bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_0128_orf1
Length=305
Score E
Sequences producing significant alignments: (Bits) Value
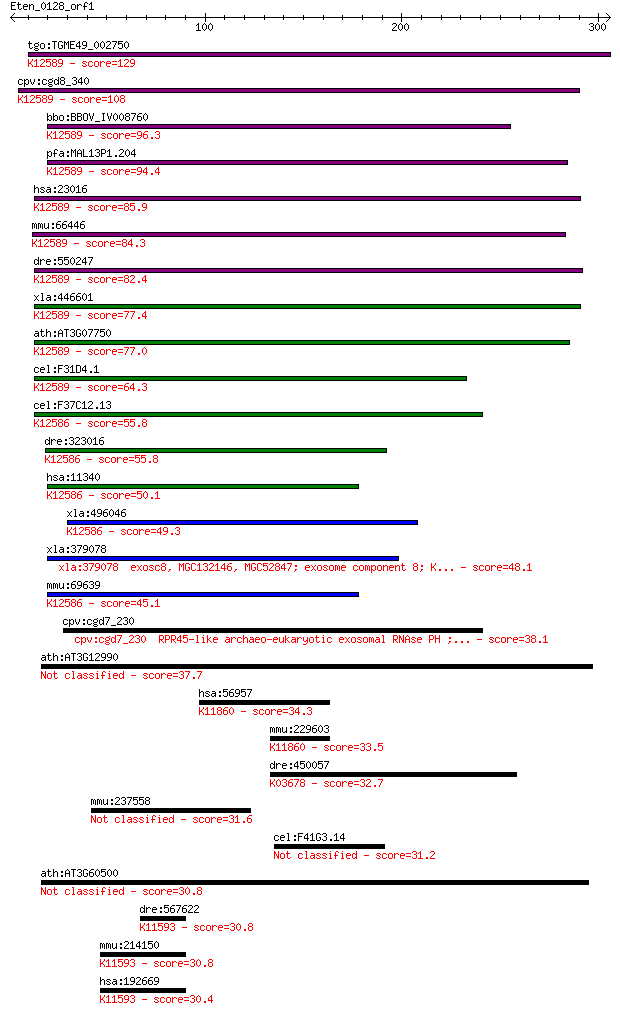
tgo:TGME49_002750 exosome complex exonuclease, putative (EC:3.... 129 1e-29
cpv:cgd8_340 exoribonuclease PH ; K12589 exosome complex compo... 108 2e-23
bbo:BBOV_IV008760 23.m05910; hypothetical protein; K12589 exos... 96.3 1e-19
pfa:MAL13P1.204 exoribonuclease PH, putative; K12589 exosome c... 94.4 5e-19
hsa:23016 EXOSC7, EAP1, FLJ26543, KIAA0116, RRP42, Rrp42p, hRr... 85.9 2e-16
mmu:66446 Exosc7, 2610002K22Rik, AV212732, mKIAA0116; exosome ... 84.3 5e-16
dre:550247 zgc:110717; K12589 exosome complex component RRP42 82.4 2e-15
xla:446601 exosc7, MGC81766; exosome component 7; K12589 exoso... 77.4 6e-14
ath:AT3G07750 3' exoribonuclease family domain 1-containing pr... 77.0 8e-14
cel:F31D4.1 exos-7; EXOSome (multiexonuclease complex) compone... 64.3 6e-10
cel:F37C12.13 exos-9; EXOSome (multiexonuclease complex) compo... 55.8 2e-07
dre:323016 exosc8, MGC174638, wu:fb79a10, zgc:110381; exosome ... 55.8 2e-07
hsa:11340 EXOSC8, CIP3, EAP2, OIP2, RP11-421P11.3, RRP43, Rrp4... 50.1 1e-05
xla:496046 hypothetical LOC496046; K12586 exosome complex comp... 49.3 2e-05
xla:379078 exosc8, MGC132146, MGC52847; exosome component 8; K... 48.1 4e-05
mmu:69639 Exosc8, 2310032N20Rik, CIP3, KIAA4013, mKIAA4013; ex... 45.1 3e-04
cpv:cgd7_230 RPR45-like archaeo-eukaryotic exosomal RNAse PH ;... 38.1 0.038
ath:AT3G12990 RRP45a; RRP45a (Ribonuclease PH45a); 3'-5'-exori... 37.7 0.054
hsa:56957 OTUD7B, CEZANNE, ZA20D1; OTU domain containing 7B (E... 34.3 0.56
mmu:229603 Otud7b, 2900060B22Rik, 4930463P07Rik, AI462125, CEZ... 33.5 0.90
dre:450057 exosc9, im:6909054, zgc:101680; exosome component 9... 32.7 1.7
mmu:237558 Gm239; predicted gene 239 31.6
cel:F41G3.14 exos-8; EXOSome (multiexonuclease complex) compon... 31.2 4.9
ath:AT3G60500 CER7; CER7 (ECERIFERUM 7); 3'-5'-exoribonuclease... 30.8 6.1
dre:567622 eif2c1; eukaryotic translation initiation factor 2C... 30.8 6.5
mmu:214150 Eif2c3, AW048688, Ago3, C130014L07Rik; eukaryotic t... 30.8 6.6
hsa:192669 EIF2C3, AGO3, FLJ12765, MGC86946; eukaryotic transl... 30.4 9.5
> tgo:TGME49_002750 exosome complex exonuclease, putative (EC:3.1.13.-);
K12589 exosome complex component RRP42
Length=297
Score = 129 bits (325), Expect = 1e-29, Method: Compositional matrix adjust.
Identities = 100/308 (32%), Positives = 150/308 (48%), Gaps = 65/308 (21%)
Query 10 QGAISRAETAFVREGVACGVRGDGRGRAELRPRVLAVDLLPFSCSSAAIQTEENTVIATV 69
+G + AE ++ EGVAC VR DGR E+RP + LP +SA +++ ENTV+ V
Sbjct 43 RGLLCGAELLYLEEGVACNVREDGRACVEVRPSQVETFALPKCSASAIVRSSENTVLCGV 102
Query 70 DADVILPGGGGNQEPYSVSVGCGAAEQELADVTG--------GDGSSLSSLLQAMLEQLL 121
+ P ++ ++V C AA G G+GSS SSL QA+L ++
Sbjct 103 SMQLARPTAAPDEGEVFLTVDCSAALGVGGVGGGVALESFGLGEGSSNSSL-QALLSDMV 161
Query 122 LPHL--KPLTDPDGRLQWRISIDVIVLRAGGCLLDAVSLAIWGALKALKLPNVMVEEEDE 179
L + K L G+L WR+ +D +VL+AGGCLLDAVSLA+ AL+ LP V V+EE+
Sbjct 162 LSAVDKKKLCLVPGKLIWRVFVDCMVLKAGGCLLDAVSLAVHAALRTTVLPCVFVDEEEG 221
Query 180 KDQGLAITEYKVSCDERIAAGTPYPIDTVPILITAAQIGDTYVWDLTEIEATCGDSFLVA 239
++ A+ + + CDER AAG +P++ +PI++T +
Sbjct 222 EEGDGALGQISIQCDEREAAGECFPVEDIPIIVTVGE----------------------- 258
Query 240 AFLPSGACVGLQKFGHVLADCRTLQTTLTTSQRLSREILEDLQRQAEMKKSRLNG-NLDS 298
SQR+ REI E L+RQ +K++ N L +
Sbjct 259 -----------------------------NSQRVCREIFERLERQIASQKTKKNSQQLSA 289
Query 299 FL-SLASL 305
F+ S+A L
Sbjct 290 FMRSMADL 297
> cpv:cgd8_340 exoribonuclease PH ; K12589 exosome complex component
RRP42
Length=295
Score = 108 bits (271), Expect = 2e-23, Method: Compositional matrix adjust.
Identities = 74/297 (24%), Positives = 130/297 (43%), Gaps = 26/297 (8%)
Query 5 FFSSFQGAISRAETAFVREGVACGVRGDGRGRAELRPRVLAVDLLPFSCSSAAIQTEENT 64
F S +IS +E F+ +GV G+R DGR + RP V+A+D++ + S+ I+ +E
Sbjct 12 FLKSMTKSISDSEKLFLEDGVDQGIRNDGRDLTDFRPIVIALDVISTANGSSRIRNDELD 71
Query 65 VIATVDADVILPGGGGNQEPYSVSVGCGAAEQELADVTGGDGSSL-----SSLLQAMLEQ 119
+I V + + + + D S S + +++
Sbjct 72 IIVAVKVSFL-------NNLFFFIIALILFSSNIMDKISNVAPSFTEEDYSFYITNIVKD 124
Query 120 LLLPH--LKPLTDPDGRLQWRISIDVIVLRAGGCLLDAVSLAIWGALKALKLPNVMVE-- 175
+ + L LT + +L W I ID +L GG ++D V++AI AL+ ++P + V
Sbjct 125 MCFKNFDLSKLTILENKLYWNIYIDATILSFGGNMVDWVTIAIHTALRTTRIPKIHVSPS 184
Query 176 ---EEDEKDQGLAITEYKVSCDERIAAGTPYPIDTVPILITAAQIGDTYVWDLTEIEATC 232
+E+ KD Y VS + +GTP+P +P +I+A I +WD+ E C
Sbjct 185 ISTKENNKD-----VSYTVS--PSVCSGTPFPFQDIPFIISAGCIRGKVIWDMNMQEQIC 237
Query 233 GDSFLVAAFLPSGACVGLQKFGHVLADCRTLQTTLTTSQRLSREILEDLQRQAEMKK 289
+ + + G CV + K D + T + + +S EI + L K+
Sbjct 238 SKTIIALSINSKGECVAMNKVYSNTLDLNLIPTIINKAAEISSEISKSLDEFFSNKQ 294
> bbo:BBOV_IV008760 23.m05910; hypothetical protein; K12589 exosome
complex component RRP42
Length=267
Score = 96.3 bits (238), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 65/237 (27%), Positives = 117/237 (49%), Gaps = 4/237 (1%)
Query 20 FVREGVACGVRGDGRGRAELRPRVL-AVDLLPFSCSSAAIQTEENTVIATVDADVILPGG 78
F+R +A R DGR R + P++L ++ + SA + ++ VI T+ V P
Sbjct 3 FIRNALAANCRLDGR-RLDDNPKILIEPNVSTGAHGSAKVILDDTVVITTIMFHVTSPNT 61
Query 79 GGNQEPYSVSVGCGAA-EQELADVTGGDGSSLSSLLQAMLEQLLLPHLKPLTDPDGRLQW 137
+E +V + G+ E +D++ ++ +L+ + L + K L G+ W
Sbjct 62 NSPEEG-NVEITLGSPFTLEDSDISQKRDERMAHILEMLQFHKQLFNRKLLCILSGQYVW 120
Query 138 RISIDVIVLRAGGCLLDAVSLAIWGALKALKLPNVMVEEEDEKDQGLAITEYKVSCDERI 197
+ I VL+ GG ++DAVS+A+ A+ + +PN+ +DE + T ++ E
Sbjct 121 TMRIHTTVLQRGGGVMDAVSVAVICAMLTVSVPNIRGVIKDELESSRTATNVRLRTMEGY 180
Query 198 AAGTPYPIDTVPILITAAQIGDTYVWDLTEIEATCGDSFLVAAFLPSGACVGLQKFG 254
+P+L+T A+IG+++VW + + EA C D FL A P+G C G++ G
Sbjct 181 NTDIEKLAREIPVLVTVARIGESHVWSVNQEEAACADGFLTVAVFPNGNCAGIRAVG 237
> pfa:MAL13P1.204 exoribonuclease PH, putative; K12589 exosome
complex component RRP42
Length=278
Score = 94.4 bits (233), Expect = 5e-19, Method: Compositional matrix adjust.
Identities = 64/270 (23%), Positives = 126/270 (46%), Gaps = 12/270 (4%)
Query 20 FVREGVACGVRGDGRGRAELRPRVLAVDLLPFSCSSAAIQTEENTVIATVDADVILPGGG 79
++ + + C +R DGR R + ++L S+++ EEN VI + +I P
Sbjct 6 YIEDSINCNIRIDGRTLLTYRSIEINKNILVSGDGSSSVINEENNVICGIKLSLITPDVD 65
Query 80 GNQE-PYSVSVGCGAAEQELADVTGGDGSSLSSLLQAMLEQLLLPHL--KPLTDPDGRLQ 136
E ++ + C A+ A+ D L ++ + + ++ K L G+
Sbjct 66 TYDEGNVNLQIDCPASVA--ANRIKKD--HLQTMTTIIYNLCIKNNINKKILCILPGKFA 121
Query 137 WRISIDVIVLRAGGCLLDAVSLAIWGALKALKLPNVMVEEEDEKD---QGLAITEYKVSC 193
W + I+V+VL AGG LLD +S+AI+ ALK P V ++E E++ ++ +Y++
Sbjct 122 WSVDINVMVLNAGGGLLDIISIAIYVALKDTFFPIVKPKKEIEEENTFHEISNKDYQLEI 181
Query 194 DERIAAGTPYPIDTVPILITAAQIGDTYVWDLTEIEATCGDSFLVAAFLPSGACVGLQKF 253
E PY + +PI I+ +I + Y++D++++E + V A G CV K
Sbjct 182 VENEHVNFPY--ENIPICISIGEINNKYIYDMSKVEEELVQNIFVVAITSKGKCVAFHKL 239
Query 254 GHVLADCRTLQTTLTTSQRLSREILEDLQR 283
+ + ++ S ++S + + + +
Sbjct 240 YGISMEISSILNITENSSKISHTLFDKIHQ 269
> hsa:23016 EXOSC7, EAP1, FLJ26543, KIAA0116, RRP42, Rrp42p, hRrp42p,
p8; exosome component 7 (EC:3.1.13.-); K12589 exosome
complex component RRP42
Length=291
Score = 85.9 bits (211), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 69/281 (24%), Positives = 127/281 (45%), Gaps = 10/281 (3%)
Query 13 ISRAETAFVREGVACGVRGDGRGRAELRPRVLAVDLLPFSCSSAAIQTEENTVIATVDAD 72
+S AE ++ GV +R DGRG + R + D++ + SA ++ ++ V A+
Sbjct 6 LSEAEKVYIVHGVQEDLRVDGRGCEDYRCVEVETDVVSNTSGSARVKLGHTDILVGVKAE 65
Query 73 VILPGGGGNQEPY-SVSVGCGAAEQELADVTGGD--GSSLSSLLQAMLEQLLLPHLKPLT 129
+ P E Y V C A+ + GGD G+ +++ L + LK L
Sbjct 66 MGTPKLEKPNEGYLEFFVDCSASATPEFEGRGGDDLGTEIANTLYRIFNNKSSVDLKTLC 125
Query 130 DPDGRLQWRISIDVIVLRAGGCLLDAVSLAIWGALKALKLPNVMVEEEDEKDQGLAITEY 189
W + +DV++L GG L DA+S+A+ AL ++P V V E++E + + +++
Sbjct 126 ISPREHCWVLYVDVLLLECGGNLFDAISIAVKAALFNTRIPRVRVLEDEEGSKDIELSDD 185
Query 190 KVSCDERIAAGTPYPIDTVPILITAAQIGDTYVWDLTEIEATCGDSFLVAAFLPSGACVG 249
C ++ VP ++T +IG +V D T E C + L+ + G
Sbjct 186 PYDC-------IRLSVENVPCIVTLCKIGYRHVVDATLQEEACSLASLLVSVTSKGVVTC 238
Query 250 LQKFGHVLADCRTLQTTLTTSQRLSREILEDLQRQAEMKKS 290
++K G D ++ + T +R+ + + LQ ++S
Sbjct 239 MRKVGKGSLDPESIFEMMETGKRVGKVLHASLQSVVHKEES 279
> mmu:66446 Exosc7, 2610002K22Rik, AV212732, mKIAA0116; exosome
component 7; K12589 exosome complex component RRP42
Length=291
Score = 84.3 bits (207), Expect = 5e-16, Method: Compositional matrix adjust.
Identities = 68/274 (24%), Positives = 125/274 (45%), Gaps = 10/274 (3%)
Query 12 AISRAETAFVREGVACGVRGDGRGRAELRPRVLAVDLLPFSCSSAAIQTEENTVIATVDA 71
A+S AE ++ GV +R DGRG + R + D++ + SA ++ ++ V A
Sbjct 5 ALSEAEKVYIVHGVQEDLRVDGRGCEDYRCVEVETDVVSNTSGSARVKLGHTDILVGVKA 64
Query 72 DVILPGGGGNQEPY-SVSVGCGAAEQELADVTGGD--GSSLSSLLQAMLEQLLLPHLKPL 128
++ P E Y V C A + GGD G+ +++ L + L+ L
Sbjct 65 EMGTPKLEKPNEGYLEFFVDCSANATPEFEGRGGDDLGTEIANTLYRIFNNKSSVDLRSL 124
Query 129 TDPDGRLQWRISIDVIVLRAGGCLLDAVSLAIWGALKALKLPNVMVEEEDEKDQGLAITE 188
W + +DV++L GG L DA+S+A+ AL ++P V V E++E + + +++
Sbjct 125 CISPREHCWVLYVDVLLLECGGNLFDAISIAVKAALFNTRIPRVRVLEDEEGAKDIELSD 184
Query 189 YKVSCDERIAAGTPYPIDTVPILITAAQIGDTYVWDLTEIEATCGDSFLVAAFLPSGACV 248
C ++ VP ++T +IG +V D T E C + L+ + G
Sbjct 185 DPYDC-------IRLSVENVPCIVTLCKIGCRHVVDATLQEEACSLASLLVSVTSKGVVT 237
Query 249 GLQKFGHVLADCRTLQTTLTTSQRLSREILEDLQ 282
++K G D ++ + +S+R+ + + LQ
Sbjct 238 CMRKVGKGSLDPESIFEMMESSKRVGKVLHVSLQ 271
> dre:550247 zgc:110717; K12589 exosome complex component RRP42
Length=291
Score = 82.4 bits (202), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 70/286 (24%), Positives = 127/286 (44%), Gaps = 14/286 (4%)
Query 13 ISRAETAFVREGVACGVRGDGRGRAELRPRVLAVDLLPFSCSSAAIQTEENTVIATVDAD 72
+ AE ++ GV +R DGRG + R + D++ + SA I V+ + A+
Sbjct 6 VCEAEKVYIMHGVRDDLRLDGRGCEDYRHMEIETDVVSNTDGSAKISLGHTDVLVGIKAE 65
Query 73 VILPGGGGNQEPY-SVSVGCGAAEQELADVTGGD--GSSLSSLLQAMLEQLLLPHLKPLT 129
+ P E Y V C A + GG+ G LS+ L + L+ L
Sbjct 66 IGKPRPMVPDEGYLEFFVDCSANATPEFEGRGGEELGVELSNTLYKVFNNRHSLDLRSLC 125
Query 130 DPDGRLQWRISIDVIVLRAGGCLLDAVSLAIWGALKALKLPNVMVEEEDEKDQGLAITEY 189
G W + +DV++L+ G L DA+S+AI AL ++P V + E++E + + +++
Sbjct 126 ICPGENCWVLYVDVLLLQCDGNLFDAISIAIKAALFNTRIPKVHISEDEEGGKEIELSDD 185
Query 190 KVSCDERIAAGTPYPIDTVPILITAAQIGDTYVWDLTEIEATCGDSFLVAAFLPSGACVG 249
C ++ VP ++T +IG +V D T E C + L+ A +G
Sbjct 186 PFDC-------MRLNVENVPCIVTLCKIGHRHVVDATLQEKACSVASLLIAVTHTGMVTC 238
Query 250 LQKFGHVLADCRTLQTTLTTSQRLSRE----ILEDLQRQAEMKKSR 291
++K G D ++ +R+ + +++ LQ++ + K R
Sbjct 239 VRKMGGGSLDPESIFEMTEAGKRVGKSLHAPLMDLLQKEESLGKKR 284
> xla:446601 exosc7, MGC81766; exosome component 7; K12589 exosome
complex component RRP42
Length=291
Score = 77.4 bits (189), Expect = 6e-14, Method: Compositional matrix adjust.
Identities = 67/281 (23%), Positives = 125/281 (44%), Gaps = 10/281 (3%)
Query 13 ISRAETAFVREGVACGVRGDGRGRAELRPRVLAVDLLPFSCSSAAIQTEENTVIATVDAD 72
+S AE +V GV +R DGRG + R + D++ + SA ++ V+ V A+
Sbjct 6 LSEAEKVYVLHGVQDDLRTDGRGCEDYRCIEVESDVVSNTTGSARVKIGHTDVLVGVKAE 65
Query 73 VILPGGGGNQEPY-SVSVGCGAAEQELADVTGGD--GSSLSSLLQAMLEQLLLPHLKPLT 129
+ P E Y V C A + GG+ G+ ++ +L + + L+ L
Sbjct 66 MGTPKLERPGEGYLEFFVDCSANAAPEFEGRGGEELGTEIAFMLYKIFDNRSSIDLESLC 125
Query 130 DPDGRLQWRISIDVIVLRAGGCLLDAVSLAIWGALKALKLPNVMVEEEDEKDQGLAITEY 189
W + +DV++L GG L D +S+A+ AL ++P V V E++E + +++
Sbjct 126 IQAREHCWVLYVDVLLLECGGNLFDTISVAVKAALFNARIPKVRVLEDEEGGKDFELSDD 185
Query 190 KVSCDERIAAGTPYPIDTVPILITAAQIGDTYVWDLTEIEATCGDSFLVAAFLPSGACVG 249
C + VP ++T ++IG +V D T E C + L+ + G
Sbjct 186 PYDC-------VRLNVVNVPCIVTLSRIGHRHVVDATLQEEACSVASLLISVTSKGILTC 238
Query 250 LQKFGHVLADCRTLQTTLTTSQRLSREILEDLQRQAEMKKS 290
++K G D ++ + T + +++ + LQ ++S
Sbjct 239 MRKIGKGSLDPESIFEMIETGKHVAKSLHTSLQSALNQEES 279
> ath:AT3G07750 3' exoribonuclease family domain 1-containing
protein; K12589 exosome complex component RRP42
Length=286
Score = 77.0 bits (188), Expect = 8e-14, Method: Compositional matrix adjust.
Identities = 71/282 (25%), Positives = 129/282 (45%), Gaps = 19/282 (6%)
Query 13 ISRAETAFVREGVACGVRGDGRGRAELRPRVLAVDLLPFSCSSAAIQTEENTVIATVDAD 72
+S E F++ G+A +R DGR R RP + ++P + SA ++ VIA+V A+
Sbjct 3 LSLGEQHFIKGGIAQDLRTDGRKRLTYRPIYVETGVIPQANGSARVRIGGTDVIASVKAE 62
Query 73 VILPGG-GGNQEPYSVSVGCGAAEQELADVTGGDGSSLSSLLQAMLEQLLLP-------- 123
+ P ++ +V + C + GG+ LSS L L++ LL
Sbjct 63 IGRPSSLQPDKGKVAVFIDCSPTAEPTFGGRGGE--ELSSELALALQRCLLGGKSGAGAG 120
Query 124 -HLKPLTDPDGRLQWRISIDVIVLRAGGCLLDAVSLAIWGALKALKLPNVMVEEEDEKDQ 182
+L L +G++ W + ID +V+ + G LLDA+ AI AL +P V V E D+
Sbjct 121 INLSSLLIKEGKVCWDLYIDGLVISSDGNLLDALGAAIKAALTNTAIPKVNVSAEVADDE 180
Query 183 GLAITEYKVSCDERIAAGTPYPIDTVPILITAAQIGDTYVWDLTEIEATCGDSFLVAAFL 242
E +S +E + + +VP+++T ++G Y+ D T E + S + +
Sbjct 181 Q---PEIDISDEEYLQ----FDTSSVPVIVTLTKVGTHYIVDATAEEESQMSSAVSISVN 233
Query 243 PSGACVGLQKFGHVLADCRTLQTTLTTSQRLSREILEDLQRQ 284
+G GL K D + ++ ++ ++ ++ L +
Sbjct 234 RTGHICGLTKRSGSGLDPSVILDMISVAKHVTETLMSKLDSE 275
> cel:F31D4.1 exos-7; EXOSome (multiexonuclease complex) component
family member (exos-7); K12589 exosome complex component
RRP42
Length=312
Score = 64.3 bits (155), Expect = 6e-10, Method: Compositional matrix adjust.
Identities = 65/235 (27%), Positives = 109/235 (46%), Gaps = 19/235 (8%)
Query 13 ISRAETAFVREGVACGVRGDGRGRAELRPRVLAVDLLPFSCSSAAIQTEENT-VIATVDA 71
+S E F+ G +R DGR + R +L +L + S +Q + T V+A +
Sbjct 5 LSDDEKLFLITGAEANIRNDGRSCQDFRQIILERGVLSGTNGSCRVQLGQTTDVLAGIKL 64
Query 72 DVILPGGGGNQEP-----YSVSVGCGAAEQELADVTGGD--GSSLSSLLQAMLEQLL--L 122
++ +EP ++V A+ Q GGD LS+ QA + L +
Sbjct 65 ELESYDAETEKEPKQPINFNVDFSANASSQFAG--KGGDEYAEELSAAFQAAYSKALDIM 122
Query 123 PHLKPLTDPDGRLQWRISIDVIVLRAGGCLLDAVSLAIWGALKALKLPNVMVEEEDEKDQ 182
P+L DG +W+I++D+ VL+ G + DA+S+AI GAL L+ P V +D
Sbjct 123 PNLAKTQLADG-YRWKINVDISVLQWDGSVSDAISIAIIGALSDLEFPEGDVAPDDGGKV 181
Query 183 GLAITEYKVSC---DERIAAGTPYPIDTV--PILITAAQIGDTYVWDLTEIEATC 232
+ + KV+ DE + + +D + P+L+T ++IG + D T E C
Sbjct 182 RIVLKRPKVAGNVRDENVQVAM-WKLDVMRCPLLLTISKIGTANLVDCTPEEECC 235
> cel:F37C12.13 exos-9; EXOSome (multiexonuclease complex) component
family member (exos-9); K12586 exosome complex component
RRP43
Length=434
Score = 55.8 bits (133), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 67/237 (28%), Positives = 96/237 (40%), Gaps = 27/237 (11%)
Query 13 ISRAETAFVREGVACGVRGDGRGRAELRPRVLAVDLLPFSCSSAAIQTEENT-VIATVDA 71
++ E A + E + G R D R E R V L+ + AI T NT V+A V A
Sbjct 6 VTNCEKAVILEALKIGKRFDFRSLEEFRD----VKLVVGAEVGTAICTIGNTKVMAAVSA 61
Query 72 DVILPGGGGNQEPYS------VSVGCGAAEQELADVTGGDGSSLSSLLQAMLEQLLLPHL 125
++ P P+ V + A D G G L LL+ ++ +
Sbjct 62 EIAEPSS---MRPHKGVINIDVDLSPMANYANEHDRLGSKGMELIRLLELIIRDSRCIDV 118
Query 126 KPLTDPDGRLQWRISIDVIVLRAGGCLLDAVSLAIWGALKALKLPNVMVEEEDEKDQGLA 185
+ L G+ W+I +DV +L G LLD LA AL+ + PNV +E L
Sbjct 119 EALCIRAGKEIWKIRVDVRILDEDGSLLDCACLAAITALQHFRRPNVTLEPH----HTLI 174
Query 186 ITEYKVSCDERIAAGTPYPIDTVPILITAAQI--GDTYVWDLTEIEATCGDSFLVAA 240
+EY+ A P I +PI T + G V D T+ E C D +V A
Sbjct 175 YSEYE-------KAPVPLNIYHMPICTTIGLLDKGQIVVIDPTDKETACLDGSIVVA 224
> dre:323016 exosc8, MGC174638, wu:fb79a10, zgc:110381; exosome
component 8; K12586 exosome complex component RRP43
Length=277
Score = 55.8 bits (133), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 47/183 (25%), Positives = 81/183 (44%), Gaps = 14/183 (7%)
Query 19 AFVREGVACGVRGDGRGRAELRPRVLAVDLLPFSCSSAAIQTEENTVIATVDADVILPGG 78
+F++E R DGR E RP L ++ + + SA ++ TVI + A++ +P
Sbjct 16 SFLKENC----RPDGRELGEFRPTTLNINSISTADGSALVKIGNTTVICGIKAELAVPPS 71
Query 79 GGNQEPY---SVSVGCGAAEQELADVTGGDGSSLSSLLQAMLEQLLLPHLKPLTDPDGRL 135
+ Y +V + + + A G + S + ++E L +L+ L +L
Sbjct 72 EAQNKGYLVPNVDLPPLCSSRFRAGPPGEQAQAASQFIADVIESSDLINLEELCIEKAKL 131
Query 136 QWRISIDVIVLRAGGCLLDAVSLAIWGALKALKLPNVMVEEED-------EKDQGLAITE 188
W + D++ L G LLDA + ALK +LP V + +E EK + L I
Sbjct 132 CWVLYCDIMCLDYDGNLLDACITVLLAALKTAQLPEVTINKETDLAEVDIEKKRRLKINR 191
Query 189 YKV 191
+ V
Sbjct 192 HPV 194
> hsa:11340 EXOSC8, CIP3, EAP2, OIP2, RP11-421P11.3, RRP43, Rrp43p,
bA421P11.3, p9; exosome component 8 (EC:3.1.13.-); K12586
exosome complex component RRP43
Length=276
Score = 50.1 bits (118), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 40/161 (24%), Positives = 70/161 (43%), Gaps = 3/161 (1%)
Query 20 FVREGVACGVRGDGRGRAELRPRVLAVDLLPFSCSSAAIQTEENTVIATVDADVILPGGG 79
+ R + R DGR E R + + + + SA ++ TVI V A+ P
Sbjct 13 YYRRFLKENCRPDGRELGEFRTTTVNIGSISTADGSALVKLGNTTVICGVKAEFAAPSTD 72
Query 80 GNQEPY---SVSVGCGAAEQELADVTGGDGSSLSSLLQAMLEQLLLPHLKPLTDPDGRLQ 136
+ Y +V + + + + G + S + ++E + + L G+L
Sbjct 73 APDKGYVVPNVDLPPLCSSRFRSGPPGEEAQVASQFIADVIENSQIIQKEDLCISPGKLV 132
Query 137 WRISIDVIVLRAGGCLLDAVSLAIWGALKALKLPNVMVEEE 177
W + D+I L G +LDA + A+ ALK ++LP V + EE
Sbjct 133 WVLYCDLICLDYDGNILDACTFALLAALKNVQLPEVTINEE 173
> xla:496046 hypothetical LOC496046; K12586 exosome complex component
RRP43
Length=276
Score = 49.3 bits (116), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 47/181 (25%), Positives = 77/181 (42%), Gaps = 10/181 (5%)
Query 30 RGDGRGRAELRPRVLAVDLLPFSCSSAAIQTEENTVIATVDADVILPGGGGNQEPY---S 86
R DGR E R + V + + SA ++ TV+ V A+ P + Y +
Sbjct 23 RPDGRELTEFRTTTINVGSITTADGSALVKLGNCTVLCGVKAEFAAPPADAPNKGYVVPN 82
Query 87 VSVGCGAAEQELADVTGGDGSSLSSLLQAMLEQLLLPHLKPLTDPDGRLQWRISIDVIVL 146
V + + Q G + S + +++ + + L G+L W + D+I L
Sbjct 83 VDLSPLCSSQFRPGPPGEEAQVASQFMADVIDNSQMLLKEDLCIRHGKLAWVLYCDLICL 142
Query 147 RAGGCLLDAVSLAIWGALKALKLPNVMVEEEDEKDQGLAITEYKVSCDERIAAGTPYPID 206
G LLD + A+ AL+ ++LP+V + EE GLA K C +I +PI
Sbjct 143 DYDGNLLDTCTCALVAALQNVRLPSVKINEE----TGLAEVNLKTKCALKILK---HPIA 195
Query 207 T 207
T
Sbjct 196 T 196
> xla:379078 exosc8, MGC132146, MGC52847; exosome component 8;
K01149 [EC:3.1.13.-]
Length=276
Score = 48.1 bits (113), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 48/181 (26%), Positives = 79/181 (43%), Gaps = 11/181 (6%)
Query 20 FVREGVACGVRGDGRGRAELRPRVLAVDLLPFSCSSAAIQTEENTVIATVDADVILPGGG 79
F++E AC R DGR E R + V + + SA ++ TV+ V A++ P
Sbjct 17 FLKE--AC--RPDGRELTEFRTTTINVGSITTADGSALVKLGNCTVLCGVKAELAAPPAD 72
Query 80 GNQEPY---SVSVGCGAAEQELADVTGGDGSSLSSLLQAMLEQLLLPHLKPLTDPDGRLQ 136
+ Y +V + + Q G + S + +++ + + L G+L
Sbjct 73 SPDKGYVVPNVDLTPLCSSQFRPGPPGEEAQVASQFMADVIDNSQMLLKEDLCIAHGKLA 132
Query 137 WRISIDVIVLRAGGCLLDAVSLAIWGALKALKLPNVMVEEEDEKDQGLAITEYKVSCDER 196
W + D+I L G LLD A+ AL+ ++LP+V + EE GLA K C +
Sbjct 133 WVLYCDLICLDYDGNLLDTCMCALVAALQNVRLPSVKINEE----TGLAEVNLKTKCALK 188
Query 197 I 197
I
Sbjct 189 I 189
> mmu:69639 Exosc8, 2310032N20Rik, CIP3, KIAA4013, mKIAA4013;
exosome component 8; K12586 exosome complex component RRP43
Length=280
Score = 45.1 bits (105), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 39/165 (23%), Positives = 67/165 (40%), Gaps = 7/165 (4%)
Query 20 FVREGVACGVRGDGRGRAELRPRVLAVDLLPFSCSSAAIQTEENTVIATVDADVILPGGG 79
+ R + R DGR E R + + + + SA ++ TVI V A+ P
Sbjct 13 YYRRFLKENCRPDGRELGEFRATTVNIGSISTADGSALVKLGNTTVICGVKAEFAAPPVD 72
Query 80 GNQEPYSVSVGCG-------AAEQELADVTGGDGSSLSSLLQAMLEQLLLPHLKPLTDPD 132
Y V + + + G + S + +++ + + L
Sbjct 73 APDRGYVVFLSVPNVDLPPLCSSRFRTGPPGEEAQVTSQFIADVVDNSQVIKKEDLCISP 132
Query 133 GRLQWRISIDVIVLRAGGCLLDAVSLAIWGALKALKLPNVMVEEE 177
G+L W + D+I L G +LDA + A+ ALK ++LP V + EE
Sbjct 133 GKLAWVLYCDLICLDYDGNILDACTFALLAALKNVQLPEVTINEE 177
> cpv:cgd7_230 RPR45-like archaeo-eukaryotic exosomal RNAse PH
; K01149 [EC:3.1.13.-]
Length=291
Score = 38.1 bits (87), Expect = 0.038, Method: Compositional matrix adjust.
Identities = 48/224 (21%), Positives = 95/224 (42%), Gaps = 11/224 (4%)
Query 28 GVRGDGRGRAELRPRVLAVDLLPFSCSSA-AIQTEENTVIATVDADVILPG--GGGNQEP 84
GVR DGR R L ++ L S A +++ ++ IA +I G N
Sbjct 22 GVRCDGRKPEVCRNFKLELNELSSSNYGAVSVRIGNSSYIACCSPQIIHTGLDQESNNAI 81
Query 85 YSVSVGCGAAEQELADVTGGDGSS---LSSLLQAMLEQLLLPHLKPLTDPDGR-LQWRIS 140
+ + + TG S +S+ + + + ++ +T G+ L W I
Sbjct 82 TNTRINVSLELPTICGFTGNPNHSNFVSNSITECLNDSRIINKEAFITKFQGKELHWLID 141
Query 141 IDVIVLRAGGCLLDAVSLAIWGALKALKLPNVMVEEEDEKDQGLA---ITEYKVSCDERI 197
I V+ L G LD +A +L+ LP +V ++D ++ I++ +V ++ +
Sbjct 142 IKVVCLSYDGNPLDYTLIASIASLRNTILPKNLVWDDDYNWFRISSDPISKDEVKREQMM 201
Query 198 AAGTPYPIDTVPILITAAQIGDT-YVWDLTEIEATCGDSFLVAA 240
+ + + VPI ++ + + D+ +V D +E + G S +
Sbjct 202 QDSSSFFLKKVPIFVSFSYLNDSIWVCDPNYMEDSIGSSVTICC 245
> ath:AT3G12990 RRP45a; RRP45a (Ribonuclease PH45a); 3'-5'-exoribonuclease/
RNA binding
Length=307
Score = 37.7 bits (86), Expect = 0.054, Method: Compositional matrix adjust.
Identities = 66/295 (22%), Positives = 114/295 (38%), Gaps = 29/295 (9%)
Query 17 ETAFVREGVACGVRGDGRGRAELRPRVLAVDLLPFSCSSAAIQTEENTVIATVDADVILP 76
E+ FV + +R DGRG + R + S+ +Q + V+A V A ++ P
Sbjct 15 ESKFVESALQSELRVDGRGLYDYRKLTIKFGK---EYGSSQVQLGQTHVMAFVTAQLVQP 71
Query 77 GGGGNQEPYSVSVGCGAAEQELADVT---GGDGSS---LSSLLQAMLEQLLLPHLKPLTD 130
P S +AD + G G S L ++ L + + L
Sbjct 72 Y---KDRPSEGSFSIFTEFSPMADPSFEPGHPGESAVELGRIIDRALRESRAVDTESLCV 128
Query 131 PDGRLQWRISIDVIVLRAGGCLLDAVSLAIWGALKALKLPNVMVEEEDEKDQGLAITEYK 190
G+L W + ID+ +L GG L+DA ++A AL + P+ V ++ +D + E +
Sbjct 129 LAGKLVWSVRIDLHILDNGGNLVDAANVAALAALMTFRRPDCTVGGDNSQDVIIHPPEER 188
Query 191 VSCDERIAAGTPYPIDTVPILITAA--QIGDTYVWDLTEIEATCGDSFLVAAFLPSGACV 248
P I +PI T G V D T +E + +G
Sbjct 189 --------EPLPLIIHHLPIAFTFGFFNKGSILVMDPTYVEEAVMCGRMTVTVNANGDIC 240
Query 249 GLQKFGH------VLADC-RTLQTTLTTSQRLSREILEDLQRQAEMKKSRLNGNL 296
+QK G V+ C R + + + ++ R+ +E R+ +K + + L
Sbjct 241 AIQKPGEEGVNQSVILHCLRLASSRASATTKIIRDAVEAYNRERSSQKVKRHHTL 295
> hsa:56957 OTUD7B, CEZANNE, ZA20D1; OTU domain containing 7B
(EC:3.4.19.12); K11860 OTU domain-containing protein 7 [EC:3.-.-.-]
Length=843
Score = 34.3 bits (77), Expect = 0.56, Method: Compositional matrix adjust.
Identities = 24/74 (32%), Positives = 36/74 (48%), Gaps = 12/74 (16%)
Query 97 ELADVTGGDGSSLSSLLQAMLEQLLLPHLKPLTDPDGRLQWRISIDVIVLR--------A 148
+L D+T + S + + ++EQ +L L+ GRL W +S+D R
Sbjct 136 QLPDLTVYNEDFRSFIERDLIEQSMLVALEQA----GRLNWWVSVDPTSQRLLPLATTGD 191
Query 149 GGCLLDAVSLAIWG 162
G CLL A SL +WG
Sbjct 192 GNCLLHAASLGMWG 205
> mmu:229603 Otud7b, 2900060B22Rik, 4930463P07Rik, AI462125, CEZANNE,
Trabid2, Za20d1; OTU domain containing 7B; K11860 OTU
domain-containing protein 7 [EC:3.-.-.-]
Length=840
Score = 33.5 bits (75), Expect = 0.90, Method: Compositional matrix adjust.
Identities = 16/38 (42%), Positives = 19/38 (50%), Gaps = 8/38 (21%)
Query 133 GRLQWRISIDVIVLR--------AGGCLLDAVSLAIWG 162
GRL W +S+D R G CLL A SL +WG
Sbjct 168 GRLNWWVSMDSTCQRLLPLATTGDGNCLLHAASLGMWG 205
> dre:450057 exosc9, im:6909054, zgc:101680; exosome component
9; K03678 exosome complex component RRP45
Length=393
Score = 32.7 bits (73), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 37/127 (29%), Positives = 54/127 (42%), Gaps = 13/127 (10%)
Query 133 GRLQWRISIDVIVLRAGGCLLDAVSLAIWGALKALKLPNVMVEEEDEKDQGLAITEYKVS 192
G W+I +DV VL G L+DA S+A AL + P+V + QG +T +
Sbjct 126 GEKVWQIRVDVHVLNHDGNLMDAASIAAISALSHFRRPDVAI-------QGRDVTVF--G 176
Query 193 CDERIAAGTPYPIDTVPILITAAQI--GDTYVWDLTEIEATCGDSFLVAAFLPSGACVGL 250
+ER P I +PI ++ A G + D E E D LV A +
Sbjct 177 PEER--DPIPLSIYHMPICVSFAFFLQGSFLLVDPCEREERVKDGLLVIAMNKHREICSI 234
Query 251 QKFGHVL 257
Q G ++
Sbjct 235 QSSGGIM 241
> mmu:237558 Gm239; predicted gene 239
Length=904
Score = 31.6 bits (70), Expect = 4.2, Method: Compositional matrix adjust.
Identities = 21/81 (25%), Positives = 34/81 (41%), Gaps = 16/81 (19%)
Query 42 RVLAVDLLPFSCSSAAIQTEENTVIATVDADVILPGGGGNQEPYSVSVGCGAAEQELADV 101
R++ D P S+ I T T + + ILP A +E+ DV
Sbjct 470 RIVQYDYKPEFASAMGINTAHQTGMIAQEVQEILP----------------RAVREVGDV 513
Query 102 TGGDGSSLSSLLQAMLEQLLL 122
TGG+G +L + L +Q+ +
Sbjct 514 TGGNGETLENFLMVDKDQIFM 534
> cel:F41G3.14 exos-8; EXOSome (multiexonuclease complex) component
family member (exos-8)
Length=277
Score = 31.2 bits (69), Expect = 4.9, Method: Compositional matrix adjust.
Identities = 17/56 (30%), Positives = 30/56 (53%), Gaps = 0/56 (0%)
Query 135 LQWRISIDVIVLRAGGCLLDAVSLAIWGALKALKLPNVMVEEEDEKDQGLAITEYK 190
L+W++ I V VL G LDA A+ AL KLP++++ + + + T+ +
Sbjct 124 LEWQLHIIVKVLSLEGNFLDAAVCALGAALVDAKLPSIVLNHSEGDESTIEKTQIQ 179
> ath:AT3G60500 CER7; CER7 (ECERIFERUM 7); 3'-5'-exoribonuclease/
RNA binding
Length=438
Score = 30.8 bits (68), Expect = 6.1, Method: Compositional matrix adjust.
Identities = 65/302 (21%), Positives = 117/302 (38%), Gaps = 38/302 (12%)
Query 17 ETAFVREGVACGVRGDGRGRAELRPRVLAVDLLPFSCSSAAIQTEENTVIATVDADVILP 76
E+ FV + +R DGRG + R + S+ +Q + V+ V A ++ P
Sbjct 15 ESKFVETALQSELRVDGRGLYDYRKLTIK---FGKEYGSSEVQLGQTHVMGFVTAQLVQP 71
Query 77 GGGGNQEPYSVSVGCGAAEQELADVT------GGDGSSLSSLLQAMLEQLLLPHLKPLTD 130
P S+ +AD + G L ++ L + + L
Sbjct 72 Y---KDRPNEGSLSIFTEFSPMADPSFEPGRPGESAVELGRIIDRGLRESRAVDTESLCV 128
Query 131 PDGRLQWRISIDVIVLRAGGCLLDAVSLAIWGALKALKLPNVMVEEEDEKDQGLAITEYK 190
G++ W + ID+ +L GG L+DA ++A AL + P+ V E+ G + +
Sbjct 129 LAGKMVWSVRIDLHILDNGGNLVDAANIAALAALMTFRRPDCTVGGEN----GQEVIIHP 184
Query 191 VSCDERIAAGTPYPIDTVPILITAA--QIGDTYVWDLTEIEATCGDSFLVAAFLPSGACV 248
+ E + P I +PI T G+ V D T +E + +G
Sbjct 185 LEEREPL----PLIIHHLPIAFTFGFFNKGNIVVMDPTYVEEAVMCGRMTVTVNANGDIC 240
Query 249 GLQKFGH------VLADCRTLQTT--LTTSQRLSREI--------LEDLQRQAEMKKSRL 292
+QK G V+ C L ++ T++ + E+ L+ ++R + KS +
Sbjct 241 AIQKPGEEGVNQSVILHCLRLASSRAAATTKIIREEVEAYNCERSLQKVKRHPTLAKSEV 300
Query 293 NG 294
+G
Sbjct 301 SG 302
> dre:567622 eif2c1; eukaryotic translation initiation factor
2C, 1; K11593 eukaryotic translation initiation factor 2C
Length=895
Score = 30.8 bits (68), Expect = 6.5, Method: Compositional matrix adjust.
Identities = 13/23 (56%), Positives = 15/23 (65%), Gaps = 0/23 (0%)
Query 67 ATVDADVILPGGGGNQEPYSVSV 89
A VD DV LPG GG P+ VS+
Sbjct 124 AGVDLDVTLPGEGGKDRPFKVSI 146
> mmu:214150 Eif2c3, AW048688, Ago3, C130014L07Rik; eukaryotic
translation initiation factor 2C, 3; K11593 eukaryotic translation
initiation factor 2C
Length=860
Score = 30.8 bits (68), Expect = 6.6, Method: Compositional matrix adjust.
Identities = 18/45 (40%), Positives = 22/45 (48%), Gaps = 2/45 (4%)
Query 47 DLLPFSCSSAAIQTEENTVIAT--VDADVILPGGGGNQEPYSVSV 89
D P ++ T +AT VD DV LPG GG P+ VSV
Sbjct 81 DRRPVYDGKRSLYTANPLPVATTGVDLDVTLPGEGGKDRPFKVSV 125
> hsa:192669 EIF2C3, AGO3, FLJ12765, MGC86946; eukaryotic translation
initiation factor 2C, 3; K11593 eukaryotic translation
initiation factor 2C
Length=860
Score = 30.4 bits (67), Expect = 9.5, Method: Compositional matrix adjust.
Identities = 17/45 (37%), Positives = 22/45 (48%), Gaps = 2/45 (4%)
Query 47 DLLPFSCSSAAIQTEENTVIAT--VDADVILPGGGGNQEPYSVSV 89
D P ++ T +AT VD DV LPG GG P+ VS+
Sbjct 81 DRRPVYDGKRSLYTANPLPVATTGVDLDVTLPGEGGKDRPFKVSI 125
Lambda K H
0.318 0.135 0.391
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 12284032348
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40