIASAFSPD 395
Query 143 AVVLQCGADSLSGDRLGKFNLTVKGHAACVQFVKSF-NLPLLLLGGGGYTIRNVSRCWVY 201
V++ G D+ GD LG ++T G++ Q + +L++ GGY +R++S
Sbjct 396 FVIISAGFDAARGDPLGCCDVTPAGYSRMTQMLGDLCGGKMLVILEGGYNLRSISASATA 455
Query 202 ETAVALDRHRE------LSPQVP-LNDYYDYYAPDFQLHLPPSPVPNFNSPDHLQRLKAR 254
V L + E +P V L D + QL PS +++ L L+AR
Sbjct 456 VIKVLLGENPENELPIATTPSVAGLQTVLDVL--NIQLEFWPSLAISYSK--LLSELEAR 511
Query 255 VLQN 258
+++N
Sbjct 512 LIEN 515
> tgo:TGME49_081420 histone deacetylase (EC:4.1.1.70 2.7.1.151)
Length=2363
Score = 95.1 bits (235), Expect = 3e-19, Method: Compositional matrix adjust.
Identities = 56/177 (31%), Positives = 83/177 (46%), Gaps = 44/177 (24%)
Query 27 HHAKRAEASGFCYVNDIVLGILELLKYH--PRVMYIDIDVHHGDGVEEAFYVSHRVLTVS 84
HHA EA+GFC N++ L LL H RV +D DVHHG+G ++ FY S VL +S
Sbjct 764 HHACSHEAAGFCIYNNVALAANYLLANHGLSRVAILDWDVHHGNGTQDLFYNSSSVLYLS 823
Query 85 FHKF------------------------------------------GDFFPGTGDITDVG 102
H+F F+PGTG +T+ G
Sbjct 824 VHRFEKAKPVSASHGEALHASPSPGAGAAGPSPESPRASDSPAASGAAFYPGTGSLTETG 883
Query 103 AAQGKYFSVNVPLNDGMDDESFIGLFKPIIAKCVEVYRPGAVVLQCGADSLSGDRLG 159
+ G+ +++NVPL+ G D +F +IA + +RP A+++ G D+ +GD LG
Sbjct 884 VSVGRGYTINVPLSRGYTDGDMFWVFCGVIAPALTAFRPQAILVSAGFDAAAGDPLG 940
> sce:YNL021W HDA1; Hda1p; K11407 histone deacetylase 6/10 [EC:3.5.1.98]
Length=706
Score = 95.1 bits (235), Expect = 3e-19, Method: Compositional matrix adjust.
Identities = 56/184 (30%), Positives = 101/184 (54%), Gaps = 16/184 (8%)
Query 27 HHAKRAEASGFCYVNDIVLGILELLKYHP----RVMYIDIDVHHGDGVEEAFYVSHRVLT 82
HHA+ A GFC +++ + +LK +P R+M +D D+HHG+G +++FY +VL
Sbjct 205 HHAEPQAAGGFCLFSNVAVAAKNILKNYPESVRRIMILDWDIHHGNGTQKSFYQDDQVLY 264
Query 83 VSFHKF--GDFFPGT--GDITDVGAAQGKYFSVNV--PLNDGMDDESFIGLFKPIIAKCV 136
VS H+F G ++PGT G G +G+ F+ N+ P+ G+ D ++ F+ ++
Sbjct 265 VSLHRFEMGKYYPGTIQGQYDQTGEGKGEGFNCNITWPVG-GVGDAEYMWAFEQVVMPMG 323
Query 137 EVYRPGAVVLQCGADSLSGDRLGKFNLTVKGHAACVQFVKSF---NLPLLLLGGGGYTIR 193
++P V++ G D+ GD +G+ ++T + +KS NL ++L GGY +
Sbjct 324 REFKPDLVIISSGFDAADGDTIGQCHVTPSCYGHMTHMLKSLARGNLCVVL--EGGYNLD 381
Query 194 NVSR 197
++R
Sbjct 382 AIAR 385
> xla:380178 hdac6, MGC53140, hd6, jm21; histone deacetylase 6
(EC:3.5.1.98); K11407 histone deacetylase 6/10 [EC:3.5.1.98]
Length=1286
Score = 94.7 bits (234), Expect = 5e-19, Method: Compositional matrix adjust.
Identities = 61/189 (32%), Positives = 96/189 (50%), Gaps = 12/189 (6%)
Query 27 HHAKRAEASGFCYVNDIVLGI-----LELLKYHP-RVMYIDIDVHHGDGVEEAFYVSHRV 80
HHA+ EA GFC+ N + L L+ P RVM +D DVHHG+G + F V
Sbjct 611 HHAEPGEACGFCFFNTVALAARYAQRLQSQSEDPLRVMILDWDVHHGNGTQHIFQEDASV 670
Query 81 LTVSFHKF--GDFFPGTGDIT--DVGAAQGKYFSVNVPLNDG-MDDESFIGLFKPIIAKC 135
L +S H++ G FFP + D + VG +G ++VN+P N G M D ++ F ++
Sbjct 671 LYMSLHRYDEGLFFPNSEDASHDKVGIGKGAGYNVNIPWNGGKMGDVEYLMAFHRVVMPI 730
Query 136 VEVYRPGAVVLQCGADSLSGDRLGKFNLTVKGHAACVQFVKSF-NLPLLLLGGGGYTIRN 194
+ P V++ G D+ GD LG ++ +G+A + S ++L+ GGY + +
Sbjct 731 AYEFNPQLVLISAGFDAARGDPLGGCQVSPEGYAHMTHLLMSLAGGRVILVLEGGYNLTS 790
Query 195 VSRCWVYET 203
+S V T
Sbjct 791 ISESMVMCT 799
Score = 73.9 bits (180), Expect = 7e-13, Method: Compositional matrix adjust.
Identities = 52/178 (29%), Positives = 85/178 (47%), Gaps = 8/178 (4%)
Query 27 HHAKRAEASGFCYVNDIVLGI--LELLKYHPRVMYIDIDVHHGDGVEEAFYVSHRVLTVS 84
HHA + +G+C N + + +L RV+ +D DVHHG G + F VL S
Sbjct 210 HHAHTDQMNGYCMFNQLAIAARYAQLTYGAKRVLIVDWDVHHGQGTQFIFESDPSVLYFS 269
Query 85 FHKF--GDFFPGTGDITD--VGAAQGKYFSVNVPLNDG-MDDESFIGLFKPIIAKCVEVY 139
H++ G F+P + VG +G+ F+VNV N M D +I +F ++ +
Sbjct 270 VHRYDNGGFWPHLKESASSAVGKERGERFNVNVAWNKTRMSDADYIHVFLNVLLPVAYEF 329
Query 140 RPGAVVLQCGADSLSGDRLGKFNLTVKGHAACVQFVKSFNLPLLLLG-GGGYTIRNVS 196
+P V++ G DS+ GD G+ + T + + S L+L GGY R+++
Sbjct 330 QPHLVLVAAGFDSVVGDPKGEMSATPACFSHLTHLLMSLAQGRLILSLEGGYNQRSLA 387
> hsa:9759 HDAC4, AHO3, BDMR, HA6116, HD4, HDAC-A, HDACA, KIAA0288;
histone deacetylase 4 (EC:3.5.1.98); K11406 histone deacetylase
4/5 [EC:3.5.1.98]
Length=1084
Score = 94.4 bits (233), Expect = 5e-19, Method: Compositional matrix adjust.
Identities = 48/150 (32%), Positives = 83/150 (55%), Gaps = 10/150 (6%)
Query 27 HHAKRAEASGFCYVNDIVLG--ILELLKYHPRVMYIDIDVHHGDGVEEAFYVSHRVLTVS 84
HHA+ + GFCY N + + +L+ +++ +D DVHHG+G ++AFY VL +S
Sbjct 802 HHAEESTPMGFCYFNSVAVAAKLLQQRLSVSKILIVDWDVHHGNGTQQAFYSDPSVLYMS 861
Query 85 FHKF--GDFFPGTGDITDVGAAQGKYFSVNVPLNDGMD----DESFIGLFKPIIAKCVEV 138
H++ G+FFPG+G +VG G F+VN+ G+D D ++ F+ ++
Sbjct 862 LHRYDDGNFFPGSGAPDEVGTGPGVGFNVNMAFTGGLDPPMGDAEYLAAFRTVVMPIASE 921
Query 139 YRPGAVVLQCGADSLSG--DRLGKFNLTVK 166
+ P V++ G D++ G LG +NL+ +
Sbjct 922 FAPDVVLVSSGFDAVEGHPTPLGGYNLSAR 951
> hsa:51564 HDAC7, DKFZp586J0917, FLJ99588, HD7A, HDAC7A; histone
deacetylase 7 (EC:3.5.1.98); K11408 histone deacetylase
7 [EC:3.5.1.98]
Length=954
Score = 94.0 bits (232), Expect = 8e-19, Method: Compositional matrix adjust.
Identities = 48/150 (32%), Positives = 83/150 (55%), Gaps = 10/150 (6%)
Query 27 HHAKRAEASGFCYVNDIVLGILELLKYHP--RVMYIDIDVHHGDGVEEAFYVSHRVLTVS 84
HHA + A GFC+ N + + +L + +++ +D DVHHG+G ++ FY VL +S
Sbjct 671 HHADHSTAMGFCFFNSVAIACRQLQQQSKASKILIVDWDVHHGNGTQQTFYQDPSVLYIS 730
Query 85 FHKF--GDFFPGTGDITDVGAAQGKYFSVNVPLNDGMD----DESFIGLFKPIIAKCVEV 138
H+ G+FFPG+G + +VGA G+ F+VNV G+D D ++ F+ ++
Sbjct 731 LHRHDDGNFFPGSGAVDEVGAGSGEGFNVNVAWAGGLDPPMGDPEYLAAFRIVVMPIARE 790
Query 139 YRPGAVVLQCGADSLSGD--RLGKFNLTVK 166
+ P V++ G D+ G LG ++++ K
Sbjct 791 FSPDLVLVSAGFDAAEGHPAPLGGYHVSAK 820
> mmu:208727 Hdac4, 4932408F19Rik, AI047285; histone deacetylase
4 (EC:3.5.1.98); K11406 histone deacetylase 4/5 [EC:3.5.1.98]
Length=1076
Score = 93.2 bits (230), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 49/150 (32%), Positives = 82/150 (54%), Gaps = 10/150 (6%)
Query 27 HHAKRAEASGFCYVNDIVLG--ILELLKYHPRVMYIDIDVHHGDGVEEAFYVSHRVLTVS 84
HHA+ + GFCY N + + +L+ +++ +D DVHHG+G ++AFY VL +S
Sbjct 794 HHAEESTPMGFCYFNSVAVAAKLLQQRLNVSKILIVDWDVHHGNGTQQAFYNDPNVLYMS 853
Query 85 FHKF--GDFFPGTGDITDVGAAQGKYFSVNVPLNDG----MDDESFIGLFKPIIAKCVEV 138
H++ G+FFPG+G +VG G F+VN+ G M D ++ F+ ++
Sbjct 854 LHRYDDGNFFPGSGAPDEVGTGPGVGFNVNMAFTGGLEPPMGDAEYLAAFRTVVMPIANE 913
Query 139 YRPGAVVLQCGADSLSG--DRLGKFNLTVK 166
+ P V++ G D++ G LG +NL+ K
Sbjct 914 FAPDVVLVSSGFDAVEGHPTPLGGYNLSAK 943
> mmu:100504541 histone deacetylase 9-like
Length=314
Score = 93.2 bits (230), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 49/154 (31%), Positives = 85/154 (55%), Gaps = 12/154 (7%)
Query 27 HHAKRAEASGFCYVNDIVLGILELLKYH--PRVMYIDIDVHHGDGVEEAFYVSHRVLTVS 84
HHA+ + A GFC+ N + + L +++ +D+DVHHG+G ++AFY +L +S
Sbjct 30 HHAEESAAMGFCFFNSVAITAKYLRDQLNISKILIVDLDVHHGNGTQQAFYADPSILYIS 89
Query 85 FHKF--GDFFPGTGDITDVGAAQGKYFSVNVPLNDGMD----DESFIGLFKPIIAKCVEV 138
H++ G+FFPG+G +VG G+ ++VN+ G+D D ++ F+ ++
Sbjct 90 LHRYDEGNFFPGSGAPNEVGVGLGEGYNVNIAWTGGLDPPMGDVEYLEAFRTVVMPVARE 149
Query 139 YRPGAVVLQCGADSLSGDR--LGKFNLTVK--GH 168
+ P V++ G D+L G LG + +T K GH
Sbjct 150 FDPDMVLVSAGFDALEGHTPPLGGYKVTAKCFGH 183
> mmu:56233 Hdac7, 5830434K02Rik, HD7, HD7a, Hdac7a, mFLJ00062;
histone deacetylase 7 (EC:3.5.1.98); K11408 histone deacetylase
7 [EC:3.5.1.98]
Length=953
Score = 92.8 bits (229), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 47/150 (31%), Positives = 83/150 (55%), Gaps = 10/150 (6%)
Query 27 HHAKRAEASGFCYVNDIVLGILELLKY--HPRVMYIDIDVHHGDGVEEAFYVSHRVLTVS 84
HHA + A GFC+ N + + +L ++ +++ +D DVHHG+G ++ FY VL +S
Sbjct 671 HHADHSTAMGFCFFNSVAIACRQLQQHGKASKILIVDWDVHHGNGTQQTFYQDPSVLYIS 730
Query 85 FHKF--GDFFPGTGDITDVGAAQGKYFSVNVPLNDGMD----DESFIGLFKPIIAKCVEV 138
H+ G+FFPG+G + +VG G+ F+VNV G+D D ++ F+ ++
Sbjct 731 LHRHDDGNFFPGSGAVDEVGTGSGEGFNVNVAWAGGLDPPMGDPEYLAAFRIVVMPIARE 790
Query 139 YRPGAVVLQCGADSLSGD--RLGKFNLTVK 166
+ P V++ G D+ G LG ++++ K
Sbjct 791 FAPDLVLVSAGFDAAEGHPAPLGGYHVSAK 820
> dre:568877 hdac4, KIAA0288, wu:fa96d08, wu:fc56f08, zgc:152701;
histone deacetylase 4 (EC:3.5.1.98); K11406 histone deacetylase
4/5 [EC:3.5.1.98]
Length=1023
Score = 92.4 bits (228), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 49/150 (32%), Positives = 81/150 (54%), Gaps = 10/150 (6%)
Query 27 HHAKRAEASGFCYVNDIVLG--ILELLKYHPRVMYIDIDVHHGDGVEEAFYVSHRVLTVS 84
HHA+ + GFCY N + + +L+ +++ +D DVHHG+G ++AFY VL +S
Sbjct 741 HHAEESTPMGFCYFNSVAIAAKLLQQRLNVSKILIVDWDVHHGNGTQQAFYSDPNVLYLS 800
Query 85 FHKF--GDFFPGTGDITDVGAAQGKYFSVNVPLNDG----MDDESFIGLFKPIIAKCVEV 138
H++ G+FFPG+G +VG G F+VN+ G M D ++ F+ ++
Sbjct 801 LHRYDDGNFFPGSGAPDEVGIGPGVGFNVNMAFTGGLEPPMGDADYLAAFRSVVMPIANE 860
Query 139 YRPGAVVLQCGADSLSGD--RLGKFNLTVK 166
+ P V++ G D++ G LG + LT K
Sbjct 861 FSPDVVLVSSGFDAVEGHPPPLGGYKLTAK 890
> cel:F41H10.6 hypothetical protein; K11407 histone deacetylase
6/10 [EC:3.5.1.98]
Length=955
Score = 91.3 bits (225), Expect = 4e-18, Method: Compositional matrix adjust.
Identities = 62/203 (30%), Positives = 100/203 (49%), Gaps = 13/203 (6%)
Query 27 HHAKRAEASGFCYVNDIVLGILELLKYHP--RVMYIDIDVHHGDGVEEAFYVSHRVLTVS 84
HHA +++SGFC N++ + + H RV+ +D DVHHG+G +E FY V+ +S
Sbjct 560 HHASASKSSGFCIFNNVAVAAKYAQRRHKAKRVLILDWDVHHGNGTQEIFYEDSNVMYMS 619
Query 85 FHKF--GDFFP--GTGDITDVGAAQGKYFSVNVPLND-GMDDESFIGLFKPIIAKCVEVY 139
H+ G+F+P D +DVG G+ SVNVP + M D + F+ +I +
Sbjct 620 IHRHDKGNFYPIGEPKDYSDVGEGAGEGMSVNVPFSGVQMGDNEYQMAFQRVIMPIAYQF 679
Query 140 RPGAVVLQCGADSLSGDRLGKFNLTVKGHAACVQFVKSF-NLPLLLLGGGGYTIRNVSR- 197
P V++ G D+ D LG++ +T + A + S ++ + GGY + ++S
Sbjct 680 NPDLVLISAGFDAAVDDPLGEYKVTPETFALMTYQLSSLAGGRIITVLEGGYNLTSISNS 739
Query 198 ----CWVYETAVALDRHRELSPQ 216
C V + L R RE Q
Sbjct 740 AQAVCEVLQNRSMLRRLREEKEQ 762
Score = 81.3 bits (199), Expect = 5e-15, Method: Compositional matrix adjust.
Identities = 55/183 (30%), Positives = 89/183 (48%), Gaps = 11/183 (6%)
Query 27 HHAKRAEASGFCYVNDIVLGILE-LLKYHPRVMYIDIDVHHGDGVEEAFYVSHRVLTVSF 85
HHA GFC N++ E R++ +D+DVHHG G + FY RVL S
Sbjct 145 HHADSVSPCGFCLFNNVAQAAEEAFFSGAERILIVDLDVHHGHGTQRIFYDDKRVLYFSI 204
Query 86 HK--FGDFFPG--TGDITDVGAAQGKYFSVNVPLND-GMDDESFIGLFKPIIAKCVEVYR 140
H+ G F+P D +G+ +G ++ N+ LN+ G D ++ + ++ +
Sbjct 205 HRHEHGLFWPHLPESDFDHIGSGKGLGYNANLALNETGCTDSDYLSIIFHVLLPLATQFD 264
Query 141 PGAVVLQCGADSLSGDRLGKFNLTVKGHAACVQFVKSF---NLPLLLLGGGGYTIRNVS- 196
P V++ G D+L GD LG LT G++ + +KS + ++L GG + I V+
Sbjct 265 PHFVIISAGFDALLGDPLGGMCLTPDGYSHILYHLKSLAQGRMLVVLEGGYNHQISAVAV 324
Query 197 -RC 198
RC
Sbjct 325 QRC 327
> tpv:TP03_0788 hypothetical protein
Length=900
Score = 90.9 bits (224), Expect = 6e-18, Method: Compositional matrix adjust.
Identities = 53/182 (29%), Positives = 91/182 (50%), Gaps = 11/182 (6%)
Query 27 HHAKRAEASGFCYVNDIVLGILELLKYH--PRVMYIDIDVHHGDGVEEAFYVSHRVLTVS 84
HHA + GFC N++ + L RV +D DVHHG+G ++ FY + V +S
Sbjct 225 HHATPDKMMGFCIYNNVAIAARYLQHKFGLKRVAIVDWDVHHGNGTQDIFYDDNSVCFIS 284
Query 85 FHKFG----DFFPGTGDITDVGAAQGKYFSVNVPLNDGMDDESFIGLFKPIIAKCVEVYR 140
H++G F+P TG ++G +G ++VN+PL + + F ++ +E++
Sbjct 285 LHRYGTDEDSFYPYTGYCDEIGVGKGSKYNVNIPLEKSFTNADLVHSFNKVVIPVLELFE 344
Query 141 PGAVVLQCGADSLSGDRLGKFNLTVKGHAACV----QFVKSFNLPLLLLG-GGGYTIRNV 195
P +++ G DS GD LG L +G + + + ++ LLL GGYT+ +
Sbjct 345 PEFILVSAGFDSGRGDLLGGCRLDWEGFSWATFKLCELAEKYSKGRLLLSLEGGYTLSRL 404
Query 196 SR 197
S+
Sbjct 405 SQ 406
Lambda K H
0.320 0.138 0.433
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 13585434760
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40