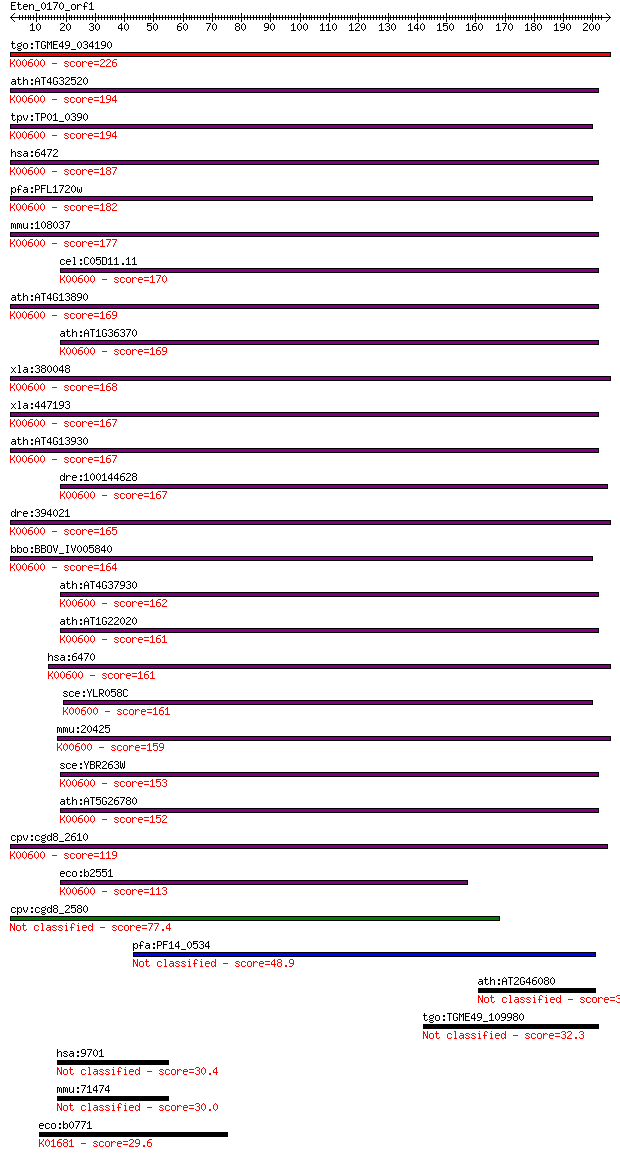
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_0170_orf1
Length=205
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_034190 glycine hydroxymethyltransferase, putative (... 226 3e-59
ath:AT4G32520 SHM3; SHM3 (SERINE HYDROXYMETHYLTRANSFERASE 3); ... 194 2e-49
tpv:TP01_0390 serine hydroxymethyltransferase; K00600 glycine ... 194 2e-49
hsa:6472 SHMT2, GLYA, SHMT; serine hydroxymethyltransferase 2 ... 187 3e-47
pfa:PFL1720w serine hydroxymethyltransferase (EC:2.1.2.1); K00... 182 9e-46
mmu:108037 Shmt2, 2700043D08Rik, AA408223, AA986903; serine hy... 177 2e-44
cel:C05D11.11 mel-32; Maternal Effect Lethal family member (me... 170 3e-42
ath:AT4G13890 EDA36; EDA36 (EMBRYO SAC DEVELOPMENT ARREST 37);... 169 4e-42
ath:AT1G36370 SHM7; SHM7 (serine hydroxymethyltransferase 7); ... 169 7e-42
xla:380048 shmt1, MGC53442; serine hydroxymethyltransferase 1 ... 168 2e-41
xla:447193 shmt2, MGC79128; serine hydroxymethyltransferase 2 ... 167 2e-41
ath:AT4G13930 SHM4; SHM4 (serine hydroxymethyltransferase 4); ... 167 2e-41
dre:100144628 shmt2; serine hydroxymethyltransferase 2 (mitoch... 167 3e-41
dre:394021 shmt1, MGC66171, zgc:66171, zgc:77524; serine hydro... 165 1e-40
bbo:BBOV_IV005840 23.m06426; serine hydroxymethyltransferase (... 164 1e-40
ath:AT4G37930 SHM1; SHM1 (SERINE TRANSHYDROXYMETHYLTRANSFERASE... 162 8e-40
ath:AT1G22020 SHM6; SHM6 (serine hydroxymethyltransferase 6); ... 161 1e-39
hsa:6470 SHMT1, CSHMT, MGC15229, MGC24556, SHMT; serine hydrox... 161 2e-39
sce:YLR058C SHM2, SHMT2; Cytosolic serine hydroxymethyltransfe... 161 2e-39
mmu:20425 Shmt1, AI324848, AI385541, C81125, Shmt, mshmt, mshm... 159 5e-39
sce:YBR263W SHM1, SHMT1, TMP3; Mitochondrial serine hydroxymet... 153 3e-37
ath:AT5G26780 SHM2; SHM2 (SERINE HYDROXYMETHYLTRANSFERASE 2); ... 152 9e-37
cpv:cgd8_2610 cytosolic serine hydroxymethyl transferase ; K00... 119 6e-27
eco:b2551 glyA, ECK2548, JW2535; serine hydroxymethyltransfera... 113 4e-25
cpv:cgd8_2580 mitochondrial serine hydroxymethyl transferase 77.4 3e-14
pfa:PF14_0534 serine hydroxymethyltransferase, putative (EC:2.... 48.9 1e-05
ath:AT2G46080 hypothetical protein 32.3 1.3
tgo:TGME49_109980 dynein-1-alpha heavy chain, flagellar inner ... 32.3 1.3
hsa:9701 PPP6R2, KIAA0685, PP6R2, SAP190, SAPS2, dJ579N16.1; p... 30.4 4.2
mmu:71474 Ppp6r2, 1110033O10Rik, 8430411H09Rik, B230107H12Rik,... 30.0 5.2
eco:b0771 ybhJ, ECK0760, JW5103; predicted hydratase; K01681 a... 29.6 8.5
> tgo:TGME49_034190 glycine hydroxymethyltransferase, putative
(EC:2.1.2.1); K00600 glycine hydroxymethyltransferase [EC:2.1.2.1]
Length=595
Score = 226 bits (577), Expect = 3e-59, Method: Compositional matrix adjust.
Identities = 110/206 (53%), Positives = 148/206 (71%), Gaps = 2/206 (0%)
Query 1 RSGIIFVNTRRVPNGASRIDSAVFPGLQGGPHENHIAAVAHQLKQTCTPEWRAFSRCVLE 60
RSG+IF+N RRVP+G I+S VFP LQGGPH + IAA+A QLK+ +P W ++ V+
Sbjct 391 RSGMIFINKRRVPDGEGLINSGVFPSLQGGPHNHQIAALACQLKEVMSPSWATYASQVIR 450
Query 61 NCKVLARELQNHGLQLVTGGTDNHLLLLDLKPQGLTGAKLQLICDMANITLNKNSIAGDS 120
N LA LQ+HG +L T GTDNHLLL+DL+P G+TG K+QL CD A+ITLNKN++ GD+
Sbjct 451 NSNALAARLQHHGHRLTTDGTDNHLLLMDLRPDGITGTKMQLCCDEASITLNKNTVPGDT 510
Query 121 SGI-PTGVRIGTPAMTTRGFGAEEFKLVADFIREAVDICCEIQEKSGKKLAEFRKHVSPS 179
S P+GVRIG+PA+TTRGF ++F+ +AD++ E V I EIQ GKKL +F+K V P
Sbjct 511 SAANPSGVRIGSPALTTRGFKEKDFEQIADWLHEIVLIAQEIQTNYGKKLVDFKKGV-PG 569
Query 180 HPRISNLRDRVKALARSFPMPGRPDI 205
+PR+ ++ + A SF MPG+ DI
Sbjct 570 NPRLLEIKQAITDWACSFSMPGQADI 595
> ath:AT4G32520 SHM3; SHM3 (SERINE HYDROXYMETHYLTRANSFERASE 3);
catalytic/ glycine hydroxymethyltransferase/ pyridoxal phosphate
binding (EC:2.1.2.1); K00600 glycine hydroxymethyltransferase
[EC:2.1.2.1]
Length=529
Score = 194 bits (494), Expect = 2e-49, Method: Compositional matrix adjust.
Identities = 102/211 (48%), Positives = 146/211 (69%), Gaps = 12/211 (5%)
Query 1 RSGIIFVNTRRVP-NGA---SRIDSAVFPGLQGGPHENHIAAVAHQLKQTCTPEWRAFSR 56
R G+IF R+ P NG S +++AVFPGLQGGPH + I +A LK +PE++A+ +
Sbjct 320 RGGMIFF--RKDPINGVDLESAVNNAVFPGLQGGPHNHTIGGLAVCLKHAQSPEFKAYQK 377
Query 57 CVLENCKVLARELQNHGLQLVTGGTDNHLLLLDLKPQGLTGAKLQLICDMANITLNKNSI 116
V+ NC+ LA L G +LV+GG+DNHL+L+DL+P G+ GA+++ I DMA+ITLNKNS+
Sbjct 378 RVVSNCRALANRLVELGFKLVSGGSDNHLVLVDLRPMGMDGARVEKILDMASITLNKNSV 437
Query 117 AGDSSG-IPTGVRIGTPAMTTRGFGAEEFKLVADFIREAVDICCEIQEKS-GKKLAEFRK 174
GD S +P G+RIG+PAMTTRG ++F +VADFI+E V+I E ++ + G KL +F K
Sbjct 438 PGDKSALVPGGIRIGSPAMTTRGLSEKDFVVVADFIKEGVEITMEAKKAAPGSKLQDFNK 497
Query 175 HV-SPSHP---RISNLRDRVKALARSFPMPG 201
V SP P R+ +L++RV+ FP+PG
Sbjct 498 FVTSPEFPLKERVKSLKERVETFTSRFPIPG 528
> tpv:TP01_0390 serine hydroxymethyltransferase; K00600 glycine
hydroxymethyltransferase [EC:2.1.2.1]
Length=503
Score = 194 bits (492), Expect = 2e-49, Method: Compositional matrix adjust.
Identities = 90/200 (45%), Positives = 140/200 (70%), Gaps = 2/200 (1%)
Query 1 RSGIIFVNTRRVPNGASRIDSAVFPGLQGGPHENHIAAVAHQLKQTCTPEWRAFSRCVLE 60
RSGIIF N + +P+ I+ +VFP LQGGPH N+IAA+A QLKQ PEW+ +++ +++
Sbjct 303 RSGIIFFNKKLLPDFGECINQSVFPTLQGGPHNNNIAALAVQLKQLSKPEWKTYAQRIVD 362
Query 61 NCKVLARELQNHGLQLVTGGTDNHLLLLDLKPQGLTGAKLQLICDMANITLNKNSIAGDS 120
N +VLA EL+ + +VTGGTDNH +++ L+P G+TG+K +L+CD+ NI+++K++I GD
Sbjct 363 NARVLAAELEKRDMPVVTGGTDNHTVIVSLRPFGVTGSKAELVCDLVNISISKSTIPGDK 422
Query 121 SGI-PTGVRIGTPAMTTRGFGAEEFKLVADFIREAVDICCEIQEKSGKKLAEFRKHVSPS 179
S P+G+R+GTP++T+RG ++ VAD IR+ VDIC ++QE+ GKKL +F+ + +
Sbjct 423 SAFNPSGIRLGTPSLTSRGAFPQDMVFVADVIRKVVDICVKVQEEKGKKLVDFKVGLDVN 482
Query 180 HPRISNLRDRVKALARSFPM 199
I L++ V FP
Sbjct 483 E-DIIKLKNEVVEWISKFPY 501
> hsa:6472 SHMT2, GLYA, SHMT; serine hydroxymethyltransferase
2 (mitochondrial) (EC:2.1.2.1); K00600 glycine hydroxymethyltransferase
[EC:2.1.2.1]
Length=494
Score = 187 bits (474), Expect = 3e-47, Method: Compositional matrix adjust.
Identities = 100/216 (46%), Positives = 140/216 (64%), Gaps = 16/216 (7%)
Query 1 RSGIIFVNT----------RRVPNG-ASRIDSAVFPGLQGGPHENHIAAVAHQLKQTCTP 49
RSG+IF R +P RI+ AVFP LQGGPH + IAAVA LKQ CTP
Sbjct 276 RSGLIFYRKGVKAVDPKTGREIPYTFEDRINFAVFPSLQGGPHNHAIAAVAVALKQACTP 335
Query 50 EWRAFSRCVLENCKVLARELQNHGLQLVTGGTDNHLLLLDLKPQGLTGAKLQLICDMANI 109
+R +S VL+N + +A L G LV+GGTDNHL+L+DL+P+GL GA+ + + ++ +I
Sbjct 336 MFREYSLQVLKNARAMADALLERGYSLVSGGTDNHLVLVDLRPKGLDGARAERVLELVSI 395
Query 110 TLNKNSIAGDSSGI-PTGVRIGTPAMTTRGFGAEEFKLVADFIREAVDICCEIQEKSGKK 168
T NKN+ GD S I P G+R+G PA+T+R F ++F+ V DFI E V+I E++ K+ K
Sbjct 396 TANKNTCPGDRSAITPGGLRLGAPALTSRQFREDDFRRVVDFIDEGVNIGLEVKSKTA-K 454
Query 169 LAEFRKHV---SPSHPRISNLRDRVKALARSFPMPG 201
L +F+ + S + R++NLR RV+ AR+FPMPG
Sbjct 455 LQDFKSFLLKDSETSQRLANLRQRVEQFARAFPMPG 490
> pfa:PFL1720w serine hydroxymethyltransferase (EC:2.1.2.1); K00600
glycine hydroxymethyltransferase [EC:2.1.2.1]
Length=442
Score = 182 bits (461), Expect = 9e-46, Method: Compositional matrix adjust.
Identities = 91/200 (45%), Positives = 133/200 (66%), Gaps = 2/200 (1%)
Query 1 RSGIIFVNTRRVPNGASRIDSAVFPGLQGGPHENHIAAVAHQLKQTCTPEWRAFSRCVLE 60
RS +IF N +R P +I+S+VFP QGGPH N IAAVA QLK+ TP ++ +++ VL
Sbjct 243 RSALIFFNKKRNPGIDQKINSSVFPSFQGGPHNNKIAAVACQLKEVNTPFFKEYTKQVLL 302
Query 61 NCKVLARELQNHGLQLVTGGTDNHLLLLDLKPQGLTGAKLQLICDMANITLNKNSIAGDS 120
N K LA L L LVT GTDNHL+++DL+ +TG+KLQ C+ NI LNKN+I D
Sbjct 303 NSKALAECLLKRNLDLVTNGTDNHLIVVDLRKYNITGSKLQETCNAINIALNKNTIPSDV 362
Query 121 SGI-PTGVRIGTPAMTTRGFGAEEFKLVADFIREAVDICCEIQEKSGKKLAEFRKHVSPS 179
+ P+G+RIGTPA+TTRG ++ + +AD + +A+ + E+Q+K GKKL +F+K + +
Sbjct 363 DCVSPSGIRIGTPALTTRGCKEKDMEFIADMLLKAILLTDELQQKYGKKLVDFKKGLV-N 421
Query 180 HPRISNLRDRVKALARSFPM 199
+P+I L+ V A++ P
Sbjct 422 NPKIDELKKEVVQWAKNLPF 441
> mmu:108037 Shmt2, 2700043D08Rik, AA408223, AA986903; serine
hydroxymethyltransferase 2 (mitochondrial) (EC:2.1.2.1); K00600
glycine hydroxymethyltransferase [EC:2.1.2.1]
Length=504
Score = 177 bits (450), Expect = 2e-44, Method: Compositional matrix adjust.
Identities = 96/216 (44%), Positives = 136/216 (62%), Gaps = 16/216 (7%)
Query 1 RSGIIFVNT----------RRVPNG-ASRIDSAVFPGLQGGPHENHIAAVAHQLKQTCTP 49
RSG+IF + +P RI+ AVFP LQGGPH + IAAVA LKQ CTP
Sbjct 286 RSGLIFYRKGVRTVDPKTGKEIPYTFEDRINFAVFPSLQGGPHNHAIAAVAVALKQACTP 345
Query 50 EWRAFSRCVLENCKVLARELQNHGLQLVTGGTDNHLLLLDLKPQGLTGAKLQLICDMANI 109
+R +S VL N + +A L G LV+GGTD HL+L+DL+P+GL GA+ + + ++ +I
Sbjct 346 MFREYSLQVLRNAQAMADALLKRGYSLVSGGTDTHLVLVDLRPKGLDGARAERVLELVSI 405
Query 110 TLNKNSIAGDSSGI-PTGVRIGTPAMTTRGFGAEEFKLVADFIREAVDICCEIQEKSGKK 168
T NKN+ GD S I P G+R+G PA+T+R F ++F+ V DFI E V+I E++ K+ K
Sbjct 406 TANKNTCPGDRSAITPGGLRLGAPALTSRQFREDDFRRVVDFIDEGVNIGLEVKRKTA-K 464
Query 169 LAEFRKHV---SPSHPRISNLRDRVKALARSFPMPG 201
L +F+ + + R++NLR +V+ AR FPMPG
Sbjct 465 LQDFKSFLLKDPETSQRLANLRQQVEQFARGFPMPG 500
> cel:C05D11.11 mel-32; Maternal Effect Lethal family member (mel-32);
K00600 glycine hydroxymethyltransferase [EC:2.1.2.1]
Length=507
Score = 170 bits (431), Expect = 3e-42, Method: Compositional matrix adjust.
Identities = 82/188 (43%), Positives = 123/188 (65%), Gaps = 4/188 (2%)
Query 18 RIDSAVFPGLQGGPHENHIAAVAHQLKQTCTPEWRAFSRCVLENCKVLARELQNHGLQLV 77
+I+SAVFPGLQGGPH + IA +A L+Q + ++ + VL+N K LA ++ HG L
Sbjct 316 KINSAVFPGLQGGPHNHTIAGIAVALRQCLSEDFVQYGEQVLKNAKTLAERMKKHGYALA 375
Query 78 TGGTDNHLLLLDLKPQGLTGAKLQLICDMANITLNKNSIAGDSSGI-PTGVRIGTPAMTT 136
TGGTDNHLLL+DL+P G+ GA+ + + D+A+I NKN+ GD S + P G+R+GTPA+T+
Sbjct 376 TGGTDNHLLLVDLRPIGVEGARAEHVLDLAHIACNKNTCPGDVSALRPGGIRLGTPALTS 435
Query 137 RGFGAEEFKLVADFIREAVDICCEIQEKSGKKLAEFRKHVSPSHP---RISNLRDRVKAL 193
RGF ++F+ V DFI E V I + ++GK L +F+ + P +++L RV+
Sbjct 436 RGFQEQDFEKVGDFIHEGVQIAKKYNAEAGKTLKDFKSFTETNEPFKKDVADLAKRVEEF 495
Query 194 ARSFPMPG 201
+ F +PG
Sbjct 496 STKFEIPG 503
> ath:AT4G13890 EDA36; EDA36 (EMBRYO SAC DEVELOPMENT ARREST 37);
catalytic/ glycine hydroxymethyltransferase/ pyridoxal phosphate
binding (EC:2.1.2.1); K00600 glycine hydroxymethyltransferase
[EC:2.1.2.1]
Length=470
Score = 169 bits (429), Expect = 4e-42, Method: Compositional matrix adjust.
Identities = 91/215 (42%), Positives = 132/215 (61%), Gaps = 17/215 (7%)
Query 1 RSGIIFVNTRRVPNGA-------------SRIDSAVFPGLQGGPHENHIAAVAHQLKQTC 47
R+G+IF R+ P A ++I+SAVFP LQ GPH N I A+A LKQ
Sbjct 250 RAGMIFY--RKGPKPAKKGQPEGEVYDFDAKINSAVFPALQSGPHNNKIGALAVALKQVM 307
Query 48 TPEWRAFSRCVLENCKVLARELQNHGLQLVTGGTDNHLLLLDLKPQGLTGAKLQLICDMA 107
P ++ +++ V N LA L N G LVT GTDNHL+L DL+P GLTG K++ +C++
Sbjct 308 APSFKVYAKQVKANAACLASYLINKGYTLVTDGTDNHLILWDLRPLGLTGNKVEKVCELC 367
Query 108 NITLNKNSIAGDSSGI-PTGVRIGTPAMTTRGFGAEEFKLVADFIREAVDICCEIQEKSG 166
ITLN+N++ GD+S + P GVRIGTPAMT+RG ++F+ + +F+ AV I +IQE+ G
Sbjct 368 YITLNRNAVFGDTSFLAPGGVRIGTPAMTSRGLVEKDFEKIGEFLHRAVTITLDIQEQYG 427
Query 167 KKLAEFRKHVSPSHPRISNLRDRVKALARSFPMPG 201
K + +F K + ++ I ++ V+ F MPG
Sbjct 428 KVMKDFNKGLV-NNKEIDEIKADVEEFTYDFDMPG 461
> ath:AT1G36370 SHM7; SHM7 (serine hydroxymethyltransferase 7);
catalytic/ glycine hydroxymethyltransferase/ pyridoxal phosphate
binding (EC:2.1.2.1); K00600 glycine hydroxymethyltransferase
[EC:2.1.2.1]
Length=598
Score = 169 bits (427), Expect = 7e-42, Method: Compositional matrix adjust.
Identities = 93/185 (50%), Positives = 124/185 (67%), Gaps = 2/185 (1%)
Query 18 RIDSAVFPGLQGGPHENHIAAVAHQLKQTCTPEWRAFSRCVLENCKVLARELQNHGLQLV 77
+I+ AVFP LQGGPH NHIAA+A LKQ TPE++A+ + + +N + LA L +LV
Sbjct 409 KINFAVFPSLQGGPHNNHIAALAIALKQVATPEYKAYIQQMKKNAQALAAALLRRKCRLV 468
Query 78 TGGTDNHLLLLDLKPQGLTGAKLQLICDMANITLNKNSIAGDSSGI-PTGVRIGTPAMTT 136
TGGTDNHLLL DL P GLTG + +C+M +ITLNK +I GD+ I P GVRIGTPAMTT
Sbjct 469 TGGTDNHLLLWDLTPMGLTGKVYEKVCEMCHITLNKTAIFGDNGTISPGGVRIGTPAMTT 528
Query 137 RGFGAEEFKLVADFIREAVDICCEIQEKSGKKLAEFRKHVSPSHPRISNLRDRVKALARS 196
RG +F+ +ADF+ +A I +Q + GK EF K + ++ I+ LR+RV+A A
Sbjct 529 RGCIESDFETMADFLIKAAQITSALQREHGKSHKEFVKSLC-TNKDIAELRNRVEAFALQ 587
Query 197 FPMPG 201
+ MP
Sbjct 588 YEMPA 592
> xla:380048 shmt1, MGC53442; serine hydroxymethyltransferase
1 (soluble) (EC:2.1.2.1); K00600 glycine hydroxymethyltransferase
[EC:2.1.2.1]
Length=485
Score = 168 bits (425), Expect = 2e-41, Method: Compositional matrix adjust.
Identities = 88/222 (39%), Positives = 135/222 (60%), Gaps = 17/222 (7%)
Query 1 RSGIIF-----------VNTRRVPNGASRIDSAVFPGLQGGPHENHIAAVAHQLKQTCTP 49
RSG+IF + N S I+ AVFPGLQGGPH + IA VA LKQ +P
Sbjct 264 RSGMIFYRKGVRSVDPKTGKETLYNYESLINQAVFPGLQGGPHNHAIAGVAVALKQALSP 323
Query 50 EWRAFSRCVLENCKVLARELQNHGLQLVTGGTDNHLLLLDLKPQGLTGAKLQLICDMANI 109
E++ + + V+ NCK L+ ++ G +VTGG+DNHL+L++L+ + G + + + + +I
Sbjct 324 EFKLYQKQVVSNCKALSLAIEELGYHVVTGGSDNHLILVNLRDKKTDGGRAEKVLEACSI 383
Query 110 TLNKNSIAGDSSGI-PTGVRIGTPAMTTRGFGAEEFKLVADFIREAVDICCEIQE--KSG 166
NKN+ GD S + P+G+R+GTPA+T+RGF E+FK VA FI +++ EIQ+ G
Sbjct 384 ACNKNTCPGDKSALRPSGLRLGTPALTSRGFNEEDFKKVAQFIHRGIELTLEIQKSMNPG 443
Query 167 KKLAEFRKHVSPSH---PRISNLRDRVKALARSFPMPGRPDI 205
L +F++ ++ P+I LR V+ A +FP+PG PD+
Sbjct 444 ATLKDFKEKLASQDVHTPKILALRAEVEKFAGTFPIPGLPDL 485
> xla:447193 shmt2, MGC79128; serine hydroxymethyltransferase
2 (mitochondrial) (EC:2.1.2.1); K00600 glycine hydroxymethyltransferase
[EC:2.1.2.1]
Length=496
Score = 167 bits (423), Expect = 2e-41, Method: Compositional matrix adjust.
Identities = 89/216 (41%), Positives = 137/216 (63%), Gaps = 16/216 (7%)
Query 1 RSGIIFVNT----------RRVP-NGASRIDSAVFPGLQGGPHENHIAAVAHQLKQTCTP 49
RSG+IF + +P N +I+ +VFP +QGGPH + IAAVA LKQ +P
Sbjct 278 RSGLIFFRKGVKSVDKKTGKEIPYNLEDKINFSVFPSIQGGPHNHAIAAVAVALKQASSP 337
Query 50 EWRAFSRCVLENCKVLARELQNHGLQLVTGGTDNHLLLLDLKPQGLTGAKLQLICDMANI 109
+R ++ VL+N K +A L + G LV+GGTDNHL+L+DL+P+G+ GA+ + + ++ +I
Sbjct 338 MFREYAVQVLKNAKSMAAALLSKGYTLVSGGTDNHLVLVDLRPKGIDGARAERVLELVSI 397
Query 110 TLNKNSIAGDSSGI-PTGVRIGTPAMTTRGFGAEEFKLVADFIREAVDICCEIQEKSGKK 168
T NKN+ GD S + P G+R+G PA+T+R F +F+ V DFI E + I +++ K+ K
Sbjct 398 TANKNTCPGDKSALTPGGLRLGAPALTSRNFKEADFEKVVDFIDEGIRIGLDVKRKT-NK 456
Query 169 LAEFRKHV---SPSHPRISNLRDRVKALARSFPMPG 201
L +F+ + + RI +LR +V+ AR+FPMPG
Sbjct 457 LQDFKNFLLEDQETVKRIGDLRKQVEQFARAFPMPG 492
> ath:AT4G13930 SHM4; SHM4 (serine hydroxymethyltransferase 4);
catalytic/ glycine hydroxymethyltransferase/ pyridoxal phosphate
binding (EC:2.1.2.1); K00600 glycine hydroxymethyltransferase
[EC:2.1.2.1]
Length=471
Score = 167 bits (423), Expect = 2e-41, Method: Compositional matrix adjust.
Identities = 89/213 (41%), Positives = 133/213 (62%), Gaps = 13/213 (6%)
Query 1 RSGIIFVNT------RRVPNGA-----SRIDSAVFPGLQGGPHENHIAAVAHQLKQTCTP 49
R+G+IF + P GA +I+ AVFP LQGGPH + I A+A LKQ TP
Sbjct 250 RAGMIFYRKGPKPPKKGQPEGAVYDFEDKINFAVFPALQGGPHNHQIGALAVALKQANTP 309
Query 50 EWRAFSRCVLENCKVLARELQNHGLQLVTGGTDNHLLLLDLKPQGLTGAKLQLICDMANI 109
++ +++ V N L L + G Q+VT GT+NHL+L DL+P GLTG K++ +CD+ +I
Sbjct 310 GFKVYAKQVKANAVALGNYLMSKGYQIVTNGTENHLVLWDLRPLGLTGNKVEKLCDLCSI 369
Query 110 TLNKNSIAGDSSGI-PTGVRIGTPAMTTRGFGAEEFKLVADFIREAVDICCEIQEKSGKK 168
TLNKN++ GDSS + P GVRIG PAMT+RG ++F+ + +F+ AV + +IQ+ GK
Sbjct 370 TLNKNAVFGDSSALAPGGVRIGAPAMTSRGLVEKDFEQIGEFLSRAVTLTLDIQKTYGKL 429
Query 169 LAEFRKHVSPSHPRISNLRDRVKALARSFPMPG 201
L +F K + ++ + L+ V+ + S+ MPG
Sbjct 430 LKDFNKGLV-NNKDLDQLKADVEKFSASYEMPG 461
> dre:100144628 shmt2; serine hydroxymethyltransferase 2 (mitochondrial)
(EC:2.1.2.1); K00600 glycine hydroxymethyltransferase
[EC:2.1.2.1]
Length=492
Score = 167 bits (423), Expect = 3e-41, Method: Compositional matrix adjust.
Identities = 84/191 (43%), Positives = 131/191 (68%), Gaps = 5/191 (2%)
Query 18 RIDSAVFPGLQGGPHENHIAAVAHQLKQTCTPEWRAFSRCVLENCKVLARELQNHGLQLV 77
+++ +VFP LQGGPH + IA VA LKQ +P +R + VL+N K +A L + G LV
Sbjct 302 KVNFSVFPSLQGGPHNHAIAGVAVALKQATSPMFREYIAQVLKNSKAMAAALLDKGYTLV 361
Query 78 TGGTDNHLLLLDLKPQGLTGAKLQLICDMANITLNKNSIAGDSSGI-PTGVRIGTPAMTT 136
+GGTDNHL+L+DL+PQG+ GA+ + + ++ +IT NKN+ GD S + P G+R+GTPA+T+
Sbjct 362 SGGTDNHLVLVDLRPQGMDGARAERVLELVSITANKNTCPGDKSALTPGGLRLGTPALTS 421
Query 137 RGFGAEEFKLVADFIREAVDICCEIQEKSGKKLAEFRKHV---SPSHPRISNLRDRVKAL 193
R +F+ V +FI + + I ++++K+ KKL++F+ + + + RI++LR RV+A
Sbjct 422 RQLKECDFQKVVEFIHQGIQIGQDVKKKT-KKLSDFKSFLLEDAETVSRIADLRSRVEAF 480
Query 194 ARSFPMPGRPD 204
AR FPMPG D
Sbjct 481 ARPFPMPGFHD 491
> dre:394021 shmt1, MGC66171, zgc:66171, zgc:77524; serine hydroxymethyltransferase
1 (soluble) (EC:2.1.2.1); K00600 glycine
hydroxymethyltransferase [EC:2.1.2.1]
Length=481
Score = 165 bits (417), Expect = 1e-40, Method: Compositional matrix adjust.
Identities = 89/222 (40%), Positives = 130/222 (58%), Gaps = 17/222 (7%)
Query 1 RSGIIF-----------VNTRRVPNGASRIDSAVFPGLQGGPHENHIAAVAHQLKQTCTP 49
R+G+IF + N S I+ AVFPGLQGGPH + IA VA LKQ TP
Sbjct 260 RAGVIFFRKGVRSVDAKTGKETMYNLESLINQAVFPGLQGGPHNHAIAGVAVALKQALTP 319
Query 50 EWRAFSRCVLENCKVLARELQNHGLQLVTGGTDNHLLLLDLKPQGLTGAKLQLICDMANI 109
E++ + VL NCK LA L + G ++VTGG+DNHL+L+DL+ G G + + + + I
Sbjct 320 EFKTYQLQVLANCKALASALMDKGYKVVTGGSDNHLILVDLRSNGTDGGRAEKVLEACAI 379
Query 110 TLNKNSIAGDSSGI-PTGVRIGTPAMTTRGFGAEEFKLVADFIREAVDICCEIQEKSGKK 168
NKN+ GD S + P+G+R+G+PA+T+RG E F VA+FI + + + EIQ+ K
Sbjct 380 ACNKNTCPGDKSALRPSGLRLGSPALTSRGLLEEHFHKVAEFIHQGIVLTLEIQKNMNPK 439
Query 169 --LAEFRKHVSPSHP---RISNLRDRVKALARSFPMPGRPDI 205
L EF++ +S + + +R V+ A FPMPG P++
Sbjct 440 ATLKEFKEELSQNEKYQLKTKEIRKEVEDFAGKFPMPGLPEL 481
> bbo:BBOV_IV005840 23.m06426; serine hydroxymethyltransferase
(EC:2.1.2.1); K00600 glycine hydroxymethyltransferase [EC:2.1.2.1]
Length=453
Score = 164 bits (416), Expect = 1e-40, Method: Compositional matrix adjust.
Identities = 80/200 (40%), Positives = 131/200 (65%), Gaps = 2/200 (1%)
Query 1 RSGIIFVNTRRVPNGASRIDSAVFPGLQGGPHENHIAAVAHQLKQTCTPEWRAFSRCVLE 60
R+G+IF N + P +I++AVFP +QGGPH N IA++A QLK +PEW+ +++ ++E
Sbjct 253 RAGMIFFNKKIDPTIEDKINNAVFPTVQGGPHNNAIASLAVQLKTVMSPEWKVYAKNIVE 312
Query 61 NCKVLARELQNHGLQLVTGGTDNHLLLLDLKPQGLTGAKLQLICDMANITLNKNSIAGDS 120
N + LA E ++ G +VTGGTDNH ++++LKP G+ G K + IC+ N+T++K+++ GD
Sbjct 313 NARRLAIECESRGFLVVTGGTDNHTVVINLKPFGVNGNKAEHICNAINVTVSKSTVPGDV 372
Query 121 SGI-PTGVRIGTPAMTTRGFGAEEFKLVADFIREAVDICCEIQEKSGKKLAEFRKHVSPS 179
S + P+G+R+GT + RG E+ +A+ + EAV I IQE G+ +F++ +
Sbjct 373 SAMNPSGLRLGTAMIVARGAVPEDMAFIAEALLEAVKITQSIQESHGENHEDFKRG-AEG 431
Query 180 HPRISNLRDRVKALARSFPM 199
+ RI+ LR +V R FP+
Sbjct 432 NERIAALRKKVVDWIRQFPI 451
> ath:AT4G37930 SHM1; SHM1 (SERINE TRANSHYDROXYMETHYLTRANSFERASE
1); glycine hydroxymethyltransferase/ poly(U) binding (EC:2.1.2.1);
K00600 glycine hydroxymethyltransferase [EC:2.1.2.1]
Length=517
Score = 162 bits (410), Expect = 8e-40, Method: Compositional matrix adjust.
Identities = 80/189 (42%), Positives = 122/189 (64%), Gaps = 5/189 (2%)
Query 18 RIDSAVFPGLQGGPHENHIAAVAHQLKQTCTPEWRAFSRCVLENCKVLARELQNHGLQLV 77
+I+ AVFPGLQGGPH + I +A LKQ T E++A+ VL N A+ L G +LV
Sbjct 319 KINQAVFPGLQGGPHNHTITGLAVALKQATTSEYKAYQEQVLSNSAKFAQTLMERGYELV 378
Query 78 TGGTDNHLLLLDLKPQGLTGAKLQLICDMANITLNKNSIAGD-SSGIPTGVRIGTPAMTT 136
+GGTDNHL+L++LKP+G+ G++++ + + +I NKN++ GD S+ +P G+R+GTPA+T+
Sbjct 379 SGGTDNHLVLVNLKPKGIDGSRVEKVLEAVHIASNKNTVPGDVSAMVPGGIRMGTPALTS 438
Query 137 RGFGAEEFKLVADFIREAVDICCEIQ-EKSGKKLAEFRKHVSPS---HPRISNLRDRVKA 192
RGF E+F VA++ +AV I +++ E G KL +F + S I+ LR V+
Sbjct 439 RGFVEEDFAKVAEYFDKAVTIALKVKSEAQGTKLKDFVSAMESSSTIQSEIAKLRHEVEE 498
Query 193 LARSFPMPG 201
A+ FP G
Sbjct 499 FAKQFPTIG 507
> ath:AT1G22020 SHM6; SHM6 (serine hydroxymethyltransferase 6);
catalytic/ glycine hydroxymethyltransferase/ pyridoxal phosphate
binding (EC:2.1.2.1); K00600 glycine hydroxymethyltransferase
[EC:2.1.2.1]
Length=599
Score = 161 bits (408), Expect = 1e-39, Method: Compositional matrix adjust.
Identities = 87/185 (47%), Positives = 119/185 (64%), Gaps = 2/185 (1%)
Query 18 RIDSAVFPGLQGGPHENHIAAVAHQLKQTCTPEWRAFSRCVLENCKVLARELQNHGLQLV 77
+I+ +VFP LQGGPH NHIAA+A LKQ +PE++ + R V +N K LA L + +L+
Sbjct 413 KINFSVFPSLQGGPHNNHIAALAIALKQAASPEYKLYMRQVKKNAKALASALISRKCKLI 472
Query 78 TGGTDNHLLLLDLKPQGLTGAKLQLICDMANITLNKNSIAGDSSGI-PTGVRIGTPAMTT 136
TGGTDNHLLL DL P GLTG + +C+M +IT+NK +I ++ I P GVRIG+PAMT+
Sbjct 473 TGGTDNHLLLWDLTPLGLTGKVYEKVCEMCHITVNKVAIFSENGVISPGGVRIGSPAMTS 532
Query 137 RGFGAEEFKLVADFIREAVDICCEIQEKSGKKLAEFRKHVSPSHPRISNLRDRVKALARS 196
RG EF+ +ADF+ A I Q + GK E K + I++LR++V+A A
Sbjct 533 RGCLEPEFETMADFLYRAAQIASAAQREHGKLQKEPLKSIYHCK-EIADLRNQVEAFATQ 591
Query 197 FPMPG 201
F MP
Sbjct 592 FAMPA 596
> hsa:6470 SHMT1, CSHMT, MGC15229, MGC24556, SHMT; serine hydroxymethyltransferase
1 (soluble) (EC:2.1.2.1); K00600 glycine
hydroxymethyltransferase [EC:2.1.2.1]
Length=483
Score = 161 bits (407), Expect = 2e-39, Method: Compositional matrix adjust.
Identities = 81/197 (41%), Positives = 126/197 (63%), Gaps = 5/197 (2%)
Query 14 NGASRIDSAVFPGLQGGPHENHIAAVAHQLKQTCTPEWRAFSRCVLENCKVLARELQNHG 73
N S I+SAVFPGLQGGPH + IA VA LKQ T E++ + V+ NC+ L+ L G
Sbjct 287 NLESLINSAVFPGLQGGPHNHAIAGVAVALKQAMTLEFKVYQHQVVANCRALSEALTELG 346
Query 74 LQLVTGGTDNHLLLLDLKPQGLTGAKLQLICDMANITLNKNSIAGDSSGI-PTGVRIGTP 132
++VTGG+DNHL+L+DL+ +G G + + + + +I NKN+ GD S + P+G+R+GTP
Sbjct 347 YKIVTGGSDNHLILVDLRSKGTDGGRAEKVLEACSIACNKNTCPGDRSALRPSGLRLGTP 406
Query 133 AMTTRGFGAEEFKLVADFIREAVDICCEIQEKSGKK--LAEFRKHVS--PSHPRISNLRD 188
A+T+RG ++F+ VA FI +++ +IQ +G + L EF++ ++ + LR+
Sbjct 407 ALTSRGLLEKDFQKVAHFIHRGIELTLQIQSDTGVRATLKEFKERLAGDKYQAAVQALRE 466
Query 189 RVKALARSFPMPGRPDI 205
V++ A FP+PG PD
Sbjct 467 EVESFASLFPLPGLPDF 483
> sce:YLR058C SHM2, SHMT2; Cytosolic serine hydroxymethyltransferase,
converts serine to glycine plus 5,10 methylenetetrahydrofolate;
major isoform involved in generating precursors
for purine, pyrimidine, amino acid, and lipid biosynthesis (EC:2.1.2.1);
K00600 glycine hydroxymethyltransferase [EC:2.1.2.1]
Length=469
Score = 161 bits (407), Expect = 2e-39, Method: Compositional matrix adjust.
Identities = 78/185 (42%), Positives = 118/185 (63%), Gaps = 4/185 (2%)
Query 19 IDSAVFPGLQGGPHENHIAAVAHQLKQTCTPEWRAFSRCVLENCKVLARELQNHGLQLVT 78
I+ +VFPG QGGPH + IAA+A LKQ TPE++ + VL+N K L E +N G +LV+
Sbjct 283 INFSVFPGHQGGPHNHTIAALATALKQAATPEFKEYQTQVLKNAKALESEFKNLGYRLVS 342
Query 79 GGTDNHLLLLDLKPQGLTGAKLQLICDMANITLNKNSIAGDSSG-IPTGVRIGTPAMTTR 137
GTD+H++L+ L+ +G+ GA+++ IC+ NI LNKNSI GD S +P GVRIG PAMTTR
Sbjct 343 NGTDSHMVLVSLREKGVDGARVEYICEKINIALNKNSIPGDKSALVPGGVRIGAPAMTTR 402
Query 138 GFGAEEFKLVADFIREAVDICCEIQE---KSGKKLAEFRKHVSPSHPRISNLRDRVKALA 194
G G E+F + +I +AV+ ++Q+ K +L +F+ V ++ + + A
Sbjct 403 GMGEEDFHRIVQYINKAVEFAQQVQQSLPKDACRLKDFKAKVDEGSDVLNTWKKEIYDWA 462
Query 195 RSFPM 199
+P+
Sbjct 463 GEYPL 467
> mmu:20425 Shmt1, AI324848, AI385541, C81125, Shmt, mshmt, mshmt1,
mshmt2; serine hydroxymethyltransferase 1 (soluble) (EC:2.1.2.1);
K00600 glycine hydroxymethyltransferase [EC:2.1.2.1]
Length=478
Score = 159 bits (403), Expect = 5e-39, Method: Compositional matrix adjust.
Identities = 81/195 (41%), Positives = 123/195 (63%), Gaps = 6/195 (3%)
Query 17 SRIDSAVFPGLQGGPHENHIAAVAHQLKQTCTPEWRAFSRCVLENCKVLARELQNHGLQL 76
S I+SAVFPGLQGGPH + IA VA LKQ T E++ + VL NC+ L+ L G ++
Sbjct 284 SLINSAVFPGLQGGPHNHAIAGVAVALKQAMTTEFKIYQLQVLANCRALSDALTELGYKI 343
Query 77 VTGGTDNHLLLLDLKPQGLTGAKLQLICDMANITLNKNSIAGDSSGI-PTGVRIGTPAMT 135
VTGG+DNHL+L+DL+ +G G + + + + +I NKN+ GD S + P+G+R+GTPA+T
Sbjct 344 VTGGSDNHLILMDLRSKGTDGGRAEKVLEACSIACNKNTCPGDKSALRPSGLRLGTPALT 403
Query 136 TRGFGAEEFKLVADFIREAVDICCEIQEKSGKK--LAEFRKHVSPS---HPRISNLRDRV 190
+RG E+F+ VA FI +++ +IQ K L EF++ ++ ++ LR+ V
Sbjct 404 SRGLLEEDFQKVAHFIHRGIELTLQIQSHMATKATLKEFKEKLAGDEKIQSAVATLREEV 463
Query 191 KALARSFPMPGRPDI 205
+ A +F +PG PD
Sbjct 464 ENFASNFSLPGLPDF 478
> sce:YBR263W SHM1, SHMT1, TMP3; Mitochondrial serine hydroxymethyltransferase,
converts serine to glycine plus 5,10 methylenetetrahydrofolate;
involved in generating precursors for
purine, pyrimidine, amino acid, and lipid biosynthesis; reverse
reaction generates serine (EC:2.1.2.1); K00600 glycine hydroxymethyltransferase
[EC:2.1.2.1]
Length=490
Score = 153 bits (387), Expect = 3e-37, Method: Compositional matrix adjust.
Identities = 81/192 (42%), Positives = 121/192 (63%), Gaps = 9/192 (4%)
Query 18 RIDSAVFPGLQGGPHENHIAAVAHQLKQTCTPEWRAFSRCVLENCKVLARELQNHGLQLV 77
+I+ +VFPG QGGPH + I A+A LKQ +PE++ + + +++N K A+EL G +LV
Sbjct 298 KINFSVFPGHQGGPHNHTIGAMAVALKQAMSPEFKEYQQKIVDNSKWFAQELTKMGYKLV 357
Query 78 TGGTDNHLLLLDLKPQGLTGAKLQLICDMANITLNKNSIAGDSSGI-PTGVRIGTPAMTT 136
+GGTDNHL+++DL + GA+++ I NI NKN+I GD S + P+G+RIGTPAMTT
Sbjct 358 SGGTDNHLIVIDLSGTQVDGARVETILSALNIAANKNTIPGDKSALFPSGLRIGTPAMTT 417
Query 137 RGFGAEEFKLVADFIREAVDICCEIQ--EKSGK-----KLAEFRKHVSPSHPRISNLRDR 189
RGFG EEF VA +I AV + ++ E + K +L EF+K + S ++ L
Sbjct 418 RGFGREEFSQVAKYIDSAVKLAENLKTLEPTTKLDARSRLNEFKKLCNES-SEVAALSGE 476
Query 190 VKALARSFPMPG 201
+ +P+PG
Sbjct 477 ISKWVGQYPVPG 488
> ath:AT5G26780 SHM2; SHM2 (SERINE HYDROXYMETHYLTRANSFERASE 2);
catalytic/ glycine hydroxymethyltransferase/ pyridoxal phosphate
binding (EC:2.1.2.1); K00600 glycine hydroxymethyltransferase
[EC:2.1.2.1]
Length=533
Score = 152 bits (383), Expect = 9e-37, Method: Compositional matrix adjust.
Identities = 82/205 (40%), Positives = 123/205 (60%), Gaps = 21/205 (10%)
Query 18 RIDSAVFPGLQGGPHENHIAAVAHQLKQTCTPEWRAFSRCVLENCKVLA----------- 66
RI+ AVFPGLQGGPH + I +A LKQ TPE++A+ VL NC A
Sbjct 319 RINQAVFPGLQGGPHNHTITGLAVALKQARTPEYKAYQDQVLRNCSKFAELDIRPTVIIS 378
Query 67 -----RELQNHGLQLVTGGTDNHLLLLDLKPQGLTGAKLQLICDMANITLNKNSIAGD-S 120
+ L G LV+GGTDNHL+L++LK +G+ G++++ + ++ +I NKN++ GD S
Sbjct 379 YGLSMQTLLAKGYDLVSGGTDNHLVLVNLKNKGIDGSRVEKVLELVHIAANKNTVPGDVS 438
Query 121 SGIPTGVRIGTPAMTTRGFGAEEFKLVADFIREAVDICCEIQ-EKSGKKLAEFRKHVSPS 179
+ +P G+R+GTPA+T+RGF E+F VA++ AV I +I+ E G KL +F + +
Sbjct 439 AMVPGGIRMGTPALTSRGFIEEDFAKVAEYFDLAVKIALKIKAESQGTKLKDFVATMQSN 498
Query 180 ---HPRISNLRDRVKALARSFPMPG 201
+S LR+ V+ A+ FP G
Sbjct 499 EKLQSEMSKLREMVEEYAKQFPTIG 523
> cpv:cgd8_2610 cytosolic serine hydroxymethyl transferase ; K00600
glycine hydroxymethyltransferase [EC:2.1.2.1]
Length=445
Score = 119 bits (299), Expect = 6e-27, Method: Compositional matrix adjust.
Identities = 68/205 (33%), Positives = 114/205 (55%), Gaps = 3/205 (1%)
Query 1 RSGIIFVNTRRVPNGASRIDSAVFPGLQGGPHENHIAAVAHQLKQTCTPEWRAFSRCVLE 60
RS IIF +ID +V +Q H N +AA+ QLKQ + W ++ VLE
Sbjct 241 RSAIIFYRKDVESKIRVKIDESVSKEIQSSIHFNQVAALCFQLKQVVSASWVKYASRVLE 300
Query 61 NCKVLARELQNHGLQLVTGGTDNHLLLLDLKPQGLTGAKLQLICDMANITLNKNSIAGDS 120
+ ++L + L+ G++++T GTD+H +L+D + ++GAK + ++ I+ +++S+ D
Sbjct 301 SSQLLCKLLEESGIKILTNGTDSHKILIDTRSLNISGAKAEKALEVCEISTSRSSLPCDG 360
Query 121 SGIP-TGVRIGTPAMTTRGFGAEEFKLVADFIREAVDICCEIQEKSGKKLAEFRKHVSPS 179
+ +GVR+GT A+ +RG ++FK V+ I E + +IQ K G+ LAEF ++ S
Sbjct 361 RTMNCSGVRLGTAALASRGMELDDFKFVSRIIVEVLTTARDIQ-KDGETLAEFVSNIKAS 419
Query 180 HPRISNLRDRVKALARSFPMPGRPD 204
P I N+R V+ A SF D
Sbjct 420 -PAIENIRQTVRKFASSFEFVSIVD 443
> eco:b2551 glyA, ECK2548, JW2535; serine hydroxymethyltransferase
(EC:2.1.2.1); K00600 glycine hydroxymethyltransferase [EC:2.1.2.1]
Length=417
Score = 113 bits (283), Expect = 4e-25, Method: Compositional matrix adjust.
Identities = 59/141 (41%), Positives = 88/141 (62%), Gaps = 2/141 (1%)
Query 18 RIDSAVFPGLQGGPHENHIAAVAHQLKQTCTPEWRAFSRCVLENCKVLARELQNHGLQLV 77
+++SAVFPG QGGP + IA A LK+ PE++ + + V +N K + G ++V
Sbjct 251 KLNSAVFPGGQGGPLMHVIAGKAVALKEAMEPEFKTYQQQVAKNAKAMVEVFLERGYKVV 310
Query 78 TGGTDNHLLLLDLKPQGLTGAKLQLICDMANITLNKNSIAGD--SSGIPTGVRIGTPAMT 135
+GGTDNHL L+DL + LTG + ANIT+NKNS+ D S + +G+R+GTPA+T
Sbjct 311 SGGTDNHLFLVDLVDKNLTGKEADAALGRANITVNKNSVPNDPKSPFVTSGIRVGTPAIT 370
Query 136 TRGFGAEEFKLVADFIREAVD 156
RGF E K +A ++ + +D
Sbjct 371 RRGFKEAEAKELAGWMCDVLD 391
> cpv:cgd8_2580 mitochondrial serine hydroxymethyl transferase
Length=438
Score = 77.4 bits (189), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 47/167 (28%), Positives = 79/167 (47%), Gaps = 4/167 (2%)
Query 1 RSGIIFVNTRRVPNGASRIDSAVFPGLQGGPHENHIAAVAHQLKQTCTPEWRAFSRCVLE 60
+ G + +N + P +++SAVFPGLQGGPH + I + A Q++ T F L+
Sbjct 234 KGGFLMLNNSKNPGLFQKVNSAVFPGLQGGPHNHQIGSFAVQIQGMLTSRTSEFVAAALD 293
Query 61 NCKVLARELQNHGLQLVTGGTDNHLLLLDLKPQGLTGAKLQLICDMANITLNKNSIAGDS 120
N VLA+ + + G+ L+ GTD HL+ +D + G+ + I I +
Sbjct 294 NSAVLAQTMLDSGIPLLGDGTDTHLVSVDCERLGVPCELISKILTECKIRHTYRKFGENR 353
Query 121 SGIPTGVRIGTPAMTTRGFGAEEFKLVADFIREAVDICCEIQEKSGK 167
S + GT T R + ++ + I + +++ EI K GK
Sbjct 354 SSL----MFGTLVYTFREGSVSQMTILGNLISDCINVGVEIFNKVGK 396
> pfa:PF14_0534 serine hydroxymethyltransferase, putative (EC:2.1.2.1)
Length=462
Score = 48.9 bits (115), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 36/162 (22%), Positives = 67/162 (41%), Gaps = 10/162 (6%)
Query 43 LKQTCTPEWRAFSRCVLENCKVLARELQNHGLQLVTGGTDNHLLLLDLKPQGLTG--AKL 100
K E++ + + + EN +L + + + ++ +L P T +
Sbjct 303 FKLMKNEEFKEYVKQIKENTYILYKYINRKYFHIQYSQNNS---FFNLNPSSCTFNIQEF 359
Query 101 QLICDMANITLNKNSIAGDSSGIPTGVRIGTPAMTTRGFGAEEFKLVADFIREAVDICCE 160
L+C+ NI I D S IGT +T+ G + K VA+F E+V +
Sbjct 360 YLLCNKLNIYF---DILKDKSSNQKSFNIGTNNLTSLGLLTHDIKNVAEFFNESVVLYFY 416
Query 161 IQEKSGKKLAEFRKHV--SPSHPRISNLRDRVKALARSFPMP 200
++EKS F +++ + S I +L + + S+P P
Sbjct 417 LKEKSKLTNMSFIQYIEDNSSASDIYSLAVDISSFISSYPSP 458
> ath:AT2G46080 hypothetical protein
Length=347
Score = 32.3 bits (72), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 16/47 (34%), Positives = 27/47 (57%), Gaps = 7/47 (14%)
Query 161 IQEKSGKK-------LAEFRKHVSPSHPRISNLRDRVKALARSFPMP 200
+Q SG+K L +R+HV+ ++PRI N R + +L +S +P
Sbjct 135 LQSDSGEKYLQARSSLDSWRQHVNTNNPRIENCRAVLDSLVKSLSLP 181
> tgo:TGME49_109980 dynein-1-alpha heavy chain, flagellar inner
arm I1 complex, putative
Length=4629
Score = 32.3 bits (72), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 21/60 (35%), Positives = 30/60 (50%), Gaps = 4/60 (6%)
Query 142 EEFKLVADFIREAVDICCEIQEKSGKKLAEFRKHVSPSHPRISNLRDRVKALARSFPMPG 201
E V ++RE V++ + Q G +L K V+ H I L V+ALARSF + G
Sbjct 542 EYMSKVCTYLREVVEVINQFQRFLGPEL----KTVTRYHKGIDRLNQDVQALARSFDVLG 597
> hsa:9701 PPP6R2, KIAA0685, PP6R2, SAP190, SAPS2, dJ579N16.1;
protein phosphatase 6, regulatory subunit 2
Length=932
Score = 30.4 bits (67), Expect = 4.2, Method: Compositional matrix adjust.
Identities = 14/38 (36%), Positives = 20/38 (52%), Gaps = 0/38 (0%)
Query 17 SRIDSAVFPGLQGGPHENHIAAVAHQLKQTCTPEWRAF 54
+RI +AV L+ GP + HI+ V L C W +F
Sbjct 481 TRIANAVVQNLERGPVQTHISEVIRGLPADCRGRWESF 518
> mmu:71474 Ppp6r2, 1110033O10Rik, 8430411H09Rik, B230107H12Rik,
Pp6r2, Saps2; protein phosphatase 6, regulatory subunit 2
Length=923
Score = 30.0 bits (66), Expect = 5.2, Method: Compositional matrix adjust.
Identities = 14/38 (36%), Positives = 20/38 (52%), Gaps = 0/38 (0%)
Query 17 SRIDSAVFPGLQGGPHENHIAAVAHQLKQTCTPEWRAF 54
+RI +AV L+ GP + HI+ V L C W +F
Sbjct 483 TRIANAVVQNLEQGPVQAHISEVIRGLPADCRGRWESF 520
> eco:b0771 ybhJ, ECK0760, JW5103; predicted hydratase; K01681
aconitate hydratase 1 [EC:4.2.1.3]
Length=753
Score = 29.6 bits (65), Expect = 8.5, Method: Compositional matrix adjust.
Identities = 18/64 (28%), Positives = 30/64 (46%), Gaps = 2/64 (3%)
Query 11 RVPNGASRIDSAVFPGLQGGPHENHIAAVAHQLKQTCTPEWRAFSRCVLENCKVLARELQ 70
+V NG ++ + G GG +EN IAA Q+C + FS V + + + +L
Sbjct 344 KVENGRLKVQQGIIAGCSGGNYENVIAAANALRGQSCGND--TFSLAVYPSSQPVFMDLA 401
Query 71 NHGL 74
G+
Sbjct 402 KKGV 405
Lambda K H
0.321 0.137 0.413
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 6318090968
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40