bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_0185_orf2
Length=161
Score E
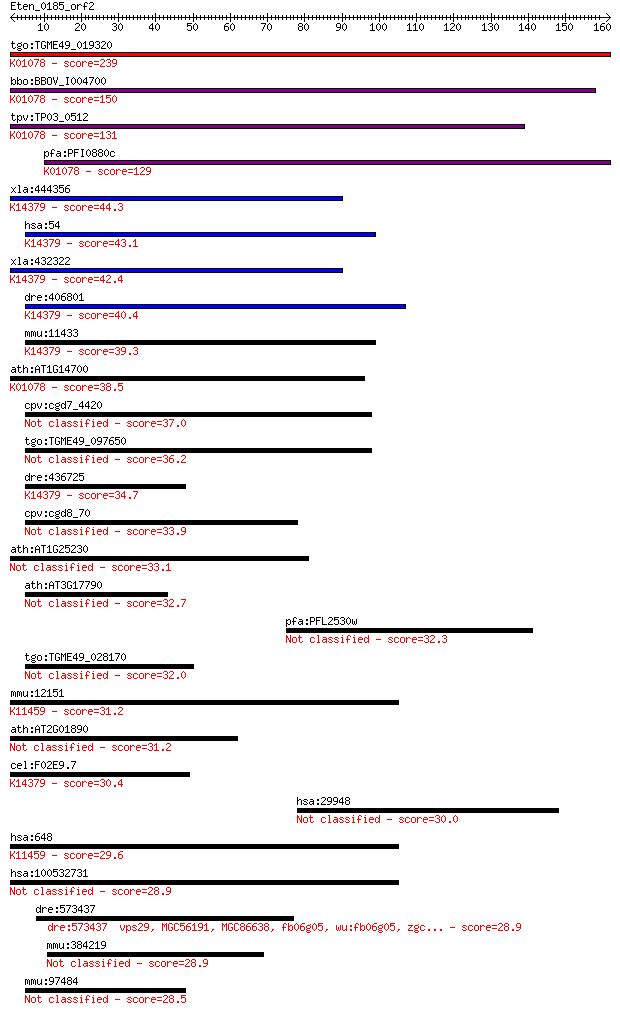
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_019320 acid phosphatase, putative (EC:3.1.3.2); K01... 239 3e-63
bbo:BBOV_I004700 19.m02068; acid phosphatase (EC:3.1.3.2); K01... 150 1e-36
tpv:TP03_0512 acid phosphatase (EC:3.1.3.2); K01078 acid phosp... 131 1e-30
pfa:PFI0880c GAP50; glideosome-associated protein 50 (EC:3.1.3... 129 3e-30
xla:444356 MGC82831 protein; K14379 tartrate-resistant acid ph... 44.3 2e-04
hsa:54 ACP5, MGC117378, TRAP; acid phosphatase 5, tartrate res... 43.1 3e-04
xla:432322 acp5, MGC78938; acid phosphatase 5, tartrate resist... 42.4 7e-04
dre:406801 acp5a, acp5, sb:cb576, wu:fb19f01, wu:fb30b03, wu:f... 40.4 0.002
mmu:11433 Acp5, TRACP, TRAP; acid phosphatase 5, tartrate resi... 39.3 0.006
ath:AT1G14700 PAP3; PAP3 (PURPLE ACID PHOSPHATASE 3); acid pho... 38.5 0.010
cpv:cgd7_4420 secreted acid phosphatase (calcineurin family),s... 37.0 0.026
tgo:TGME49_097650 serine/threonine protein phosphatase, putati... 36.2 0.046
dre:436725 acp5b, zgc:92339; acid phosphatase 5b, tartrate res... 34.7 0.14
cpv:cgd8_70 hypothetical protein 33.9 0.25
ath:AT1G25230 purple acid phosphatase family protein 33.1 0.39
ath:AT3G17790 PAP17; PAP17; acid phosphatase/ phosphatase/ pro... 32.7 0.54
pfa:PFL2530w lysophospholipase, putative 32.3 0.67
tgo:TGME49_028170 serine/threonine protein phosphatase, putati... 32.0 0.87
mmu:12151 Bmi1, AW546694, Bmi-1, Pcgf4; Bmi1 polycomb ring fin... 31.2 1.6
ath:AT2G01890 PAP8; PAP8 (PURPLE ACID PHOSPHATASE 8); acid pho... 31.2 1.6
cel:F02E9.7 hypothetical protein; K14379 tartrate-resistant ac... 30.4 2.5
hsa:29948 OSGIN1, BDGI, OKL38; oxidative stress induced growth... 30.0 3.3
hsa:648 BMI1, FLVI2/BMI1, MGC12685, PCGF4, RNF51; BMI1 polycom... 29.6 3.9
hsa:100532731 COMMD3-BMI1, DKFZp434I153; COMMD3-BMI1 readthrough 28.9 6.7
dre:573437 vps29, MGC56191, MGC86638, fb06g05, wu:fb06g05, zgc... 28.9 6.9
mmu:384219 Vmn2r11, EG384219; vomeronasal 2, receptor 11 28.9
mmu:97484 Cog8, BB235941, C87832; component of oligomeric golg... 28.5 9.2
> tgo:TGME49_019320 acid phosphatase, putative (EC:3.1.3.2); K01078
acid phosphatase [EC:3.1.3.2]
Length=431
Score = 239 bits (610), Expect = 3e-63, Method: Compositional matrix adjust.
Identities = 105/161 (65%), Positives = 136/161 (84%), Gaps = 0/161 (0%)
Query 1 LQYYLQPLLQKAQVDAYISGYDRDMELIQDEDISYINCGTGSVSGSSALVTASGSKFFSG 60
LQYYLQPLL+KA VDAYISGYD +E+I D++IS+++CG GS + S +V SGS +++G
Sbjct 271 LQYYLQPLLKKANVDAYISGYDFSLEVISDDNISHVSCGAGSKAAGSPIVKHSGSLYYAG 330
Query 61 EKGFCLFELTADGLITKFINGTNGEVMFEHKQPLKSRPERSTIDQFNYLSEMPEVVYYPV 120
E GFCLFELTA+GL+T+ ++GT GE ++ HKQPLK+RPER +ID FN++S++PEV YYPV
Sbjct 331 ETGFCLFELTAEGLVTRLVSGTTGETLYTHKQPLKNRPERKSIDAFNFVSQLPEVRYYPV 390
Query 121 PEMGKLPGKDVFVRVVGTIGLCILTFLASLSLASGISRAMK 161
PEMGK+PG+DVFVRVVGTIGLCI T SLS+A+G+SR MK
Sbjct 391 PEMGKMPGRDVFVRVVGTIGLCIATIFLSLSVANGLSRYMK 431
> bbo:BBOV_I004700 19.m02068; acid phosphatase (EC:3.1.3.2); K01078
acid phosphatase [EC:3.1.3.2]
Length=395
Score = 150 bits (380), Expect = 1e-36, Method: Compositional matrix adjust.
Identities = 74/161 (45%), Positives = 99/161 (61%), Gaps = 4/161 (2%)
Query 1 LQYYLQPLLQKAQVDAYISGYDRDMELIQDEDISYINCGTGSVSGSSALVTASGSKFFSG 60
L YYL PLL+ AQVDAYI+GYD +ME+I I+ I G G ++ + S FFS
Sbjct 234 LSYYLLPLLRDAQVDAYIAGYDHNMEVIDSNGIAMIVTGNAGTGGRKPIMKTTNSAFFSE 293
Query 61 EKGFCLFELTADGLITKFINGTNGEVMFEHKQPLKSRPERSTIDQFNYLSEMPEVVYYPV 120
+ GFC+ EL ADG+ TKFING G+VM+ HKQ +K RP+R ++ ++S +P V YP+
Sbjct 294 KAGFCIHELGADGMETKFINGETGDVMYTHKQAIKKRPQRQYGNEVQHVSALPTVSLYPI 353
Query 121 PEMGKLPGKDVFVRVVGTIGLCI----LTFLASLSLASGIS 157
EM D FV++VGTIGL I LT L+ +L S
Sbjct 354 GEMASPTQMDAFVKIVGTIGLIIAGLHLTLLSGTTLGKAAS 394
> tpv:TP03_0512 acid phosphatase (EC:3.1.3.2); K01078 acid phosphatase
[EC:3.1.3.2]
Length=404
Score = 131 bits (329), Expect = 1e-30, Method: Compositional matrix adjust.
Identities = 57/138 (41%), Positives = 91/138 (65%), Gaps = 0/138 (0%)
Query 1 LQYYLQPLLQKAQVDAYISGYDRDMELIQDEDISYINCGTGSVSGSSALVTASGSKFFSG 60
L Y L PLL++AQVDAY++GYD+DMEL+ E + + CG+ G +++ + SKF++
Sbjct 246 LSYKLLPLLKQAQVDAYVAGYDQDMELLDYEGTALVVCGSSGNKGRKSVIKSPHSKFYTE 305
Query 61 EKGFCLFELTADGLITKFINGTNGEVMFEHKQPLKSRPERSTIDQFNYLSEMPEVVYYPV 120
GFCL EL A+G TKF+NG GEV++ H QP K+R +R + ++++PEV ++P+
Sbjct 306 APGFCLHELNAEGFTTKFVNGNTGEVLYTHVQPKKNRKQRQHGSELKLINKLPEVTFHPL 365
Query 121 PEMGKLPGKDVFVRVVGT 138
++ + D F ++VGT
Sbjct 366 GDLEGVSYSDAFTKIVGT 383
> pfa:PFI0880c GAP50; glideosome-associated protein 50 (EC:3.1.3.2);
K01078 acid phosphatase [EC:3.1.3.2]
Length=396
Score = 129 bits (325), Expect = 3e-30, Method: Compositional matrix adjust.
Identities = 60/152 (39%), Positives = 98/152 (64%), Gaps = 0/152 (0%)
Query 10 QKAQVDAYISGYDRDMELIQDEDISYINCGTGSVSGSSALVTASGSKFFSGEKGFCLFEL 69
+ A+VD YISG+D +ME+I+D D+++I CG+GS+S + + S S FFS + GFC+ EL
Sbjct 245 KDAEVDLYISGHDNNMEVIEDNDMAHITCGSGSMSQGKSGMKNSKSLFFSSDIGFCVHEL 304
Query 70 TADGLITKFINGTNGEVMFEHKQPLKSRPERSTIDQFNYLSEMPEVVYYPVPEMGKLPGK 129
+ +G++TKF++ GEV++ HK +K + ++ + + +P V VP G + K
Sbjct 305 SNNGIVTKFVSSKKGEVIYTHKLNIKKKKTLDKVNALQHFAALPNVELTDVPSSGPMGNK 364
Query 130 DVFVRVVGTIGLCILTFLASLSLASGISRAMK 161
D FVRVVGTIG+ I + + + +S +S+ MK
Sbjct 365 DTFVRVVGTIGILIGSVIVFIGASSFLSKNMK 396
> xla:444356 MGC82831 protein; K14379 tartrate-resistant acid
phosphatase type 5 [EC:3.1.3.2]
Length=325
Score = 44.3 bits (103), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 29/98 (29%), Positives = 52/98 (53%), Gaps = 10/98 (10%)
Query 1 LQYYLQPLLQKAQVDAYISGYDRDMELIQDED-ISYINCGTGSVSGSSAL----VTASGS 55
L + L+PLL+K V AY+ G++ +M+ +QD+ I Y+ G G+ +S + V
Sbjct 221 LLHTLEPLLKKYGVTAYLCGHEHNMQYLQDDQGIGYLLSGAGNFMENSRIHEDDVPTDYL 280
Query 56 KFFSGEK----GFCLFELTADGLITKFINGTNGEVMFE 89
KFF G+ F E+T + ++ +NG+ +F+
Sbjct 281 KFFQGDPDTMGAFAYIEITPKEMTITYVQ-SNGKCLFQ 317
> hsa:54 ACP5, MGC117378, TRAP; acid phosphatase 5, tartrate resistant
(EC:3.1.3.2); K14379 tartrate-resistant acid phosphatase
type 5 [EC:3.1.3.2]
Length=325
Score = 43.1 bits (100), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 28/103 (27%), Positives = 53/103 (51%), Gaps = 10/103 (9%)
Query 5 LQPLLQKAQVDAYISGYDRDMELIQDED-ISYINCGTGSVSGSSAL----VTASGSKFFS 59
L+PLL V AY+ G+D +++ +QDE+ + Y+ G G+ S V +F
Sbjct 224 LRPLLATYGVTAYLCGHDHNLQYLQDENGVGYVLSGAGNFMDPSKRHQRKVPNGYLRFHY 283
Query 60 GEK----GFCLFELTADGLITKFINGTNGEVMFEHKQPLKSRP 98
G + GF E+++ + +I + G+ +F+ + P ++RP
Sbjct 284 GTEDSLGGFAYVEISSKEMTVTYIEAS-GKSLFKTRLPRRARP 325
> xla:432322 acp5, MGC78938; acid phosphatase 5, tartrate resistant
(EC:3.1.3.2); K14379 tartrate-resistant acid phosphatase
type 5 [EC:3.1.3.2]
Length=326
Score = 42.4 bits (98), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 28/98 (28%), Positives = 52/98 (53%), Gaps = 10/98 (10%)
Query 1 LQYYLQPLLQKAQVDAYISGYDRDMELIQDED-ISYINCGTGSVSGSSAL----VTASGS 55
L + ++PLL+K V AY+ G++ +M+ +QD+ I YI G G+ +S + V
Sbjct 222 LIHTVEPLLKKYGVTAYLCGHEHNMQYLQDDQGIGYILSGAGNFMENSRIHKDDVPKGYL 281
Query 56 KFFSGEK----GFCLFELTADGLITKFINGTNGEVMFE 89
+FF G+ F E+T + ++ +NG+ +F+
Sbjct 282 QFFQGDPETMGAFAYIEITPKEMTVTYVQ-SNGKCLFQ 318
> dre:406801 acp5a, acp5, sb:cb576, wu:fb19f01, wu:fb30b03, wu:fi14e01,
wu:fj66f03, zgc:63825; acid phosphatase 5a, tartrate
resistant (EC:3.1.3.2); K14379 tartrate-resistant acid phosphatase
type 5 [EC:3.1.3.2]
Length=339
Score = 40.4 bits (93), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 29/110 (26%), Positives = 49/110 (44%), Gaps = 11/110 (10%)
Query 5 LQPLLQKAQVDAYISGYDRDMELIQDEDISYINCGTGSVSGSSA----LVTASGSKFFSG 60
L+PLL+K + AY+ G+D +++ I++ I Y+ G G+ V KFF+G
Sbjct 233 LRPLLKKYKATAYLCGHDHNLQYIKESGIGYVVSGAGNFMDPDVRHRNRVPKGYLKFFNG 292
Query 61 EK----GFCLFELTADGLITKFINGTNGEVMFEHKQPLKSRPERSTIDQF 106
+ GF E+ + FI + ++ LK R + D F
Sbjct 293 DASTLGGFAHIEVDKKQMTVTFIQARGTSL---YRAVLKKRDDVLEDDNF 339
> mmu:11433 Acp5, TRACP, TRAP; acid phosphatase 5, tartrate resistant
(EC:3.1.3.2); K14379 tartrate-resistant acid phosphatase
type 5 [EC:3.1.3.2]
Length=327
Score = 39.3 bits (90), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 27/103 (26%), Positives = 50/103 (48%), Gaps = 10/103 (9%)
Query 5 LQPLLQKAQVDAYISGYDRDMELIQDED-ISYINCGTGSVSGSSAL----VTASGSKFFS 59
L+PLL V AY+ G+D +++ +QDE+ + Y+ G G+ S V +F
Sbjct 226 LRPLLATYGVTAYLCGHDHNLQYLQDENGVGYVLSGAGNFMDPSVRHQRKVPNGYLRFHY 285
Query 60 GEK----GFCLFELTADGLITKFINGTNGEVMFEHKQPLKSRP 98
G + GF E++ + ++ + G+ +F+ P + RP
Sbjct 286 GSEDSLGGFTHVEISPKEMTIIYVEAS-GKSLFKTSLPRRPRP 327
> ath:AT1G14700 PAP3; PAP3 (PURPLE ACID PHOSPHATASE 3); acid phosphatase/
protein serine/threonine phosphatase; K01078 acid
phosphatase [EC:3.1.3.2]
Length=364
Score = 38.5 bits (88), Expect = 0.010, Method: Compositional matrix adjust.
Identities = 28/100 (28%), Positives = 48/100 (48%), Gaps = 6/100 (6%)
Query 1 LQYYLQPLLQKAQVDAYISGYDRDMELIQ--DEDISYINCGTGSVS---GSSALVTASGS 55
L+ +L P+LQ +VD Y++G+D +E I D +I ++ G GS + G V
Sbjct 259 LEKHLLPILQANEVDLYVNGHDHCLEHISSVDSNIQFMTSGGGSKAWKGGDVNYVEPEEM 318
Query 56 KFFSGEKGFCLFELTADGLITKFINGTNGEVMFEHKQPLK 95
+F+ +GF ++ L F + G V+ K+ K
Sbjct 319 RFYYDGQGFMSVHVSEAELRVVFYD-VFGHVLHHWKKTYK 357
> cpv:cgd7_4420 secreted acid phosphatase (calcineurin family),signal
peptide
Length=826
Score = 37.0 bits (84), Expect = 0.026, Method: Compositional matrix adjust.
Identities = 27/95 (28%), Positives = 43/95 (45%), Gaps = 3/95 (3%)
Query 5 LQPLLQKAQVDAYISGYDRDMELIQDED--ISYINCGTGSVSGSSALVTASGSKFFSGEK 62
L PLL+K +VDAYI+G+D +EL + + S+ G+ F +G
Sbjct 238 LDPLLKKYKVDAYIAGHDHHLELSRPKGSCTSHFLIGSACCPKKHDYFNNKHRIFRTGRG 297
Query 63 GFCLFELTADGLITKFINGTNGEVMFEHKQPLKSR 97
GF +LT + + N G+ +F Q +R
Sbjct 298 GFASHKLTYSQFHSTYHN-IEGKPIFTTTQKRLNR 331
> tgo:TGME49_097650 serine/threonine protein phosphatase, putative
(EC:3.1.3.2)
Length=679
Score = 36.2 bits (82), Expect = 0.046, Method: Composition-based stats.
Identities = 25/95 (26%), Positives = 47/95 (49%), Gaps = 3/95 (3%)
Query 5 LQPLLQKAQVDAYISGYDRDMELIQ--DEDISYINCGTGSVSGSSALVTASGSKFFSGEK 62
L L++K +VD ++SG++ + Q D + ++I GTGS + + + F E
Sbjct 416 LTELMRKYKVDTFLSGHEHALSFFQEPDANTTHIISGTGSKLSARDPIPSKDCLFSVREH 475
Query 63 GFCLFELTADGLITKFINGTNGEVMFEHKQPLKSR 97
G + L + L F++ +G+V+F Q + R
Sbjct 476 GVAVHVLGKEELHHGFVSA-DGKVLFTASQRRQDR 509
> dre:436725 acp5b, zgc:92339; acid phosphatase 5b, tartrate resistant;
K14379 tartrate-resistant acid phosphatase type 5
[EC:3.1.3.2]
Length=327
Score = 34.7 bits (78), Expect = 0.14, Method: Compositional matrix adjust.
Identities = 17/44 (38%), Positives = 28/44 (63%), Gaps = 1/44 (2%)
Query 5 LQPLLQKAQVDAYISGYDRDMELIQDED-ISYINCGTGSVSGSS 47
L+PLL+K V Y+SG+D ++ I+++D S++ G G SS
Sbjct 225 LRPLLKKYNVSLYLSGHDHSLQFIREDDGSSFVVSGAGVEEDSS 268
> cpv:cgd8_70 hypothetical protein
Length=424
Score = 33.9 bits (76), Expect = 0.25, Method: Compositional matrix adjust.
Identities = 25/75 (33%), Positives = 36/75 (48%), Gaps = 2/75 (2%)
Query 5 LQPLLQKAQVDAYISGYDRDMELIQDEDI-SYIN-CGTGSVSGSSALVTASGSKFFSGEK 62
L PL++K +VD ISG+D E++ ED SY G S +S T S F S
Sbjct 294 LLPLIKKYRVDFIISGHDHHSEILVPEDFNSYFQIVGASSKPRTSFGATDENSIFKSNTC 353
Query 63 GFCLFELTADGLITK 77
F F + D +++
Sbjct 354 SFASFTFSKDIAVSR 368
> ath:AT1G25230 purple acid phosphatase family protein
Length=339
Score = 33.1 bits (74), Expect = 0.39, Method: Compositional matrix adjust.
Identities = 24/84 (28%), Positives = 40/84 (47%), Gaps = 4/84 (4%)
Query 1 LQYYLQPLLQKAQVDAYISGYDRDMELIQDED--ISYINCGTGSVS--GSSALVTASGSK 56
L+ L P+L+ +VD Y++G+D ++ I I ++ G GS + G T K
Sbjct 233 LESLLLPILEANKVDLYMNGHDHCLQHISTSQSPIQFLTSGGGSKAWRGYYNWTTPEDMK 292
Query 57 FFSGEKGFCLFELTADGLITKFIN 80
FF +GF ++T L F +
Sbjct 293 FFYDGQGFMSVKITRSELSVVFYD 316
> ath:AT3G17790 PAP17; PAP17; acid phosphatase/ phosphatase/ protein
serine/threonine phosphatase
Length=338
Score = 32.7 bits (73), Expect = 0.54, Method: Compositional matrix adjust.
Identities = 15/40 (37%), Positives = 26/40 (65%), Gaps = 2/40 (5%)
Query 5 LQPLLQKAQVDAYISGYDRDMELIQDED--ISYINCGTGS 42
L P+L++ VD Y++G+D ++ + DED I ++ G GS
Sbjct 237 LLPILKENGVDLYMNGHDHCLQHMSDEDSPIQFLTSGAGS 276
> pfa:PFL2530w lysophospholipase, putative
Length=453
Score = 32.3 bits (72), Expect = 0.67, Method: Compositional matrix adjust.
Identities = 21/67 (31%), Positives = 36/67 (53%), Gaps = 5/67 (7%)
Query 75 ITKFINGTNGEVMFEHKQPLKSRPERSTID-QFNYLSEMPEVVYYPVPEMGKLPGKDVFV 133
+ ++IN N V+ E + P E S D +N+ ++MP +V P+ MG G ++ +
Sbjct 132 VIQYINKINSSVIKEREDP----KEYSYSDYNYNFKNKMPNIVRSPLYIMGLSMGGNIAL 187
Query 134 RVVGTIG 140
RV+ IG
Sbjct 188 RVLELIG 194
> tgo:TGME49_028170 serine/threonine protein phosphatase, putative
(EC:3.1.3.2 3.2.1.3)
Length=1491
Score = 32.0 bits (71), Expect = 0.87, Method: Composition-based stats.
Identities = 17/45 (37%), Positives = 26/45 (57%), Gaps = 1/45 (2%)
Query 5 LQPLLQKAQVDAYISGYDRDMELIQDEDISYINCGTGSVSGSSAL 49
+Q LL QVD Y+S +D ME + ED+S N T ++ +A+
Sbjct 623 IQMLLSHYQVDLYVSAHDHFMEYVALEDLSK-NTTTAFITSGAAV 666
> mmu:12151 Bmi1, AW546694, Bmi-1, Pcgf4; Bmi1 polycomb ring finger
oncogene; K11459 polycomb group RING finger protein 4
Length=324
Score = 31.2 bits (69), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 22/104 (21%), Positives = 42/104 (40%), Gaps = 8/104 (7%)
Query 1 LQYYLQPLLQKAQVDAYISGYDRDMELIQDEDISYINCGTGSVSGSSALVTASGSKFFSG 60
L+Y ++P ++ ++ G EL D N G V +S+ + + + S
Sbjct 221 LKYRVRPTCKRMKMSHQRDGLTNAGELESDSGSDKANSPAGGVPSTSSCLPSPSTPVQSP 280
Query 61 EKGFCLFELTADGLITKFINGTNGEVMFEHKQPLKSRPERSTID 104
F I+ +NGT+ H+ SRP +S+++
Sbjct 281 HPQF--------PHISSTMNGTSNSPSANHQSSFASRPRKSSLN 316
> ath:AT2G01890 PAP8; PAP8 (PURPLE ACID PHOSPHATASE 8); acid phosphatase/
protein serine/threonine phosphatase
Length=335
Score = 31.2 bits (69), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 21/61 (34%), Positives = 32/61 (52%), Gaps = 11/61 (18%)
Query 1 LQYYLQPLLQKAQVDAYISGYDRDMELIQDEDISYINCGTGSVSGSSALVTASGSKFFSG 60
L+ L P+L+ +VD YI+G+D + E IS IN SG + + GSK + G
Sbjct 232 LEKQLLPILEANEVDLYINGHDHCL-----EHISSIN------SGIQFMTSGGGSKAWKG 280
Query 61 E 61
+
Sbjct 281 D 281
> cel:F02E9.7 hypothetical protein; K14379 tartrate-resistant
acid phosphatase type 5 [EC:3.1.3.2]
Length=419
Score = 30.4 bits (67), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 16/53 (30%), Positives = 28/53 (52%), Gaps = 5/53 (9%)
Query 1 LQYYLQPLLQKAQVDAYISGYDRDMELIQ-----DEDISYINCGTGSVSGSSA 48
L+ L PLL++ V+AY SG+D ++ + I+Y+ G S + +S
Sbjct 281 LRQRLDPLLKRFNVNAYFSGHDHSLQHFTFPGYGEHIINYVVSGAASRADAST 333
> hsa:29948 OSGIN1, BDGI, OKL38; oxidative stress induced growth
inhibitor 1
Length=560
Score = 30.0 bits (66), Expect = 3.3, Method: Composition-based stats.
Identities = 22/70 (31%), Positives = 31/70 (44%), Gaps = 5/70 (7%)
Query 78 FINGTNGEVMFEHKQPLKSRPERSTIDQFNYLSEMPEVVYYPVPEMGKLPGKDVFVRVVG 137
F+ G + + QPL ++ +D F Y S E +Y MG L G D FVR V
Sbjct 487 FLPGAGADFAVDPDQPLSAKRNPIDVDPFTYQSTRQEGLY----AMGPLAG-DNFVRFVQ 541
Query 138 TIGLCILTFL 147
L + + L
Sbjct 542 GGALAVASSL 551
> hsa:648 BMI1, FLVI2/BMI1, MGC12685, PCGF4, RNF51; BMI1 polycomb
ring finger oncogene; K11459 polycomb group RING finger
protein 4
Length=326
Score = 29.6 bits (65), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 20/104 (19%), Positives = 42/104 (40%), Gaps = 8/104 (7%)
Query 1 LQYYLQPLLQKAQVDAYISGYDRDMELIQDEDISYINCGTGSVSGSSALVTASGSKFFSG 60
L+Y ++P ++ ++ G EL D N G + +S+ + + + S
Sbjct 223 LKYRVRPTCKRMKISHQRDGLTNAGELESDSGSDKANSPAGGIPSTSSCLPSPSTPVQSP 282
Query 61 EKGFCLFELTADGLITKFINGTNGEVMFEHKQPLKSRPERSTID 104
F I+ +NGT+ H+ +RP +S+++
Sbjct 283 HPQF--------PHISSTMNGTSNSPSGNHQSSFANRPRKSSVN 318
> hsa:100532731 COMMD3-BMI1, DKFZp434I153; COMMD3-BMI1 readthrough
Length=469
Score = 28.9 bits (63), Expect = 6.7, Method: Compositional matrix adjust.
Identities = 20/104 (19%), Positives = 42/104 (40%), Gaps = 8/104 (7%)
Query 1 LQYYLQPLLQKAQVDAYISGYDRDMELIQDEDISYINCGTGSVSGSSALVTASGSKFFSG 60
L+Y ++P ++ ++ G EL D N G + +S+ + + + S
Sbjct 366 LKYRVRPTCKRMKISHQRDGLTNAGELESDSGSDKANSPAGGIPSTSSCLPSPSTPVQSP 425
Query 61 EKGFCLFELTADGLITKFINGTNGEVMFEHKQPLKSRPERSTID 104
F I+ +NGT+ H+ +RP +S+++
Sbjct 426 HPQF--------PHISSTMNGTSNSPSGNHQSSFANRPRKSSVN 461
> dre:573437 vps29, MGC56191, MGC86638, fb06g05, wu:fb06g05, zgc:56191,
zgc:86638; vacuolar protein sorting 29 (yeast) (EC:3.1.3.3);
K07095
Length=182
Score = 28.9 bits (63), Expect = 6.9, Method: Compositional matrix adjust.
Identities = 23/72 (31%), Positives = 35/72 (48%), Gaps = 13/72 (18%)
Query 8 LLQKA-QVDAYISGYDRDMELIQDEDISYINCG--TGSVSGSSALVTASGSKFFSGEKGF 64
LLQ+ VD ISG+ E ++E+ YIN G TG+ S + +T S F
Sbjct 101 LLQRQLDVDILISGHTHKFEAFENENKFYINPGSATGAYSALESNITPS----------F 150
Query 65 CLFELTADGLIT 76
L ++ A ++T
Sbjct 151 VLMDIQASTVVT 162
> mmu:384219 Vmn2r11, EG384219; vomeronasal 2, receptor 11
Length=861
Score = 28.9 bits (63), Expect = 7.8, Method: Compositional matrix adjust.
Identities = 14/59 (23%), Positives = 27/59 (45%), Gaps = 1/59 (1%)
Query 11 KAQVDAYISGYDRDMELIQDEDISYINCGTGSVSGSSALVTASG-SKFFSGEKGFCLFE 68
K ++ +Y+ Y R +L ED+ ++ G+ S ++ +G K E C F+
Sbjct 488 KVKIGSYLHCYPRSQQLHISEDLEWVTGGSSVPSSMCSMTCTAGFRKIHQNETADCCFD 546
> mmu:97484 Cog8, BB235941, C87832; component of oligomeric golgi
complex 8
Length=640
Score = 28.5 bits (62), Expect = 9.2, Method: Composition-based stats.
Identities = 13/43 (30%), Positives = 21/43 (48%), Gaps = 0/43 (0%)
Query 5 LQPLLQKAQVDAYISGYDRDMELIQDEDISYINCGTGSVSGSS 47
L P+ Q+ + + + +E QDE SY T ++ GSS
Sbjct 373 LAPVFQRVAISTFQKAVEEAVEKFQDEMTSYTLISTAAILGSS 415
Lambda K H
0.319 0.138 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3702936828
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40