bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_0206_orf2
Length=448
Score E
Sequences producing significant alignments: (Bits) Value
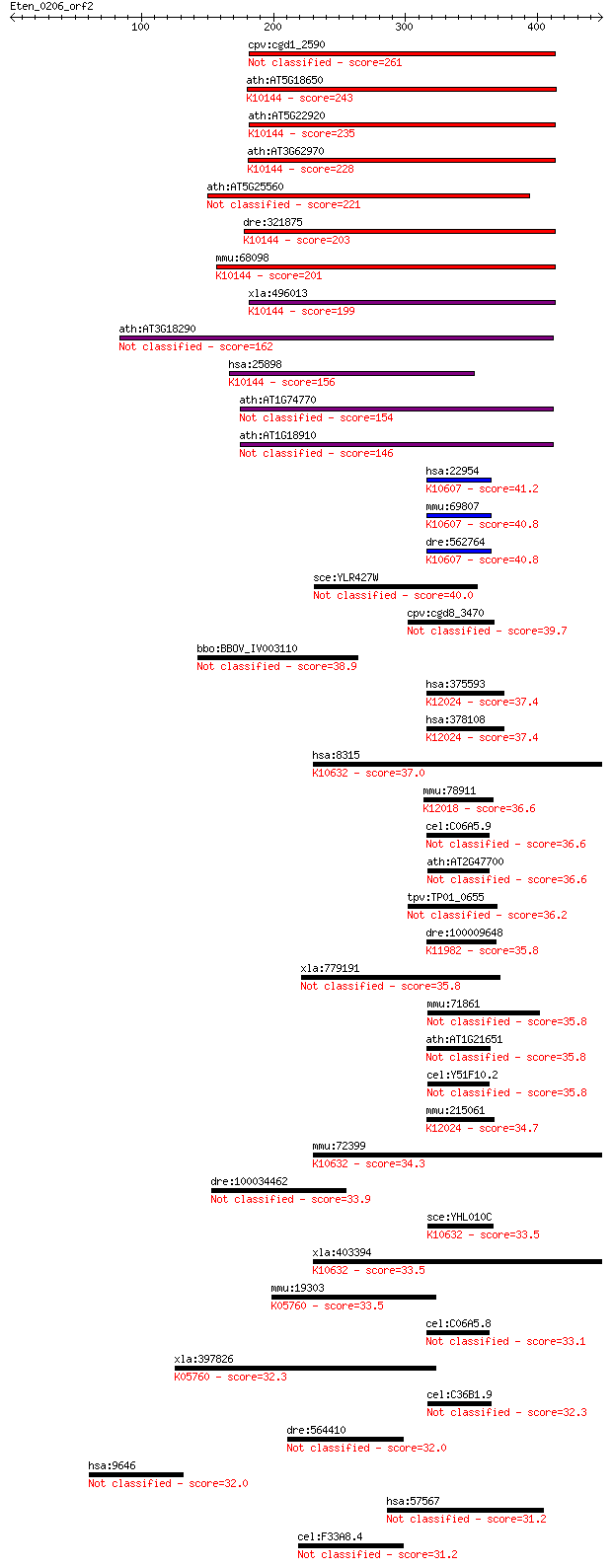
cpv:cgd1_2590 PGPD14 protein with at least one predicted RING ... 261 4e-69
ath:AT5G18650 zinc finger (C3HC4-type RING finger) family prot... 243 1e-63
ath:AT5G22920 zinc finger (C3HC4-type RING finger) family prot... 235 3e-61
ath:AT3G62970 protein binding / zinc ion binding; K10144 ring ... 228 5e-59
ath:AT5G25560 zinc finger (C3HC4-type RING finger) family protein 221 3e-57
dre:321875 rchy1, Zfp363, wu:fb37e07, zgc:101128; ring finger ... 203 1e-51
mmu:68098 Rchy1, 6720407C15Rik, ARNIP, AU042618, CHIMP, PRO199... 201 5e-51
xla:496013 rchy1; ring finger and CHY zinc finger domain conta... 199 2e-50
ath:AT3G18290 EMB2454 (embryo defective 2454); protein binding... 162 2e-39
hsa:25898 RCHY1, ARNIP, CHIMP, DKFZp586C1620, PIRH2, PRO1996, ... 156 1e-37
ath:AT1G74770 protein binding / zinc ion binding 154 6e-37
ath:AT1G18910 protein binding / zinc ion binding 146 2e-34
hsa:22954 TRIM32, BBS11, HT2A, LGMD2H, TATIP; tripartite motif... 41.2 0.009
mmu:69807 Trim32, 1810045E12Rik, 3f3, BBS11, Zfp117; tripartit... 40.8 0.010
dre:562764 trim32; tripartite motif-containing 32; K10607 trip... 40.8 0.011
sce:YLR427W MAG2; Cytoplasmic protein of unknown function; ind... 40.0 0.018
cpv:cgd8_3470 membrane associated protein with a RING finger, ... 39.7 0.025
bbo:BBOV_IV003110 21.m03046; CHY zinc finger domain containing... 38.9 0.045
hsa:375593 TRIM73, FLJ99347, MGC45477, TRIM50B; tripartite mot... 37.4 0.13
hsa:378108 TRIM74, MGC45440, TRIM50C; tripartite motif contain... 37.4 0.13
hsa:8315 BRAP, BRAP2, IMP, RNF52; BRCA1 associated protein; K1... 37.0 0.14
mmu:78911 Trim42, 4930486B16Rik; tripartite motif-containing 4... 36.6 0.19
cel:C06A5.9 rnf-1; RiNg Finger protein family member (rnf-1) 36.6 0.20
ath:AT2G47700 zinc finger (C3HC4-type RING finger) family protein 36.6 0.22
tpv:TP01_0655 hypothetical protein 36.2 0.24
dre:100009648 zgc:158807; K11982 E3 ubiquitin-protein ligase R... 35.8 0.33
xla:779191 znf234, MGC115682; zinc finger protein 234 35.8
mmu:71861 Zswim2, 1700025P14Rik, 4933437F18Rik, MEX; zinc fing... 35.8 0.37
ath:AT1G21651 protein binding / zinc ion binding 35.8 0.38
cel:Y51F10.2 hypothetical protein 35.8 0.39
mmu:215061 Trim50; tripartite motif-containing 50; K12024 trip... 34.7 0.88
mmu:72399 Brap, 3010002G07Rik; BRCA1 associated protein; K1063... 34.3 1.0
dre:100034462 si:dkey-24l11.2 33.9 1.2
sce:YHL010C ETP1; Etp1p; K10632 BRCA1-associated protein [EC:6... 33.5 1.5
xla:403394 IMP protein; K10632 BRCA1-associated protein [EC:6.... 33.5 1.9
mmu:19303 Pxn, AW108311, AW123232, Pax; paxillin; K05760 paxillin 33.5 1.9
cel:C06A5.8 hypothetical protein 33.1 2.1
xla:397826 pxn; paxillin; K05760 paxillin 32.3 3.9
cel:C36B1.9 hypothetical protein 32.3 4.4
dre:564410 zgc:172248 32.0 4.8
hsa:9646 CTR9, KIAA0155, SH2BP1, TSBP, p150, p150TSP; Ctr9, Pa... 32.0 5.5
hsa:57567 ZNF319, MGC126816, ZFP319; zinc finger protein 319 31.2
cel:F33A8.4 hypothetical protein 31.2 9.0
> cpv:cgd1_2590 PGPD14 protein with at least one predicted RING
finger, possible plant origin
Length=256
Score = 261 bits (667), Expect = 4e-69, Method: Compositional matrix adjust.
Identities = 118/243 (48%), Positives = 161/243 (66%), Gaps = 16/243 (6%)
Query 182 GCAHYRRKCKVVAPCCKEIYWCRHCHNEAYEMDLSNAHEIDRHAVEEIVCAVCEARQPVS 241
GC HY+ +C+++APCC YWCRHCHNE+ E + HE+DR +++E+VC C RQP S
Sbjct 3 GCKHYKSRCRIIAPCCNNEYWCRHCHNESQE----DHHEVDRFSIKEVVCRRCNKRQPAS 58
Query 242 NKCVG---CG---------TVFGAYFCSVCKFWDNLGEKKKVYHCDECGICRLGGRENYF 289
N C+ CG T F YFCS C WD+ G +K V+HCDECGICR+GG+E+YF
Sbjct 59 NSCINSAECGEKSGKKCDVTQFAKYFCSKCNLWDDNGIEKNVFHCDECGICRVGGQESYF 118
Query 290 HCPTCGSCYPMQLQNKHKCLENAMHRQCPVCLEDMFSSLRQSQVLQCGHTIHSDCLRLLQ 349
HC C CYP+ ++ HKC+EN+ R CP+CLED+F S++ +L CGHTIH DCL LL
Sbjct 119 HCKVCCMCYPLSIKETHKCIENSCRRPCPLCLEDLFYSIKTVSILNCGHTIHEDCLLLLG 178
Query 350 KQKGFQAIRCPMCSKSIADYSEFWKQLSEEIERTPMEEQMRRKVRIACNDCLERCTTDFH 409
+ KG ++RCP+CSKS+ D S+ W ++ + I +P+ E+ + V I CNDC +C T H
Sbjct 179 EAKGLTSLRCPICSKSLGDNSQIWNEIDKMIAESPIPEESKELVNIFCNDCNIKCNTYSH 238
Query 410 FLG 412
G
Sbjct 239 PYG 241
> ath:AT5G18650 zinc finger (C3HC4-type RING finger) family protein;
K10144 ring finger and CHY zinc finger domain-containing
protein 1
Length=267
Score = 243 bits (619), Expect = 1e-63, Method: Compositional matrix adjust.
Identities = 109/237 (45%), Positives = 162/237 (68%), Gaps = 7/237 (2%)
Query 180 ALGCAHYRRKCKVVAPCCKEIYWCRHCHNEAYEM--DLSNAHEIDRHAVEEIVCAVCEAR 237
GC HY+R+C++ APCC E++ CRHCHNE+ ++ + H++ R V++++C+VC+
Sbjct 15 GFGCKHYKRRCQIRAPCCNEVFDCRHCHNESTSTLRNIYDRHDLVRQDVKQVICSVCDTE 74
Query 238 QPVSNKCVGCGTVFGAYFCSVCKFWDNLGEKKKVYHCDECGICRLGGRENYFHCPTCGSC 297
QP + C CG G YFCS+C F+D+ EK++ +HCD+CGICR+GGREN+FHC CGSC
Sbjct 75 QPAAQVCSNCGVNMGEYFCSICIFYDDDTEKQQ-FHCDDCGICRVGGRENFFHCKKCGSC 133
Query 298 YPMQLQNKHKCLENAMHRQCPVCLEDMFSSLRQSQVLQCGHTIHSDCLRLLQKQKGFQAI 357
Y + L+N H+C+EN+M CP+C E +F SL+ + V++CGHT+H +C + K+ F
Sbjct 134 YAVGLRNNHRCVENSMRHHCPICYEYLFDSLKDTNVMKCGHTMHVECYNEMIKRDKFC-- 191
Query 358 RCPMCSKSIADYSEFWKQLSEEIERTPMEEQMR-RKVRIACNDCLERCTTDFHFLGE 413
CP+CS+S+ D S+ W++L EEIE T M R +KV I CNDC + FH +G+
Sbjct 192 -CPICSRSVIDMSKTWQRLDEEIEATAMPSDYRDKKVWILCNDCNDTTEVHFHIIGQ 247
> ath:AT5G22920 zinc finger (C3HC4-type RING finger) family protein;
K10144 ring finger and CHY zinc finger domain-containing
protein 1
Length=291
Score = 235 bits (600), Expect = 3e-61, Method: Compositional matrix adjust.
Identities = 114/235 (48%), Positives = 148/235 (62%), Gaps = 8/235 (3%)
Query 182 GCAHYRRKCKVVAPCCKEIYWCRHCHNEA---YEMDLSNAHEIDRHAVEEIVCAVCEARQ 238
GC+HYRR+CK+ APCC EI+ CRHCHNEA ++ + HE+ RH V +++C++CE Q
Sbjct 26 GCSHYRRRCKIRAPCCDEIFDCRHCHNEAKDSLHIEQHHRHELPRHEVSKVICSLCETEQ 85
Query 239 PVSNKCVGCGTVFGAYFCSVCKFWDNLGEKKKVYHCDECGICRLGGRENYFHCPTCGSCY 298
V C CG G YFCS CKF+D+ KK+ YHCDECGICR GG EN+FHC C CY
Sbjct 86 DVQQNCSNCGVCMGKYFCSKCKFFDDDLSKKQ-YHCDECGICRTGGEENFFHCKRCRCCY 144
Query 299 PMQLQNKHKCLENAMHRQCPVCLEDMFSSLRQSQVLQCGHTIHSDCLRLLQKQKGFQAIR 358
+++KH+C+E AMH CPVC E +F S R VL+CGHT+H +C + + +
Sbjct 145 SKIMEDKHQCVEGAMHHNCPVCFEYLFDSTRDITVLRCGHTMHLECTKDMGLHNRYT--- 201
Query 359 CPMCSKSIADYSEFWKQLSEEIERTPMEEQMRRK-VRIACNDCLERCTTDFHFLG 412
CP+CSKSI D S WK+L EE+ PM + K V I CNDC FH +
Sbjct 202 CPVCSKSICDMSNLWKKLDEEVAAYPMPKMYENKMVWILCNDCGSNTNVRFHLIA 256
> ath:AT3G62970 protein binding / zinc ion binding; K10144 ring
finger and CHY zinc finger domain-containing protein 1
Length=287
Score = 228 bits (580), Expect = 5e-59, Method: Compositional matrix adjust.
Identities = 103/234 (44%), Positives = 150/234 (64%), Gaps = 6/234 (2%)
Query 181 LGCAHYRRKCKVVAPCCKEIYWCRHCHNEAYEM--DLSNAHEIDRHAVEEIVCAVCEARQ 238
GC HY+R+CK+ APCC I+ CRHCHN++ D H++ R V+++VC++C+ Q
Sbjct 33 FGCEHYKRRCKIRAPCCNLIFSCRHCHNDSANSLPDPKERHDLVRQNVKQVVCSICQTEQ 92
Query 239 PVSNKCVGCGTVFGAYFCSVCKFWDNLGEKKKVYHCDECGICRLGGRENYFHCPTCGSCY 298
V+ C CG G YFC +CKF+D+ K++ +HCD+CGICR+GGR+ +FHC CG+CY
Sbjct 93 EVAKVCSNCGVNMGEYFCDICKFFDDDISKEQ-FHCDDCGICRVGGRDKFFHCQNCGACY 151
Query 299 PMQLQNKHKCLENAMHRQCPVCLEDMFSSLRQSQVLQCGHTIHSDCLRLLQKQKGFQAIR 358
M L++KH C+EN+ CPVC E +F S++ + V++CGHT+H DC + + + R
Sbjct 152 GMGLRDKHSCIENSTKNSCPVCYEYLFDSVKAAHVMKCGHTMHMDCFEQMINENQY---R 208
Query 359 CPMCSKSIADYSEFWKQLSEEIERTPMEEQMRRKVRIACNDCLERCTTDFHFLG 412
CP+C+KS+ D S W L EI T M + + +V I CNDC + FH LG
Sbjct 209 CPICAKSMVDMSPSWHLLDFEISATEMPVEYKFEVSILCNDCNKGSKAMFHILG 262
> ath:AT5G25560 zinc finger (C3HC4-type RING finger) family protein
Length=328
Score = 221 bits (564), Expect = 3e-57, Method: Compositional matrix adjust.
Identities = 111/255 (43%), Positives = 150/255 (58%), Gaps = 15/255 (5%)
Query 150 EDSDSSSSSNSSEDSPKTVADEPAADAEGNAL--------GCAHYRRKCKVVAPCCKEIY 201
E S S + +E+S + + AA++ N + GC HYRR+C + APCC EI+
Sbjct 22 EMSRHSHPHSINEESESSTLERVAAESLTNKVLDRGLMEYGCPHYRRRCCIRAPCCNEIF 81
Query 202 WCRHCHNEA---YEMDLSNAHEIDRHAVEEIVCAVCEARQPVSNKCVGCGTVFGAYFCSV 258
C HCH EA +D H+I RH VE+++C +C Q V C+ CG G YFC V
Sbjct 82 GCHHCHYEAKNNINVDQKQRHDIPRHQVEQVICLLCGTEQEVGQICIHCGVCMGKYFCKV 141
Query 259 CKFWDNLGEKKKVYHCDECGICRLGGRENYFHCPTCGSCYPMQLQNKHKCLENAMHRQCP 318
CK +D+ KK+ YHCD CGICR+GGREN+FHC CG CY + L+N H C+E AMH CP
Sbjct 142 CKLYDDDTSKKQ-YHCDGCGICRIGGRENFFHCYKCGCCYSILLKNGHPCVEGAMHHDCP 200
Query 319 VCLEDMFSSLRQSQVLQCGHTIHSDCLRLLQKQKGFQAIRCPMCSKSIADYSEFWKQLSE 378
+C E +F S VL CGHTIH CL ++ + CP+CSKS+ D S+ W++
Sbjct 201 ICFEFLFESRNDVTVLPCGHTIHQKCLEEMRDHYQYA---CPLCSKSVCDMSKVWEKFDM 257
Query 379 EIERTPMEEQMRRKV 393
EI TPM E + ++
Sbjct 258 EIAATPMPEPYQNRM 272
> dre:321875 rchy1, Zfp363, wu:fb37e07, zgc:101128; ring finger
and CHY zinc finger domain containing 1; K10144 ring finger
and CHY zinc finger domain-containing protein 1
Length=264
Score = 203 bits (516), Expect = 1e-51, Method: Compositional matrix adjust.
Identities = 102/236 (43%), Positives = 132/236 (55%), Gaps = 13/236 (5%)
Query 178 GNALGCAHYRRKCKVVAPCCKEIYWCRHCHNEAYEMDLSNAHEIDRHAVEEIVCAVCEAR 237
+GC HY R C + APCC + Y CR CH D H++DR V+E+ CAVC
Sbjct 3 ATKVGCEHYVRSCLLKAPCCGKFYVCRLCH------DAEETHQMDRFKVQEVKCAVCNTI 56
Query 238 QPVSNKCVGCGTVFGAYFCSVCKFWDNLGEKKKVYHCDECGICRLGGRENYFHCPTCGSC 297
Q C C FG Y+C +C +D + KK YHC CGICR+G RE YFHC C C
Sbjct 57 QEAQQICKECEVKFGEYYCDICHLFD---KDKKQYHCQPCGICRIGPREKYFHCTKCNLC 113
Query 298 YPMQLQNKHKCLENAMHRQCPVCLEDMFSSLRQSQVLQCGHTIHSDCLRLLQKQKGFQAI 357
+L++KHKC+EN + CPVC+ED+ + + VL CGH +H C + K +
Sbjct 114 LGTELKDKHKCVENVSRQNCPVCMEDIHTFRVGAHVLPCGHLLHGTCFDDMLKTGAY--- 170
Query 358 RCPMCSKSIADYSEFWKQLSEEIERTPM-EEQMRRKVRIACNDCLERCTTDFHFLG 412
RCP+C S + E+WKQ+ EEI +TPM E V+I CNDC R T FH LG
Sbjct 171 RCPLCMHSAFNMKEYWKQMDEEISQTPMPTEYQDSTVKIICNDCQARSTVSFHVLG 226
> mmu:68098 Rchy1, 6720407C15Rik, ARNIP, AU042618, CHIMP, PRO1996,
Pirh2, Zfp363, mARNIP; ring finger and CHY zinc finger
domain containing 1; K10144 ring finger and CHY zinc finger
domain-containing protein 1
Length=261
Score = 201 bits (511), Expect = 5e-51, Method: Compositional matrix adjust.
Identities = 103/257 (40%), Positives = 143/257 (55%), Gaps = 22/257 (8%)
Query 157 SSNSSEDSPKTVADEPAADAEGNALGCAHYRRKCKVVAPCCKEIYWCRHCHNEAYEMDLS 216
++ + ED + +A P GC HY R C + APCC ++Y CR CH D +
Sbjct 2 AATAREDGVRNLAQGPR--------GCEHYDRACLLKAPCCDKLYTCRLCH------DTN 47
Query 217 NAHEIDRHAVEEIVCAVCEARQPVSNKCVGCGTVFGAYFCSVCKFWDNLGEKKKVYHCDE 276
H++DR V+E+ C CE Q C C T+FG Y+CS+C +D + K+ YHC+
Sbjct 48 EDHQLDRFKVKEVQCINCEKLQHAQQTCEDCSTLFGEYYCSICHLFD---KDKRQYHCES 104
Query 277 CGICRLGGRENYFHCPTCGSCYPMQLQNKHKCLENAMHRQCPVCLEDMFSSLRQSQVLQC 336
CGICR+G +E++FHC C C L+ KHKC+EN + CP+CLED+ +S + VL C
Sbjct 105 CGICRIGPKEDFFHCLKCNLCLTTNLRGKHKCIENVSRQNCPICLEDIHTSRVVAHVLPC 164
Query 337 GHTIHSDCLRLLQKQKGFQAIRCPMCSKSIADYSEFWKQLSEEIERTPM-EEQMRRKVRI 395
GH +H C + K + RCP+C S D + +W+QL E+ +TPM E V I
Sbjct 165 GHLLHRTCYEEMLK----EGYRCPLCMHSALDMTRYWRQLDTEVAQTPMPSEYQNVTVDI 220
Query 396 ACNDCLERCTTDFHFLG 412
CNDC R T FH LG
Sbjct 221 LCNDCNGRSTVQFHILG 237
> xla:496013 rchy1; ring finger and CHY zinc finger domain containing
1; K10144 ring finger and CHY zinc finger domain-containing
protein 1
Length=248
Score = 199 bits (506), Expect = 2e-50, Method: Compositional matrix adjust.
Identities = 99/232 (42%), Positives = 135/232 (58%), Gaps = 14/232 (6%)
Query 182 GCAHYRRKCKVVAPCCKEIYWCRHCHNEAYEMDLSNAHEIDRHAVEEIVCAVCEARQPVS 241
GC HY R C++ APCC + Y CR CH D +H++DR V ++ C C+ Q
Sbjct 6 GCEHYSRSCQLRAPCCGKFYTCRLCH------DSKESHKMDRFNVTQVQCMECKCVQKAQ 59
Query 242 NKCVGCGTVFGAYFCSVCKFWDNLGEKKKVYHCDECGICRLGGRENYFHCPTCGSCYPMQ 301
C C VFG Y+C++C +D + KK YHCD CGICR+G +E + HC C C P+
Sbjct 60 QTCEQCHMVFGDYYCNICHLFD---KDKKQYHCDGCGICRIGPKEEFEHCTKCNLCLPLS 116
Query 302 LQNKHKCLENAMHRQCPVCLEDMFSSLRQSQVLQCGHTIHSDCLRLLQKQKGFQAIRCPM 361
L+ HKC+EN + CP+CLED+ +S ++VL CGH +HS C + K Q RCP+
Sbjct 117 LRGNHKCIENVSRQDCPICLEDIHTSRVGARVLPCGHLLHSTCYEDMLK----QGYRCPL 172
Query 362 CSKSIADYSEFWKQLSEEIERTPM-EEQMRRKVRIACNDCLERCTTDFHFLG 412
C +S D + +W+QL +E+ +TPM E V I CNDC R T FH LG
Sbjct 173 CMRSALDMTRYWRQLDDEVAQTPMPSEYQNMTVEILCNDCSSRSTVPFHILG 224
> ath:AT3G18290 EMB2454 (embryo defective 2454); protein binding
/ zinc ion binding
Length=1254
Score = 162 bits (411), Expect = 2e-39, Method: Compositional matrix adjust.
Identities = 109/353 (30%), Positives = 171/353 (48%), Gaps = 44/353 (12%)
Query 84 GSHDNNSA-TLGPAPQSEGAVEEEIEGEGEAAGE-----LRGTEVEYSDNGSQHWEEAVS 137
GS D++S T P+PQ + +E ++ GE R + E + ++++
Sbjct 893 GSPDSSSTETSKPSPQKDNDHQEILDQSGELFKPGWKDIFRMNQNELEAEIRKVYQDSTL 952
Query 138 SPEEGSILFQSDEDSDSSSSSNSSEDSPKTVAD-------EPAA-DAEGNALGCAHYRRK 189
P L Q+ S ++ +T + P+ D E GC HY+R
Sbjct 953 DPRRKDYLVQNWRTSRWIAAQQKLPKEAETAVNGDVELGCSPSFRDPEKQIYGCEHYKRN 1012
Query 190 CKVVAPCCKEIYWCRHCHNEAYEMDLSNAHEIDRHAVEEIVCAVCEARQPVSNKCV--GC 247
CK+ A CC +++ CR CH++ + H +DR V E++C C QPV C C
Sbjct 1013 CKLRAACCDQLFTCRFCHDKVSD------HSMDRKLVTEMLCMRCLKVQPVGPICTTPSC 1066
Query 248 -GTVFGAYFCSVCKFWDNLGEKKKVYHCDECGICRLGGRE--NYFHCPTCGSCYPMQLQN 304
G ++CS+CK +D+ ++ VYHC C +CR+G ++FHC TC C M+L N
Sbjct 1067 DGFPMAKHYCSICKLFDD---ERAVYHCPFCNLCRVGEGLGIDFFHCMTCNCCLGMKLVN 1123
Query 305 KHKCLENAMHRQCPVCLEDMFSSLRQSQVLQCGHTIHSDCLRLLQKQKGFQA-----IRC 359
HKCLE ++ CP+C E +F+S + L CGH +HS C FQA C
Sbjct 1124 -HKCLEKSLETNCPICCEFLFTSSEAVRALPCGHYMHSAC---------FQAYTCSHYTC 1173
Query 360 PMCSKSIADYSEFWKQLSEEIERTPMEEQMRRKVR-IACNDCLERCTTDFHFL 411
P+C KS+ D + ++ L + + E+ + + + I CNDC + TT FH+L
Sbjct 1174 PICGKSLGDMAVYFGMLDALLAAEELPEEYKNRCQDILCNDCERKGTTRFHWL 1226
> hsa:25898 RCHY1, ARNIP, CHIMP, DKFZp586C1620, PIRH2, PRO1996,
RNF199, ZNF363, hARNIP; ring finger and CHY zinc finger domain
containing 1; K10144 ring finger and CHY zinc finger domain-containing
protein 1
Length=200
Score = 156 bits (395), Expect = 1e-37, Method: Compositional matrix adjust.
Identities = 77/185 (41%), Positives = 108/185 (58%), Gaps = 9/185 (4%)
Query 167 TVADEPAADAEGNALGCAHYRRKCKVVAPCCKEIYWCRHCHNEAYEMDLSNAHEIDRHAV 226
T ++ A+ E GC HY R C + APCC ++Y CR CH D + H++DR V
Sbjct 4 TAREDGASGQERGQRGCEHYDRGCLLKAPCCDKLYTCRLCH------DNNEDHQLDRFKV 57
Query 227 EEIVCAVCEARQPVSNKCVGCGTVFGAYFCSVCKFWDNLGEKKKVYHCDECGICRLGGRE 286
+E+ C CE Q C C T+FG Y+C +C +D + KK YHC+ CGICR+G +E
Sbjct 58 KEVQCINCEKIQHAQQTCEECSTLFGEYYCDICHLFD---KDKKQYHCENCGICRIGPKE 114
Query 287 NYFHCPTCGSCYPMQLQNKHKCLENAMHRQCPVCLEDMFSSLRQSQVLQCGHTIHSDCLR 346
++FHC C C M LQ +HKC+EN + CP+CLED+ +S + VL CGH +H C
Sbjct 115 DFFHCLKCNLCLAMNLQGRHKCIENVSRQNCPICLEDIHTSRVVAHVLPCGHLLHRTCYE 174
Query 347 LLQKQ 351
+ K+
Sbjct 175 EMLKE 179
> ath:AT1G74770 protein binding / zinc ion binding
Length=1259
Score = 154 bits (389), Expect = 6e-37, Method: Compositional matrix adjust.
Identities = 83/242 (34%), Positives = 127/242 (52%), Gaps = 19/242 (7%)
Query 175 DAEGNALGCAHYRRKCKVVAPCCKEIYWCRHCHNEAYEMDLSNAHEIDRHAVEEIVCAVC 234
D GC HY+R CK++APCC +++ C CH+E E D H +DR + +++C C
Sbjct 1017 DPHSLIFGCNHYKRNCKLLAPCCDKLFTCIRCHDE--EAD----HSVDRKQITKMMCMKC 1070
Query 235 EARQPVSNKC--VGCGTVFGAYFCSVCKFWDNLGEKKKVYHCDECGICRLGGRE--NYFH 290
QP+ C C + G YFC +CK +D+ ++K+YHC C +CR+G +YFH
Sbjct 1071 LLIQPIGANCSNTSCKSSMGKYFCKICKLYDD---ERKIYHCPYCNLCRVGKGLGIDYFH 1127
Query 291 CPTCGSCYPMQLQNKHKCLENAMHRQCPVCLEDMFSSLRQSQVLQCGHTIHSDCLRLLQK 350
C C +C L +H C E + CP+C E +F+S + L CGH +HS C +
Sbjct 1128 CMKCNACMSRTLV-EHVCREKCLEDNCPICHEYIFTSSSPVKALPCGHLMHSTCFQ---- 1182
Query 351 QKGFQAIRCPMCSKSIADYSEFWKQLSEEIERTPMEEQMRRKVR-IACNDCLERCTTDFH 409
+ CP+CSKS+ D ++K L + M ++ K + I CNDC + +H
Sbjct 1183 EYTCSHYTCPVCSKSLGDMQVYFKMLDALLAEEKMPDEYSNKTQVILCNDCGRKGNAPYH 1242
Query 410 FL 411
+L
Sbjct 1243 WL 1244
> ath:AT1G18910 protein binding / zinc ion binding
Length=1254
Score = 146 bits (368), Expect = 2e-34, Method: Compositional matrix adjust.
Identities = 83/242 (34%), Positives = 129/242 (53%), Gaps = 19/242 (7%)
Query 175 DAEGNALGCAHYRRKCKVVAPCCKEIYWCRHCHNEAYEMDLSNAHEIDRHAVEEIVCAVC 234
D GC HY+R CK++APCC ++Y C CH+E E+D H +DR + +++C C
Sbjct 1012 DPHKLIFGCKHYKRSCKLLAPCCNKLYTCIRCHDE--EVD----HLLDRKQITKMMCMKC 1065
Query 235 EARQPV--SNKCVGCGTVFGAYFCSVCKFWDNLGEKKKVYHCDECGICRLGG--RENYFH 290
QPV S + C + G Y+C +CK +D+ +++YHC C +CRLG +YFH
Sbjct 1066 MIIQPVGASCSNISCSSSMGKYYCKICKLFDD---DREIYHCPYCNLCRLGKGLSIDYFH 1122
Query 291 CPTCGSCYPMQLQNKHKCLENAMHRQCPVCLEDMFSSLRQSQVLQCGHTIHSDCLRLLQK 350
C C +C +L +H C E + CP+C E +F+S + L CGH +HS C +
Sbjct 1123 CMKCNACMS-RLIVEHVCREKCLEDNCPICHEYIFTSNSPVKALPCGHVMHSTCF----Q 1177
Query 351 QKGFQAIRCPMCSKSIADYSEFWKQLSEEIERTPM-EEQMRRKVRIACNDCLERCTTDFH 409
+ CP+CSKS+ D +++ L + M +E + + I CNDC + +H
Sbjct 1178 EYTCSHYTCPICSKSLGDMQVYFRMLDALLAEQKMPDEYLNQTQVILCNDCGRKGNAPYH 1237
Query 410 FL 411
+L
Sbjct 1238 WL 1239
> hsa:22954 TRIM32, BBS11, HT2A, LGMD2H, TATIP; tripartite motif
containing 32; K10607 tripartite motif-containing protein
32 [EC:6.3.2.19]
Length=653
Score = 41.2 bits (95), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 19/49 (38%), Positives = 26/49 (53%), Gaps = 1/49 (2%)
Query 316 QCPVCLEDMFSSLRQSQVLQCGHTIHSDCLRLLQKQKGFQAIRCPMCSK 364
+CP+C+E + ++L CGHTI CL L +RCP CSK
Sbjct 19 ECPICMESFTEEQLRPKLLHCGHTICRQCLEKLLASS-INGVRCPFCSK 66
> mmu:69807 Trim32, 1810045E12Rik, 3f3, BBS11, Zfp117; tripartite
motif-containing 32; K10607 tripartite motif-containing
protein 32 [EC:6.3.2.19]
Length=655
Score = 40.8 bits (94), Expect = 0.010, Method: Compositional matrix adjust.
Identities = 19/49 (38%), Positives = 26/49 (53%), Gaps = 1/49 (2%)
Query 316 QCPVCLEDMFSSLRQSQVLQCGHTIHSDCLRLLQKQKGFQAIRCPMCSK 364
+CP+C+E + ++L CGHTI CL L +RCP CSK
Sbjct 20 ECPICMESFTEEQLRPKLLHCGHTICRQCLEKLLASS-INGVRCPFCSK 67
> dre:562764 trim32; tripartite motif-containing 32; K10607 tripartite
motif-containing protein 32 [EC:6.3.2.19]
Length=663
Score = 40.8 bits (94), Expect = 0.011, Method: Compositional matrix adjust.
Identities = 19/49 (38%), Positives = 27/49 (55%), Gaps = 1/49 (2%)
Query 316 QCPVCLEDMFSSLRQSQVLQCGHTIHSDCLRLLQKQKGFQAIRCPMCSK 364
+CP+CLE + ++LQCGH++ CL L +RCP CSK
Sbjct 17 ECPICLETYNQDQLRPKLLQCGHSVCRQCLEKLLAST-INGVRCPFCSK 64
> sce:YLR427W MAG2; Cytoplasmic protein of unknown function; induced
in response to mycotoxin patulin; ubiquitinated protein
similar to the human ring finger motif protein RNF10; predicted
to be involved in repair of alkylated DNA due to interaction
with MAG1
Length=670
Score = 40.0 bits (92), Expect = 0.018, Method: Compositional matrix adjust.
Identities = 34/123 (27%), Positives = 51/123 (41%), Gaps = 25/123 (20%)
Query 231 CAVCEARQPVSNKCVGCGTVFGAYFCSVCKFWDNLGEKKKVYHCDECGICRLGGRENYFH 290
C++C + +PV+ + V CG + FC C N ++ E G + ++ Y
Sbjct 195 CSICLSEEPVAPRMVTCGHI----FCLSCLL--NFFSIEETVKNKETGYSK---KKKYKE 245
Query 291 CPTCGSCYPMQLQNKHKCLENAMHRQCPVCLEDMFSSLRQSQVLQCGHTIHSDCLRLLQK 350
CP CGS R PV ED F R +Q + G T+H L+L+ K
Sbjct 246 CPLCGSII-------------GPKRVKPVLYEDDFDVTRLNQKPEPGATVH---LQLMCK 289
Query 351 QKG 353
G
Sbjct 290 PHG 292
> cpv:cgd8_3470 membrane associated protein with a RING finger,
4xtransmembrane domain
Length=437
Score = 39.7 bits (91), Expect = 0.025, Method: Compositional matrix adjust.
Identities = 22/65 (33%), Positives = 32/65 (49%), Gaps = 9/65 (13%)
Query 302 LQNKHKCLENAMHRQCPVCLEDMFSSLRQSQVLQCGHTIHSDCLRLLQKQKGFQAIRCPM 361
L NKH L++ C +CL D FSS ++ L C H H DC+ + + CP+
Sbjct 381 LLNKHSFLQD----NCIICLND-FSSFEMARCLPCNHVFHDDCIDMWLLRNAV----CPL 431
Query 362 CSKSI 366
C S+
Sbjct 432 CQASL 436
> bbo:BBOV_IV003110 21.m03046; CHY zinc finger domain containing
protein
Length=549
Score = 38.9 bits (89), Expect = 0.045, Method: Compositional matrix adjust.
Identities = 35/135 (25%), Positives = 52/135 (38%), Gaps = 25/135 (18%)
Query 143 SILFQSDEDSDSSSSSNSSEDSPKTVADEPAADAEGNALG------------CAHYRRKC 190
++ F E +S S+ + P + +PA G C HY +
Sbjct 380 ALSFTGVEFGQDNSVSSVTSARPIRIVPKPAKPKTPGGRGLKVGTPLPDNGICKHYSKSF 439
Query 191 KVV-APCCKEIYWCRHCHNEAYEMDLSNAHEIDRHAVEEIVCAVCEARQPVSN-KCVGCG 248
+ PCC +++ C CH+E + AH IVC C +Q VSN KC CG
Sbjct 440 RWFRFPCCGKLFPCDLCHDEGTDHPYELAH--------VIVCGHCSTQQAVSNKKCQHCG 491
Query 249 TVFGAYFCSVCKFWD 263
Y S +W+
Sbjct 492 R---GYTASSSSYWE 503
> hsa:375593 TRIM73, FLJ99347, MGC45477, TRIM50B; tripartite motif
containing 73; K12024 tripartite motif-containing protein
50/73/74
Length=250
Score = 37.4 bits (85), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 24/64 (37%), Positives = 33/64 (51%), Gaps = 11/64 (17%)
Query 316 QCPVCLEDMFSSLRQSQVLQCGHTIHSDCLRLLQKQKGFQAIRCPMC------SKSIADY 369
QCP+CLE ++S +LQCGH+ CL L + +RCPMC S S+ +
Sbjct 15 QCPICLE----VFKESLMLQCGHSYCKGCLVSLSYHLDTK-VRCPMCWQVVDGSSSLPNV 69
Query 370 SEFW 373
S W
Sbjct 70 SLAW 73
> hsa:378108 TRIM74, MGC45440, TRIM50C; tripartite motif containing
74; K12024 tripartite motif-containing protein 50/73/74
Length=250
Score = 37.4 bits (85), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 24/64 (37%), Positives = 33/64 (51%), Gaps = 11/64 (17%)
Query 316 QCPVCLEDMFSSLRQSQVLQCGHTIHSDCLRLLQKQKGFQAIRCPMC------SKSIADY 369
QCP+CLE ++S +LQCGH+ CL L + +RCPMC S S+ +
Sbjct 15 QCPICLE----VFKESLMLQCGHSYCKGCLVSLSYHLDTK-VRCPMCWQVVDGSSSLPNV 69
Query 370 SEFW 373
S W
Sbjct 70 SLAW 73
> hsa:8315 BRAP, BRAP2, IMP, RNF52; BRCA1 associated protein;
K10632 BRCA1-associated protein [EC:6.3.2.19]
Length=592
Score = 37.0 bits (84), Expect = 0.14, Method: Compositional matrix adjust.
Identities = 51/226 (22%), Positives = 87/226 (38%), Gaps = 48/226 (21%)
Query 230 VCAVCEARQPVS-NKCVGCGTVFGAYFCSVCKFWDNLGEKKKVYHCDECGICRLGGRENY 288
VC C+ +PV NKC CG + C +C ++G CG R R Y
Sbjct 302 VCRYCQTPEPVEENKCFECGVQENLWICLIC---GHIG----------CG--RYVSRHAY 346
Query 289 FHCPTCGSCYPMQLQNKHKCL----ENAMHRQCPVCLEDMFSSLRQSQVLQC---GHTIH 341
H Y MQL N H+ +N +HR + +S +++Q G T
Sbjct 347 KHFEETQHTYAMQLTN-HRVWDYAGDNYVHR--------LVASKTDGKIVQYECEGDTCQ 397
Query 342 SDCLRLLQKQKGFQAIRCPMCSKSIADYSEFWKQLSEEIERTPMEEQMRRKVRIACNDCL 401
+ + LQ + + + + + +W+ IE+ EE K + + +
Sbjct 398 EEKIDALQLEYSY------LLTSQLESQRIYWENKIVRIEKDTAEEINNMKTKF--KETI 449
Query 402 ERCTTDFHFLGELIHPVNAATVRDLRSRPYCCPRLTTQTASTASTL 447
E+C H L +L+ ++ +S C +L T+ A + L
Sbjct 450 EKCDNLEHKLNDLL--------KEKQSVERKCTQLNTKVAKLTNEL 487
> mmu:78911 Trim42, 4930486B16Rik; tripartite motif-containing
42; K12018 tripartite motif-containing protein 42
Length=723
Score = 36.6 bits (83), Expect = 0.19, Method: Compositional matrix adjust.
Identities = 23/57 (40%), Positives = 31/57 (54%), Gaps = 10/57 (17%)
Query 314 HRQCPVCLEDMFSSLR-QSQVLQCGHTIHSDCLRLLQKQ----KGFQAIRCPMCSKS 365
H CP+C + LR S +L C H++ CLR LQK + F + CPMCS+S
Sbjct 143 HLTCPMC-----NRLRLHSFMLPCNHSLCEKCLRQLQKHAEVTENFFILICPMCSRS 194
> cel:C06A5.9 rnf-1; RiNg Finger protein family member (rnf-1)
Length=381
Score = 36.6 bits (83), Expect = 0.20, Method: Compositional matrix adjust.
Identities = 15/47 (31%), Positives = 25/47 (53%), Gaps = 0/47 (0%)
Query 316 QCPVCLEDMFSSLRQSQVLQCGHTIHSDCLRLLQKQKGFQAIRCPMC 362
QC VC + + + ++ L CGHT ++C+R +Q + CP C
Sbjct 21 QCQVCYQPFNETTKLARSLHCGHTFCTECIRNVQNYGNSPHLECPTC 67
> ath:AT2G47700 zinc finger (C3HC4-type RING finger) family protein
Length=358
Score = 36.6 bits (83), Expect = 0.22, Method: Compositional matrix adjust.
Identities = 19/47 (40%), Positives = 26/47 (55%), Gaps = 3/47 (6%)
Query 317 CPVCLEDMFSSLRQSQV-LQCGHTIHSDCLRLLQKQKGFQAIRCPMC 362
C +CLE + +S+ LQCGH H DC+ KG A++CP C
Sbjct 38 CSICLESVLDDGTRSKAKLQCGHQFHLDCIGSAFNMKG--AMQCPNC 82
> tpv:TP01_0655 hypothetical protein
Length=618
Score = 36.2 bits (82), Expect = 0.24, Method: Compositional matrix adjust.
Identities = 23/67 (34%), Positives = 34/67 (50%), Gaps = 7/67 (10%)
Query 302 LQNKHKCLENAMHRQCPVCLEDMFSSLRQSQVLQCGHTIHSDCLRLLQKQKGFQAIRCPM 361
L K+KC ++ + C +C D+ + S+ L+CGH H +CL K FQ CP
Sbjct 281 LYTKYKCADDEKNLNCIIC-RDVITV--NSRKLECGHVFHLNCL----KSWLFQHNNCPS 333
Query 362 CSKSIAD 368
C K I +
Sbjct 334 CRKLIYN 340
> dre:100009648 zgc:158807; K11982 E3 ubiquitin-protein ligase
RNF115/126 [EC:6.3.2.19]
Length=309
Score = 35.8 bits (81), Expect = 0.33, Method: Compositional matrix adjust.
Identities = 19/52 (36%), Positives = 29/52 (55%), Gaps = 5/52 (9%)
Query 316 QCPVCLEDMFSSLRQSQVLQCGHTIHSDCLRLLQKQKGFQAIRCPMCSKSIA 367
+CPVC ED +S+ + L C H H+DC+ +Q CP+C KS++
Sbjct 225 ECPVCKED-YSAGENVRQLPCNHLFHNDCIVPWLEQHD----TCPVCRKSLS 271
> xla:779191 znf234, MGC115682; zinc finger protein 234
Length=585
Score = 35.8 bits (81), Expect = 0.37, Method: Compositional matrix adjust.
Identities = 48/178 (26%), Positives = 63/178 (35%), Gaps = 39/178 (21%)
Query 221 IDRHAVEEIVCAVCEARQPVSNKCVGCGTVFGAYFCSVCKFWDNLGEKKKVYHCDECGIC 280
++RH + V +P S C CG F F + GEK + C+ECG C
Sbjct 401 LERHQI------VHTGEKPYS--CSVCGKQFARVFTLTVHQRTHTGEKP--FPCNECGKC 450
Query 281 -----------RLGGRENYFHCPTCGSCYPMQ--LQNKHKCLENAMHRQCPVCLEDMFSS 327
R+ E F CP CG C+ + L + K CP C + FS
Sbjct 451 FSSLAHMVSHRRIHTGEKPFSCPDCGKCFAHRSTLSSHQKIHTGQKPYTCPECGK-CFSR 509
Query 328 L-------------RQSQVLQCGHTI-HSDCLRLLQK-QKGFQAIRCPMCSKSIADYS 370
L +Q +CG H LR K G CP C K A +S
Sbjct 510 LYLLSVHRKIHITEKQYTCTECGEGFSHVQQLRQHHKIHTGENRFWCPECGKGFAHHS 567
> mmu:71861 Zswim2, 1700025P14Rik, 4933437F18Rik, MEX; zinc finger,
SWIM domain containing 2
Length=631
Score = 35.8 bits (81), Expect = 0.37, Method: Compositional matrix adjust.
Identities = 21/92 (22%), Positives = 41/92 (44%), Gaps = 8/92 (8%)
Query 317 CPVCLEDMFSSLRQSQV--LQCGHTIHSDCLRLLQKQKGF----QAIRCPMCSKSIADYS 370
CP+C E + CG+ +H C+R+L + +RCP+C + A
Sbjct 147 CPICQEVLLEKKLPVTFCRFGCGNNVHIKCMRILANYQDTGSDSSVLRCPLCREEFAPLK 206
Query 371 EFWKQL--SEEIERTPMEEQMRRKVRIACNDC 400
++ S ++ +E++ + + I CN+C
Sbjct 207 VILEEFKNSNKLITISEKERLDKHLGIPCNNC 238
> ath:AT1G21651 protein binding / zinc ion binding
Length=811
Score = 35.8 bits (81), Expect = 0.38, Method: Compositional matrix adjust.
Identities = 21/48 (43%), Positives = 26/48 (54%), Gaps = 1/48 (2%)
Query 316 QCPVCLEDMFSSLRQSQVLQCGHTIHSDCLRLLQKQKGFQAIRCPMCS 363
+CPVCL+ +VL CGHT +CL L K K IRCP C+
Sbjct 5 ECPVCLQSYDGESTVPRVLACGHTACEECLTNLPK-KFPDTIRCPACT 51
> cel:Y51F10.2 hypothetical protein
Length=305
Score = 35.8 bits (81), Expect = 0.39, Method: Compositional matrix adjust.
Identities = 19/57 (33%), Positives = 27/57 (47%), Gaps = 11/57 (19%)
Query 317 CPVCLEDMFSSLRQSQVLQCGHTIHSDCLRLLQKQ-----------KGFQAIRCPMC 362
C +CLE ++LQC HT+ C+ LL +Q + F IRCP+C
Sbjct 19 CRICLEPFDEGQHLPKILQCAHTVCERCIGLLDEQSRINHNRPPIDRSFVHIRCPVC 75
> mmu:215061 Trim50; tripartite motif-containing 50; K12024 tripartite
motif-containing protein 50/73/74
Length=483
Score = 34.7 bits (78), Expect = 0.88, Method: Compositional matrix adjust.
Identities = 19/51 (37%), Positives = 30/51 (58%), Gaps = 5/51 (9%)
Query 316 QCPVCLEDMFSSLRQSQVLQCGHTIHSDCLRLLQKQKGFQAIRCPMCSKSI 366
QCP+CLE ++ +LQCGH+ DCL L + + + CP+C +S+
Sbjct 15 QCPICLE----VFKEPLMLQCGHSYCKDCLDNLSQHLDSE-LCCPVCRQSV 60
> mmu:72399 Brap, 3010002G07Rik; BRCA1 associated protein; K10632
BRCA1-associated protein [EC:6.3.2.19]
Length=591
Score = 34.3 bits (77), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 50/226 (22%), Positives = 88/226 (38%), Gaps = 48/226 (21%)
Query 230 VCAVCEARQPVS-NKCVGCGTVFGAYFCSVCKFWDNLGEKKKVYHCDECGICRLGGRENY 288
VC C+ +PV NKC CG + C +C ++G CG R R Y
Sbjct 301 VCRYCQTPEPVEENKCFECGVQENLWICLIC---GHIG----------CG--RYVSRHAY 345
Query 289 FHCPTCGSCYPMQLQNKHKCL----ENAMHRQCPVCLEDMFSSLRQSQVLQC---GHTIH 341
H Y MQL N H+ +N +HR + +S +++Q G T
Sbjct 346 KHFEETQHTYAMQLTN-HRVWDYAGDNYVHR--------LVASKTDGKIVQYECEGDTCQ 396
Query 342 SDCLRLLQKQKGFQAIRCPMCSKSIADYSEFWKQLSEEIERTPMEEQMRRKVRIACNDCL 401
+ + LQ + + + + + +W+ IE+ EE K + + +
Sbjct 397 EEKIDALQLEYSY------LLTSQLESQRIYWENKIVRIEKDTAEEINNMKTKF--KETI 448
Query 402 ERCTTDFHFLGELIHPVNAATVRDLRSRPYCCPRLTTQTASTASTL 447
E+C + L +L+ ++ +S C +L T+ A ++ L
Sbjct 449 EKCDSLELRLSDLL--------KEKQSVERKCTQLNTRVAKLSTEL 486
> dre:100034462 si:dkey-24l11.2
Length=217
Score = 33.9 bits (76), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 30/111 (27%), Positives = 50/111 (45%), Gaps = 19/111 (17%)
Query 153 DSSSSSNSSEDSPKTVADEPAADAEGNAL----GCAHYRRKCKVV-APCCKEIYWCRHCH 207
++ + + ++ P+ D PA +G L C H+++ + + PCC Y C CH
Sbjct 77 ETKNQTGAAGQYPRRYRD-PAVQ-QGKPLPDKGACKHFKQSHRWLRFPCCGRAYPCDACH 134
Query 208 NEAYEMDLSNAHEIDRHAVEEIVCAVCEARQPVSN--KCVGCGTVF--GAY 254
+E D + E+ ++C C QP N CVGCG + GA+
Sbjct 135 DE----DQDHLMEL----ATRMICGYCAKEQPYCNGKPCVGCGGMMTRGAF 177
> sce:YHL010C ETP1; Etp1p; K10632 BRCA1-associated protein [EC:6.3.2.19]
Length=585
Score = 33.5 bits (75), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 19/49 (38%), Positives = 23/49 (46%), Gaps = 6/49 (12%)
Query 317 CPVCLEDMFSSLRQSQVLQCGHTIHSDCLRLLQKQKGFQAIRCPMCSKS 365
CPVCLE M S + C HT H CL ++ RCP+C S
Sbjct 240 CPVCLERMDSETTGLVTIPCQHTFHCQCLN------KWKNSRCPVCRHS 282
> xla:403394 IMP protein; K10632 BRCA1-associated protein [EC:6.3.2.19]
Length=585
Score = 33.5 bits (75), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 49/226 (21%), Positives = 86/226 (38%), Gaps = 48/226 (21%)
Query 230 VCAVCEARQPVS-NKCVGCGTVFGAYFCSVCKFWDNLGEKKKVYHCDECGICRLGGRENY 288
VC C+ +PV NKC CG + C +C ++G CG R R Y
Sbjct 300 VCRYCQTPEPVEENKCFECGVQENLWICLIC---GHIG----------CG--RYVSRHAY 344
Query 289 FHCPTCGSCYPMQLQNKHKCL----ENAMHRQCPVCLEDMFSSLRQSQVLQC---GHTIH 341
H Y MQL N H+ +N +HR + +S +++Q G T
Sbjct 345 KHFEETQHTYAMQLTN-HRVWDYAGDNYVHR--------LVASKTDGKIVQYECEGDTCQ 395
Query 342 SDCLRLLQKQKGFQAIRCPMCSKSIADYSEFWKQLSEEIERTPMEEQMRRKVRIACNDCL 401
+ + LQ + + + + + +W+ +E+ EE K + + +
Sbjct 396 DEKIDSLQLEYSY------LLTSQLESQRIYWENKIVRLEKDTAEEINNMKAKF--KETI 447
Query 402 ERCTTDFHFLGELIHPVNAATVRDLRSRPYCCPRLTTQTASTASTL 447
++C + H L +L A R C +L ++ A ++ L
Sbjct 448 DKCDSFEHRLNDLSKEKQAVERR--------CAQLNSKVAKLSTEL 485
> mmu:19303 Pxn, AW108311, AW123232, Pax; paxillin; K05760 paxillin
Length=557
Score = 33.5 bits (75), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 36/144 (25%), Positives = 50/144 (34%), Gaps = 39/144 (27%)
Query 199 EIYWCRHCHNEA-----YEMDLSNAHEIDRHAV-------------EEIVCAVCEARQPV 240
E + C HC E +E D E D H++ +++V A+ P
Sbjct 346 EHFVCTHCQEEIGSRNFFERDGQPYCEKDYHSLFSPRCYYCNGPILDKVVTALDRTWHPE 405
Query 241 SNKCVGCGTVFGAYFCSVCKFWDNLGEKKKVYHCDECGICRLGGRENYF--HCPTCGSCY 298
C CG FG + EK +C R++YF P CG C
Sbjct 406 HFFCAQCGAFFGP---------EGFHEKDGKAYC----------RKDYFDMFAPKCGGCA 446
Query 299 PMQLQNKHKCLENAMHRQCPVCLE 322
L+N L H +C VC E
Sbjct 447 RAILENYISALNTLWHPECFVCRE 470
> cel:C06A5.8 hypothetical protein
Length=283
Score = 33.1 bits (74), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 14/47 (29%), Positives = 26/47 (55%), Gaps = 0/47 (0%)
Query 316 QCPVCLEDMFSSLRQSQVLQCGHTIHSDCLRLLQKQKGFQAIRCPMC 362
+C VC + + + ++ L CGHT ++C++ +QK + CP C
Sbjct 37 KCQVCCTNYNETTKLARGLHCGHTFCTECIKTMQKYGNSAYLECPSC 83
> xla:397826 pxn; paxillin; K05760 paxillin
Length=548
Score = 32.3 bits (72), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 58/242 (23%), Positives = 81/242 (33%), Gaps = 64/242 (26%)
Query 126 DNGSQHWEE--AVSSPEEGSILFQSDEDSDSSSSSNSSEDSPKTVADEPAADAEG----- 178
DN S+H A S+ E L S D + S +SP + +P + +
Sbjct 239 DNSSEHQTRISASSATRELDELMASLSDFKIMAKGKSVSNSPPSNTPKPGSQLDNMLGSL 298
Query 179 ----NALGCAHYRR----KCK------VVAPCCK----EIYWCRHCHNEA-----YEMDL 215
N LG A + CK VV K E + C HC +E +E D
Sbjct 299 QSDLNKLGVATVAKGVCGACKKPIAGQVVTAMGKTWHPEHFVCTHCQDEIGSRNFFERDG 358
Query 216 SNAHEIDRH-------------AVEEIVCAVCEARQPVSNKCVGCGTVFGAYFCSVCKFW 262
E D H ++ +V A+ P C CG FG
Sbjct 359 QPYCEKDYHNLFSPRCFYCNGPILDRVVTALDRTWHPEHFFCAQCGAFFGP--------- 409
Query 263 DNLGEKKKVYHCDECGICRLGGRENYF--HCPTCGSCYPMQLQNKHKCLENAMHRQCPVC 320
+ E+ +C R++YF P CG C L+N L H +C VC
Sbjct 410 EGFHERDGKAYC----------RKDYFDMFAPKCGGCTHAILENYISALNTLWHPECFVC 459
Query 321 LE 322
E
Sbjct 460 RE 461
> cel:C36B1.9 hypothetical protein
Length=674
Score = 32.3 bits (72), Expect = 4.4, Method: Compositional matrix adjust.
Identities = 19/52 (36%), Positives = 27/52 (51%), Gaps = 4/52 (7%)
Query 317 CPVCLEDMFSSLRQSQVLQCGHTIHSDCL----RLLQKQKGFQAIRCPMCSK 364
C VC ++ SS Q++VLQCGHT + C+ + I+CP C K
Sbjct 63 CRVCYDEYHSSSNQARVLQCGHTFCTRCVVGCSSTMNNTSEEFGIKCPECRK 114
> dre:564410 zgc:172248
Length=484
Score = 32.0 bits (71), Expect = 4.8, Method: Compositional matrix adjust.
Identities = 24/87 (27%), Positives = 37/87 (42%), Gaps = 5/87 (5%)
Query 211 YEMDLSNAHEIDRHAVEEIVCAVCEARQPVSNKCVGCGTVFGAYFCSVCKFWDNLGEKKK 270
Y++D N H + +H + +S K + G FCSVC+ + + G K
Sbjct 327 YQVDYDN-HPLYKHGKTGRKQSPVRLFTNLSPKDIILPEAEGYRFCSVCERYVSAGNK-- 383
Query 271 VYHCDECGICRLGGRENYFHCPTCGSC 297
HC +C +C + HC CG C
Sbjct 384 --HCPKCDMCPSKDGREWRHCDECGRC 408
> hsa:9646 CTR9, KIAA0155, SH2BP1, TSBP, p150, p150TSP; Ctr9,
Paf1/RNA polymerase II complex component, homolog (S. cerevisiae)
Length=1173
Score = 32.0 bits (71), Expect = 5.5, Method: Compositional matrix adjust.
Identities = 28/81 (34%), Positives = 39/81 (48%), Gaps = 10/81 (12%)
Query 61 SDDPEGLTIPSGSQGSDG-----GDGGGGSHDNNSATLGPAPQSEGAVEE--EIEGEGEA 113
SD P PSGS+ SD G G +N+S P+ +S+ E + EG G+
Sbjct 1087 SDQPSRKRRPSGSEQSDNESVQSGRSHSGVSENDSRPASPSAESDHESERGSDNEGSGQG 1146
Query 114 AG---ELRGTEVEYSDNGSQH 131
+G E G+ E SD GS+H
Sbjct 1147 SGNESEPEGSNNEASDRGSEH 1167
> hsa:57567 ZNF319, MGC126816, ZFP319; zinc finger protein 319
Length=582
Score = 31.2 bits (69), Expect = 8.7, Method: Compositional matrix adjust.
Identities = 30/123 (24%), Positives = 48/123 (39%), Gaps = 29/123 (23%)
Query 286 ENYFHCPTCGSCYPMQLQN-KHKCLENAMHR--QCPVCLE--DMFSSLRQSQVLQCGHTI 340
E F CP C + + +HKCL A R +CPVC + S+L++ Q+ C
Sbjct 396 ETLFKCPVCQKGFDQSAELLRHKCLPGAAERPFKCPVCNKAYKRASALQKHQLAHCA--- 452
Query 341 HSDCLRLLQKQKGFQAIRCPMCSKSIADYSEFWKQLSEEIERTPMEEQMRRKVRIACNDC 400
+ +RC +C + SEF + + P++ C DC
Sbjct 453 -----------AAEKPLRCTLCERRFFSSSEFVQHRCDPAREKPLK----------CPDC 491
Query 401 LER 403
+R
Sbjct 492 EKR 494
> cel:F33A8.4 hypothetical protein
Length=467
Score = 31.2 bits (69), Expect = 9.0, Method: Compositional matrix adjust.
Identities = 24/79 (30%), Positives = 36/79 (45%), Gaps = 8/79 (10%)
Query 219 HEIDRHAVEEIVCAVCEARQPVSNKCVGCGTVFGAYFCSVCKFWDNLGEKKKVYHCDECG 278
H++ +H+ + IV + PV +C+ V G FC VC D ++ V HCD C
Sbjct 371 HKLYQHSEKTIVRLFTDL--PV--ECIDLKNVAGYKFCEVC---DRYVTERNV-HCDRCQ 422
Query 279 ICRLGGRENYFHCPTCGSC 297
C + + HC C C
Sbjct 423 ACTSVEQGKWNHCEKCDKC 441
Lambda K H
0.316 0.130 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 20912630864
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40