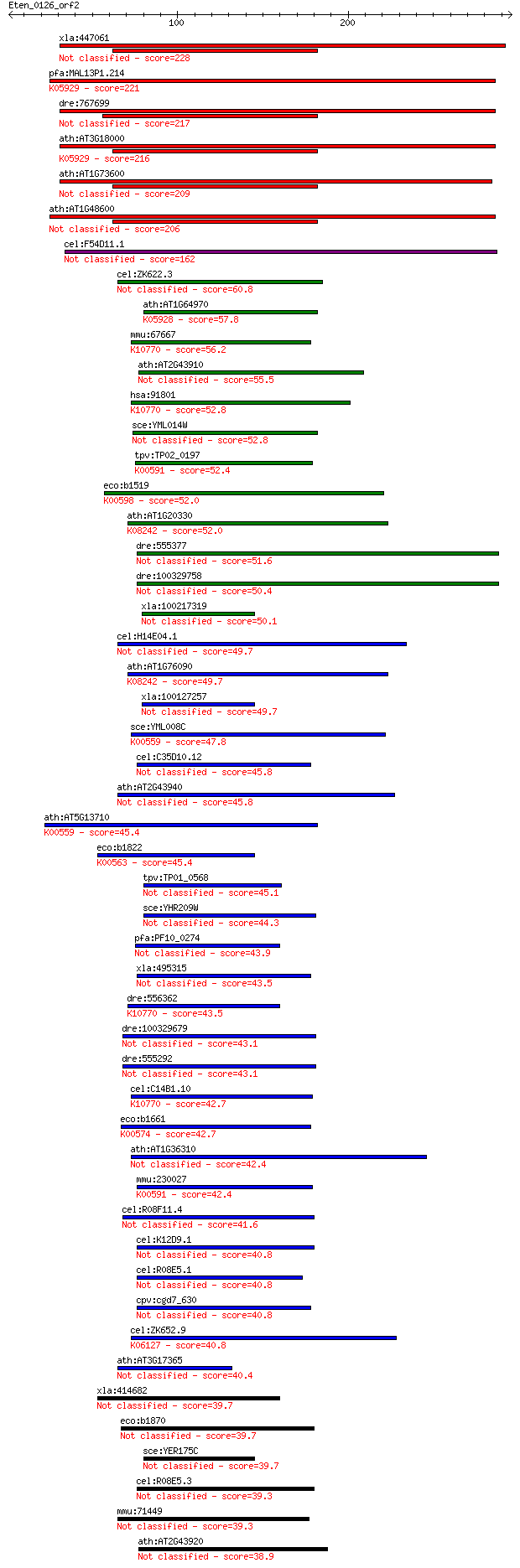
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_0126_orf2
Length=295
Score E
Sequences producing significant alignments: (Bits) Value
xla:447061 MGC83638 protein 228 2e-59
pfa:MAL13P1.214 PfPMT; phosphoethanolamine N-methyltransferase... 221 2e-57
dre:767699 pmt, MGC153034, zgc:153034; phosphoethanolamine met... 217 3e-56
ath:AT3G18000 XPL1; XPL1 (XIPOTL 1); methyltransferase/ phosph... 216 5e-56
ath:AT1G73600 methyltransferase/ phosphoethanolamine N-methylt... 209 1e-53
ath:AT1G48600 phosphoethanolamine N-methyltransferase 2, putat... 206 1e-52
cel:F54D11.1 pmt-2; Phosphoethanolamine MethyTransferase famil... 162 2e-39
cel:ZK622.3 pmt-1; Phosphoethanolamine MethyTransferase family... 60.8 6e-09
ath:AT1G64970 G-TMT; G-TMT (GAMMA-TOCOPHEROL METHYLTRANSFERASE... 57.8 5e-08
mmu:67667 Alkbh8, 4930562C03Rik, 8030431D03Rik, 9430088N01Rik,... 56.2 1e-07
ath:AT2G43910 thiol methyltransferase, putative 55.5 2e-07
hsa:91801 ALKBH8, ABH8, FLJ38204, MGC10235; alkB, alkylation r... 52.8 1e-06
sce:YML014W TRM9, KTI1; Trm9p (EC:2.1.1.-) 52.8 1e-06
tpv:TP02_0197 hexaprenyldihydroxybenzoate methyltransferase (E... 52.4 2e-06
eco:b1519 tam, ECK1512, JW1512, lsrE, yneD; trans-aconitate me... 52.0 3e-06
ath:AT1G20330 SMT2; SMT2 (STEROL METHYLTRANSFERASE 2); S-adeno... 52.0 3e-06
dre:555377 zgc:162780 51.6 4e-06
dre:100329758 hypothetical protein LOC100329758 50.4 8e-06
xla:100217319 hypothetical protein LOC100217319 50.1 1e-05
cel:H14E04.1 hypothetical protein 49.7 1e-05
ath:AT1G76090 SMT3; SMT3 (STEROL METHYLTRANSFERASE 3); S-adeno... 49.7 1e-05
xla:100127257 hypothetical protein LOC100127257 49.7 1e-05
sce:YML008C ERG6, ISE1, LIS1, SED6, VID1; Delta(24)-sterol C-m... 47.8 5e-05
cel:C35D10.12 hypothetical protein 45.8 2e-04
ath:AT2G43940 thiol methyltransferase, putative 45.8 2e-04
ath:AT5G13710 SMT1; SMT1 (STEROL METHYLTRANSFERASE 1); sterol ... 45.4 2e-04
eco:b1822 rlmA, ECK1820, JW1811, rrmA, yebH; 23S rRNA m(1)G745... 45.4 3e-04
tpv:TP01_0568 hypothetical protein 45.1 3e-04
sce:YHR209W CRG1; Crg1p 44.3 5e-04
pfa:PF10_0274 methyltransferase, putative 43.9 6e-04
xla:495315 hypothetical LOC495315 43.5 0.001
dre:556362 K1456 protein-like; K10770 alkylated DNA repair pro... 43.5 0.001
dre:100329679 hexaprenyldihydroxybenzoate methyltransferase, m... 43.1 0.001
dre:555292 fb62c07, si:ch211-31o18.2, wu:fb62c07; zgc:162396 43.1 0.001
cel:C14B1.10 hypothetical protein; K10770 alkylated DNA repair... 42.7 0.001
eco:b1661 cfa, cdfA, ECK1657, JW1653; cyclopropane fatty acyl ... 42.7 0.001
ath:AT1G36310 methyltransferase 42.4 0.002
mmu:230027 Coq3, 4732433J24, C77934; coenzyme Q3 homolog, meth... 42.4 0.002
cel:R08F11.4 hypothetical protein 41.6 0.004
cel:K12D9.1 hypothetical protein 40.8 0.006
cel:R08E5.1 hypothetical protein 40.8 0.006
cpv:cgd7_630 Ym1014wp-like, Ymb4 methylase 40.8 0.006
cel:ZK652.9 coq-5; COenzyme Q (ubiquinone) biosynthesis family... 40.8 0.006
ath:AT3G17365 catalytic/ methyltransferase 40.4 0.007
xla:414682 mettl13; methyltransferase like 13 39.7
eco:b1870 cmoA, ECK1871, JW1859, yecO; tRNA cmo(5)U34 methyltr... 39.7 0.013
sce:YER175C TMT1, TAM1; Tmt1p 39.7 0.014
cel:R08E5.3 hypothetical protein 39.3 0.017
mmu:71449 Mettl13, 5630401D24Rik; methyltransferase like 13 39.3
ath:AT2G43920 thiol methyltransferase, putative 38.9 0.023
> xla:447061 MGC83638 protein
Length=494
Score = 228 bits (582), Expect = 2e-59, Method: Compositional matrix adjust.
Identities = 120/263 (45%), Positives = 168/263 (63%), Gaps = 5/263 (1%)
Query 31 QQALDSHQYSKNGILRYEFIFGRGFVSSGGGETTAEILREIVLPKGGKAIDVGCGIGGST 90
QQ LD+ QYS+ GILRYE IFG GFVS+GG ETT E + + L G + IDVGCGIGG
Sbjct 235 QQFLDNQQYSRRGILRYEKIFGEGFVSTGGLETTKEFISMLNLRPGQRVIDVGCGIGGGD 294
Query 91 AALADKFNANVLGVDLSSNMISIAKERYSSR--PDLKFLVADALSIPIDSESVDLVYSRD 148
+A + VLG+DLSSNM+ IA ER + P ++F + DA S D+VYSRD
Sbjct 295 FYMAKTYGVEVLGMDLSSNMVEIAMERAFTEKTPLVQFEIGDATRRCFSEGSFDVVYSRD 354
Query 149 TILHLSVDEKKLLFSKAFDWLKPDGQLVITDYCCGPPEKWDDEFKAYLQDRNYKLVQLEE 208
TILH +++K+ LF + + W+KP G+L+ITDYCCG W F+ Y++ R Y L +E
Sbjct 355 TILH--INDKEALFRRFYSWIKPGGKLLITDYCCG-ERPWAPVFQEYVKQRGYILYTPQE 411
Query 209 YRQLLTEAGFDVVKAANHTQRWLKSLDEESQRLEEQKEDFMHLFTLKDFQDLRDGWQSKK 268
Y Q L +AGF V+A + T++++ L+ E R + K+ F+ F+ +D++ + DGW+ K+
Sbjct 412 YGQFLEKAGFVNVQAQDRTEQFVNVLNTELSRTRDIKQQFIENFSEEDYKYIIDGWKEKQ 471
Query 269 ERVAKGLQFWGLFIAFKPAEGVS 291
R + G Q WGLF A KP E +S
Sbjct 472 HRCSLGDQRWGLFYAEKPLEILS 494
Score = 60.5 bits (145), Expect = 8e-09, Method: Compositional matrix adjust.
Identities = 37/120 (30%), Positives = 57/120 (47%), Gaps = 1/120 (0%)
Query 62 ETTAEILREIVLPKGGKAIDVGCGIGGSTAALADKFNANVLGVDLSSNMISIAKERYSSR 121
E EI+ + G +++G G+G T LA K ++V VD N I +E R
Sbjct 37 EEKPEIILLLPCLDGLSVLELGAGMGRYTGHLA-KLASHVTAVDFMPNFIEKNREDNGFR 95
Query 122 PDLKFLVADALSIPIDSESVDLVYSRDTILHLSVDEKKLLFSKAFDWLKPDGQLVITDYC 181
++ FL AD ++ + ES D ++S ++L+ E L K WLKP G L + C
Sbjct 96 GNITFLQADVTNLDLPKESFDFIFSNWLFMYLTDAELVALTQKLLAWLKPGGYLFFRESC 155
> pfa:MAL13P1.214 PfPMT; phosphoethanolamine N-methyltransferase
(EC:2.1.1.103); K05929 phosphoethanolamine N-methyltransferase
[EC:2.1.1.103]
Length=266
Score = 221 bits (564), Expect = 2e-57, Method: Compositional matrix adjust.
Identities = 107/261 (40%), Positives = 158/261 (60%), Gaps = 0/261 (0%)
Query 25 EDIKKRQQALDSHQYSKNGILRYEFIFGRGFVSSGGGETTAEILREIVLPKGGKAIDVGC 84
E++ + L+++QY+ G+ YEFIFG ++SSGG E T +IL +I L + K +D+G
Sbjct 5 ENLNSDKTFLENNQYTDEGVKVYEFIFGENYISSGGLEATKKILSDIELNENSKVLDIGS 64
Query 85 GIGGSTAALADKFNANVLGVDLSSNMISIAKERYSSRPDLKFLVADALSIPIDSESVDLV 144
G+GG + +K+ A+ G+D+ SN++++A ER S + F D L+ + DL+
Sbjct 65 GLGGGCMYINEKYGAHTHGIDICSNIVNMANERVSGNNKIIFEANDILTKEFPENNFDLI 124
Query 145 YSRDTILHLSVDEKKLLFSKAFDWLKPDGQLVITDYCCGPPEKWDDEFKAYLQDRNYKLV 204
YSRD ILHLS++ K LF K + WLKP G L+ITDYC E WDDEFK Y++ R Y L+
Sbjct 125 YSRDAILHLSLENKNKLFQKCYKWLKPTGTLLITDYCATEKENWDDEFKEYVKQRKYTLI 184
Query 205 QLEEYRQLLTEAGFDVVKAANHTQRWLKSLDEESQRLEEQKEDFMHLFTLKDFQDLRDGW 264
+EEY +LT F V + + + W + L+ E + L E KE+F+ LF+ K F L DGW
Sbjct 185 TVEEYADILTACNFKNVVSKDLSDYWNQLLEVEHKYLHENKEEFLKLFSEKKFISLDDGW 244
Query 265 QSKKERVAKGLQFWGLFIAFK 285
K + + +Q WG F A K
Sbjct 245 SRKIKDSKRKMQRWGYFKATK 265
> dre:767699 pmt, MGC153034, zgc:153034; phosphoethanolamine methyltransferase
Length=489
Score = 217 bits (553), Expect = 3e-56, Method: Compositional matrix adjust.
Identities = 114/257 (44%), Positives = 160/257 (62%), Gaps = 5/257 (1%)
Query 31 QQALDSHQYSKNGILRYEFIFGRGFVSSGGGETTAEILREIVLPKGGKAIDVGCGIGGST 90
+Q LD+ QY++ GILRYE +FG GFVS+GG +TT E + + L G K +DVGCGIGG
Sbjct 235 RQFLDNQQYTRRGILRYEKMFGCGFVSTGGLQTTKEFVDMLNLSAGQKVLDVGCGIGGGD 294
Query 91 AALADKFNANVLGVDLSSNMISIAKERYSSR--PDLKFLVADALSIPIDSESVDLVYSRD 148
+A F VLG+DLSSNM+ IA ER P ++F V+DA + D+VYSRD
Sbjct 295 FYMAKTFGVEVLGMDLSSNMVEIAMERAVKEKLPLVQFEVSDATKRRFPDAAFDVVYSRD 354
Query 149 TILHLSVDEKKLLFSKAFDWLKPDGQLVITDYCCGPPEKWDDEFKAYLQDRNYKLVQLEE 208
TILH + +K LF+ + W+KP G+L+I+DYCCG + W F+ Y++ R Y L +
Sbjct 355 TILH--IRDKLHLFTNFYSWMKPGGKLLISDYCCG-EKPWSPAFQDYVKQRGYILYTPQR 411
Query 209 YRQLLTEAGFDVVKAANHTQRWLKSLDEESQRLEEQKEDFMHLFTLKDFQDLRDGWQSKK 268
Y Q L E GF V+A + T+++++ + E QR EE K++F+ F+ +DF + GW K
Sbjct 412 YGQFLREVGFSNVRAEDRTEQFIQVIKSELQRAEEMKDEFIQEFSKEDFDAIVSGWTEKL 471
Query 269 ERVAKGLQFWGLFIAFK 285
+R G Q WGLF A K
Sbjct 472 QRCETGDQRWGLFYATK 488
Score = 50.4 bits (119), Expect = 8e-06, Method: Compositional matrix adjust.
Identities = 37/130 (28%), Positives = 60/130 (46%), Gaps = 5/130 (3%)
Query 56 VSSGGGETTAEILREIV--LPKGGKA--IDVGCGIGGSTAALADKFNANVLGVDLSSNMI 111
+ S E T L EI+ LP ++ +++G GIG T L + +V VD +
Sbjct 19 LDSHAQELTQHELPEILDLLPALSESCVLELGAGIGRYTKHLIGR-ARHVTAVDFMEKFV 77
Query 112 SIAKERYSSRPDLKFLVADALSIPIDSESVDLVYSRDTILHLSVDEKKLLFSKAFDWLKP 171
++ ++F+ AD + S DLV+S +++LS E +LL K WL+P
Sbjct 78 EKNRQDNGHLGSVEFIQADVTKLDFPEHSFDLVFSNWLLMYLSDQELQLLAEKFLRWLRP 137
Query 172 DGQLVITDYC 181
G L + C
Sbjct 138 GGFLFFRESC 147
> ath:AT3G18000 XPL1; XPL1 (XIPOTL 1); methyltransferase/ phosphoethanolamine
N-methyltransferase (EC:2.1.1.103); K05929 phosphoethanolamine
N-methyltransferase [EC:2.1.1.103]
Length=491
Score = 216 bits (551), Expect = 5e-56, Method: Compositional matrix adjust.
Identities = 117/256 (45%), Positives = 164/256 (64%), Gaps = 4/256 (1%)
Query 31 QQALDSHQYSKNGILRYEFIFGRGFVSSGGGETTAEILREIVLPKGGKAIDVGCGIGGST 90
Q+ LD+ QY +GILRYE +FG+GFVS+GG ETT E + ++ L G K +DVGCGIGG
Sbjct 238 QRFLDNVQYKSSGILRYERVFGQGFVSTGGLETTKEFVEKMNLKPGQKVLDVGCGIGGGD 297
Query 91 AALADKFNANVLGVDLSSNMISIAKER-YSSRPDLKFLVADALSIPIDSESVDLVYSRDT 149
+A+KF+ +V+G+DLS NMIS A ER ++F VAD + S D++YSRDT
Sbjct 298 FYMAEKFDVHVVGIDLSVNMISFALERAIGLSCSVEFEVADCTTKHYPDNSFDVIYSRDT 357
Query 150 ILHLSVDEKKLLFSKAFDWLKPDGQLVITDYCCGPPEKWDDEFKAYLQDRNYKLVQLEEY 209
ILH + +K LF F WLKP G+++I+DYC P EF Y++ R Y L ++ Y
Sbjct 358 ILH--IQDKPALFRTFFKWLKPGGKVLISDYCRSPKTP-SAEFSEYIKQRGYDLHDVQAY 414
Query 210 RQLLTEAGFDVVKAANHTQRWLKSLDEESQRLEEQKEDFMHLFTLKDFQDLRDGWQSKKE 269
Q+L +AGF V A + T ++++ L E R+E++KE F+ F+ +D+ D+ GW+SK E
Sbjct 415 GQMLKDAGFTDVIAEDRTDQFMQVLKRELDRVEKEKEKFISDFSKEDYDDIVGGWKSKLE 474
Query 270 RVAKGLQFWGLFIAFK 285
R A Q WGLFIA K
Sbjct 475 RCASDEQKWGLFIANK 490
Score = 50.8 bits (120), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 34/122 (27%), Positives = 60/122 (49%), Gaps = 3/122 (2%)
Query 62 ETTAEILREIVLPKGGKAIDVGCGIGGSTAALADKFNANVLGVDLSSNMISIAKERYSSR 121
E E+L + +G +++G GIG T LA K ++ +D N+I +
Sbjct 40 EERPEVLSLLPPYEGKSVLELGAGIGRFTGELAQK-AGELIALDFIDNVIKKNESINGHY 98
Query 122 PDLKFLVADALS--IPIDSESVDLVYSRDTILHLSVDEKKLLFSKAFDWLKPDGQLVITD 179
++KF+ AD S + I S+DL++S +++LS E +LL + W+K G + +
Sbjct 99 KNVKFMCADVTSPDLKITDGSLDLIFSNWLLMYLSDKEVELLAERMVGWIKVGGYIFFRE 158
Query 180 YC 181
C
Sbjct 159 SC 160
> ath:AT1G73600 methyltransferase/ phosphoethanolamine N-methyltransferase
(EC:2.1.1.103)
Length=490
Score = 209 bits (532), Expect = 1e-53, Method: Compositional matrix adjust.
Identities = 112/254 (44%), Positives = 162/254 (63%), Gaps = 4/254 (1%)
Query 31 QQALDSHQYSKNGILRYEFIFGRGFVSSGGGETTAEILREIVLPKGGKAIDVGCGIGGST 90
Q+ LD+ QY +GILRYE +FG GFVS+GG ETT E + + L G K +DVGCGIGG
Sbjct 237 QRFLDNVQYKSSGILRYERVFGEGFVSTGGLETTKEFVDMLDLKPGQKVLDVGCGIGGGD 296
Query 91 AALADKFNANVLGVDLSSNMISIAKER-YSSRPDLKFLVADALSIPIDSESVDLVYSRDT 149
+A+ F+ +V+G+DLS NMIS A E + ++F VAD + D++YSRDT
Sbjct 297 FYMAENFDVDVVGIDLSVNMISFALEHAIGLKCSVEFEVADCTKKEYPDNTFDVIYSRDT 356
Query 150 ILHLSVDEKKLLFSKAFDWLKPDGQLVITDYCCGPPEKWDDEFKAYLQDRNYKLVQLEEY 209
ILH + +K LF + + WLKP G+++ITDYC P D F Y++ R Y L ++ Y
Sbjct 357 ILH--IQDKPALFRRFYKWLKPGGKVLITDYCRSPKTPSPD-FAIYIKKRGYDLHDVQAY 413
Query 210 RQLLTEAGFDVVKAANHTQRWLKSLDEESQRLEEQKEDFMHLFTLKDFQDLRDGWQSKKE 269
Q+L +AGF+ V A + T +++K L E +E++KE+F+ F+ +D++D+ GW+SK
Sbjct 414 GQMLRDAGFEEVIAEDRTDQFMKVLKRELDAVEKEKEEFISDFSKEDYEDIIGGWKSKLL 473
Query 270 RVAKGLQFWGLFIA 283
R + G Q WGLFIA
Sbjct 474 RSSSGEQKWGLFIA 487
Score = 50.8 bits (120), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 37/122 (30%), Positives = 59/122 (48%), Gaps = 3/122 (2%)
Query 62 ETTAEILREIVLPKGGKAIDVGCGIGGSTAALADKFNANVLGVDLSSNMISIAKERYSSR 121
E EIL + +G ++ G GIG T LA K V+ VD ++I +
Sbjct 39 EERPEILAFLPPIEGTTVLEFGAGIGRFTTELAQK-AGQVIAVDFIESVIKKNENINGHY 97
Query 122 PDLKFLVADALSIPID--SESVDLVYSRDTILHLSVDEKKLLFSKAFDWLKPDGQLVITD 179
++KFL AD S ++ +ES+DL++S +++LS E + L K W K G + +
Sbjct 98 KNVKFLCADVTSPNMNFPNESMDLIFSNWLLMYLSDQEVEDLAKKMLQWTKVGGYIFFRE 157
Query 180 YC 181
C
Sbjct 158 SC 159
> ath:AT1G48600 phosphoethanolamine N-methyltransferase 2, putative
(NMT2) (EC:2.1.1.103)
Length=475
Score = 206 bits (523), Expect = 1e-52, Method: Compositional matrix adjust.
Identities = 118/262 (45%), Positives = 169/262 (64%), Gaps = 4/262 (1%)
Query 25 EDIKKRQQALDSHQYSKNGILRYEFIFGRGFVSSGGGETTAEILREIVLPKGGKAIDVGC 84
E+ K Q+ LD+ QY +GILRYE +FG G+VS+GG ETT E + ++ L G K +DVGC
Sbjct 216 ENDKDFQRFLDNVQYKSSGILRYERVFGEGYVSTGGFETTKEFVAKMDLKPGQKVLDVGC 275
Query 85 GIGGSTAALADKFNANVLGVDLSSNMISIAKER-YSSRPDLKFLVADALSIPIDSESVDL 143
GIGG +A+ F+ +V+G+DLS NMIS A ER + ++F VAD + S D+
Sbjct 276 GIGGGDFYMAENFDVHVVGIDLSVNMISFALERAIGLKCSVEFEVADCTTKTYPDNSFDV 335
Query 144 VYSRDTILHLSVDEKKLLFSKAFDWLKPDGQLVITDYCCGPPEKWDDEFKAYLQDRNYKL 203
+YSRDTILH + +K LF F WLKP G+++ITDY C E EF Y++ R Y L
Sbjct 336 IYSRDTILH--IQDKPALFRTFFKWLKPGGKVLITDY-CRSAETPSPEFAEYIKQRGYDL 392
Query 204 VQLEEYRQLLTEAGFDVVKAANHTQRWLKSLDEESQRLEEQKEDFMHLFTLKDFQDLRDG 263
++ Y Q+L +AGFD V A + T ++++ L E +++E++KE+F+ F+ +D+ D+ G
Sbjct 393 HDVQAYGQMLKDAGFDDVIAEDRTDQFVQVLRRELEKVEKEKEEFISDFSEEDYNDIVGG 452
Query 264 WQSKKERVAKGLQFWGLFIAFK 285
W +K ER A G Q WGLFIA K
Sbjct 453 WSAKLERTASGEQKWGLFIADK 474
Score = 54.7 bits (130), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 35/122 (28%), Positives = 60/122 (49%), Gaps = 3/122 (2%)
Query 62 ETTAEILREIVLPKGGKAIDVGCGIGGSTAALADKFNANVLGVDLSSNMISIAKERYSSR 121
E E+L I +G +++G GIG T LA K V+ +D + I +
Sbjct 24 EERPEVLSLIPPYEGKSVLELGAGIGRFTGELAQK-AGEVIALDFIESAIQKNESVNGHY 82
Query 122 PDLKFLVADALS--IPIDSESVDLVYSRDTILHLSVDEKKLLFSKAFDWLKPDGQLVITD 179
++KF+ AD S + I S+DL++S +++LS E +L+ + W+KP G + +
Sbjct 83 KNIKFMCADVTSPDLKIKDGSIDLIFSNWLLMYLSDKEVELMAERMIGWVKPGGYIFFRE 142
Query 180 YC 181
C
Sbjct 143 SC 144
> cel:F54D11.1 pmt-2; Phosphoethanolamine MethyTransferase family
member (pmt-2)
Length=437
Score = 162 bits (409), Expect = 2e-39, Method: Compositional matrix adjust.
Identities = 99/256 (38%), Positives = 143/256 (55%), Gaps = 6/256 (2%)
Query 34 LDSHQYSKNGILRYEFIFGRGFVSSGGGETTAEILREIVLPKGGKA-IDVGCGIGGSTAA 92
LD QY+ GI YE++FG F+S GG + +I++ K G+ +D+G GIGG
Sbjct 182 LDKTQYTNTGIDAYEWMFGVNFISPGGYDENLKIIKRFGDFKPGQTMLDIGVGIGGGARQ 241
Query 93 LADKFNANVLGVDLSSNMISIAKERYSSRPD--LKFLVADALSIPIDSESVDLVYSRDTI 150
+AD+F +V G+DLSSNM++IA ER D +K+ + DAL + S D V+SRD I
Sbjct 242 VADEFGVHVHGIDLSSNMLAIALERLHEEKDSRVKYSITDALVYQFEDNSFDYVFSRDCI 301
Query 151 LHLSVDEKKLLFSKAFDWLKPDGQLVITDYCCGPPEKWDDEFKAYLQDRNYKLVQLEEYR 210
H+ EK LFS+ + LKP G+++IT Y G E+ D+FK Y+ R Y L L+E
Sbjct 302 QHIPDTEK--LFSRIYKALKPGGKVLITMYGKGYGEQ-SDKFKTYVAQRAYFLKNLKEIA 358
Query 211 QLLTEAGFDVVKAANHTQRWLKSLDEESQRLEEQKEDFMHLFTLKDFQDLRDGWQSKKER 270
+ + GF V+ N T R+ + L EE LE+ + +FM FT ++ L GW K
Sbjct 359 DIANKTGFVNVQTENMTPRFKEILLEERGHLEQNEAEFMSKFTQRERDSLISGWTDKLGY 418
Query 271 VAKGLQFWGLFIAFKP 286
+ K W F+A KP
Sbjct 419 IEKDNHNWNFFLAQKP 434
> cel:ZK622.3 pmt-1; Phosphoethanolamine MethyTransferase family
member (pmt-1)
Length=475
Score = 60.8 bits (146), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 35/120 (29%), Positives = 60/120 (50%), Gaps = 1/120 (0%)
Query 65 AEILREIVLPKGGKAIDVGCGIGGSTAALADKFNANVLGVDLSSNMISIAKERYSSRPDL 124
A+IL + L +D+G GIG T LA+ VL D + I +ER + ++
Sbjct 55 ADILASLPLLHNKDVVDIGAGIGRFTTVLAETARW-VLSTDFIDSFIKKNQERNAHLGNI 113
Query 125 KFLVADALSIPIDSESVDLVYSRDTILHLSVDEKKLLFSKAFDWLKPDGQLVITDYCCGP 184
+ V DA+ + ++S SVDLV++ +++LS +E WL+ G + + + C P
Sbjct 114 NYQVGDAVGLKMESNSVDLVFTNWLMMYLSDEETVEFIFNCMRWLRSHGIVHLRESCSEP 173
> ath:AT1G64970 G-TMT; G-TMT (GAMMA-TOCOPHEROL METHYLTRANSFERASE);
tocopherol O-methyltransferase (EC:2.1.1.95); K05928 tocopherol
O-methyltransferase [EC:2.1.1.95]
Length=348
Score = 57.8 bits (138), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 35/105 (33%), Positives = 54/105 (51%), Gaps = 5/105 (4%)
Query 80 IDVGCGIGGSTAALADKFNANVLGVDLSSNMISIAKERYSSRP---DLKFLVADALSIPI 136
+DVGCGIGGS+ LA KF A +G+ LS A + +++ F VADAL P
Sbjct 131 VDVGCGIGGSSRYLASKFGAECIGITLSPVQAKRANDLAAAQSLAHKASFQVADALDQPF 190
Query 137 DSESVDLVYSRDTILHLSVDEKKLLFSKAFDWLKPDGQLVITDYC 181
+ DLV+S ++ H+ +K + P G+++I +C
Sbjct 191 EDGKFDLVWSMESGEHMP--DKAKFVKELVRVAAPGGRIIIVTWC 233
> mmu:67667 Alkbh8, 4930562C03Rik, 8030431D03Rik, 9430088N01Rik,
Abh8, MGC10235; alkB, alkylation repair homolog 8 (E. coli);
K10770 alkylated DNA repair protein alkB homolog 8
Length=664
Score = 56.2 bits (134), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 34/106 (32%), Positives = 52/106 (49%), Gaps = 9/106 (8%)
Query 73 LPKGGKAIDVGCGIGGSTAALADKFNANVLGVDLSSNMISIAKERYSSRPDLKFLVADAL 132
LP G D+GCG G D + ++G D S N++ I +ER + LV DAL
Sbjct 404 LPSGSIVADIGCGNGKYLGINKDLY---MIGCDRSQNLVDICRER-----QFQALVCDAL 455
Query 133 SIPIDSESVDLVYSRDTILHLSVDEKKL-LFSKAFDWLKPDGQLVI 177
++P+ S S D S I H + E+++ + L+P GQ +I
Sbjct 456 AVPVRSGSCDACISIAVIHHFATAERRVEALQELARLLRPGGQALI 501
> ath:AT2G43910 thiol methyltransferase, putative
Length=246
Score = 55.5 bits (132), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 41/140 (29%), Positives = 68/140 (48%), Gaps = 9/140 (6%)
Query 77 GKAIDVGCGIGGSTAALADKFNANVLGVDLSSNMISIAKERYSSRPDLKF--LVADALSI 134
G+A+ GCG G A+A V+G+D+S + ++ A E Y S P ++ V + +
Sbjct 68 GRALVPGCGGGHDVVAMASP-ERFVVGLDISESALAKANETYGSSPKAEYFSFVKEDVFT 126
Query 135 PIDSESVDLVYSRDTILHLSVDEKKLLFSKAFDWLKPDGQLV-----ITDYCCGPPEKWD 189
+E DL++ + + + ++ LKPDG+L+ ITD+ GPP K D
Sbjct 127 WRPTELFDLIFDYVFFCAIEPEMRPAWAKSMYELLKPDGELITLMYPITDHVGGPPYKVD 186
Query 190 -DEFKAYLQDRNYKLVQLEE 208
F+ L +K V +EE
Sbjct 187 VSTFEEVLVPIGFKAVSVEE 206
> hsa:91801 ALKBH8, ABH8, FLJ38204, MGC10235; alkB, alkylation
repair homolog 8 (E. coli); K10770 alkylated DNA repair protein
alkB homolog 8
Length=664
Score = 52.8 bits (125), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 36/130 (27%), Positives = 63/130 (48%), Gaps = 12/130 (9%)
Query 73 LPKGGKAIDVGCGIGGSTAALADKFNANVLGVDLSSNMISIAKERYSSRPDLKFLVADAL 132
LP G D+GCG G + + ++G D S N++ I +ER + V DAL
Sbjct 404 LPSGSIVADIGCGNGKYLGINKELY---MIGCDRSQNLVDICRER-----QFQAFVCDAL 455
Query 133 SIPIDSESVDLVYSRDTILHLSVDEKKL-LFSKAFDWLKPDGQLVITDYCCGPPEKWDDE 191
++P+ S S D S I H + E+++ + L+P G+ +I Y ++++ +
Sbjct 456 AVPVRSGSCDACISIAVIHHFATAERRVAALQEIVRLLRPGGKALI--YVWAMEQEYNKQ 513
Query 192 FKAYLQ-DRN 200
YL+ +RN
Sbjct 514 KSKYLRGNRN 523
> sce:YML014W TRM9, KTI1; Trm9p (EC:2.1.1.-)
Length=279
Score = 52.8 bits (125), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 34/109 (31%), Positives = 50/109 (45%), Gaps = 9/109 (8%)
Query 74 PKGGKAIDVGCGIGGSTAALADKFNANVLGVDLSSNMISIAKERYSSRPDLKFLVADALS 133
P G IDVGCG G D + ++G D S +I A+ P LVAD L+
Sbjct 44 PMGSIGIDVGCGNGKYLGVNPDIY---IIGSDRSDGLIECAR---GINPSYNLLVADGLN 97
Query 134 IPIDSESVDLVYSRDTILHLSVDEKKL-LFSKAFDWLKPDGQLVITDYC 181
+P +E+ D S + H S E+++ + L+ GQ +I YC
Sbjct 98 LPHKNETFDFAISIAVVHHWSTRERRVEVIRHVLSKLRQGGQALI--YC 144
> tpv:TP02_0197 hexaprenyldihydroxybenzoate methyltransferase
(EC:2.1.1.114); K00591 hexaprenyldihydroxybenzoate methyltransferase
[EC:2.1.1.114]
Length=309
Score = 52.4 bits (124), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 39/121 (32%), Positives = 55/121 (45%), Gaps = 18/121 (14%)
Query 75 KGGKAIDVGCGIGGSTAALADKFNANVLGVDLSSNMISIAKER----------------- 117
KG K +DVGCG G T +LA KF + VLG+D + N+I +AK
Sbjct 105 KGLKILDVGCGGGILTESLA-KFGSKVLGIDPNENLIKVAKSHKKTHFDNYHLSLGLKND 163
Query 118 YSSRPDLKFLVADALSIPIDSESVDLVYSRDTILHLSVDEKKLLFSKAFDWLKPDGQLVI 177
YS+ D K + D+V + + I H+ EK+ F ++KP G VI
Sbjct 164 YSNNLDYKSTSVYDFLTDKTRATFDIVVASEVIEHIDNREKEQFFETLTSFVKPGGLFVI 223
Query 178 T 178
T
Sbjct 224 T 224
> eco:b1519 tam, ECK1512, JW1512, lsrE, yneD; trans-aconitate
methyltransferase (EC:2.1.1.144); K00598 trans-aconitate 2-methyltransferase
[EC:2.1.1.144]
Length=252
Score = 52.0 bits (123), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 47/176 (26%), Positives = 82/176 (46%), Gaps = 24/176 (13%)
Query 57 SSGGGETTAEILREIVLPKGGKAIDVGCGIGGSTAALADKFN-ANVLGVDLSSNMISIAK 115
S+ E+L + L D+GCG G STA L ++ A + G+D S MI+ A+
Sbjct 13 SAERSRPAVELLARVPLENVEYVADLGCGPGNSTALLQQRWPAARITGIDSSPAMIAEAR 72
Query 116 ERYSSRPDLKFLVADALSI-PIDSESVDLVYSRDTILHLSVDEKKLLFSKAFDWLKPDGQ 174
S+ PD +F+ AD + P+ +++DL+++ ++ L + LF L P G
Sbjct 73 ---SALPDCQFVEADIRNWQPV--QALDLIFANASLQWLP--DHYELFPHLVSLLNPQGV 125
Query 175 LVITDYCCGPPEKWDDEFKAYLQ----DRNY------KLVQLEEYRQLLTEAGFDV 220
L + P+ W + ++ ++NY L + Y +L+EAG +V
Sbjct 126 LAVQM-----PDNWLEPTHVLMREVAWEQNYPDRGREPLAGVHAYYDILSEAGCEV 176
> ath:AT1G20330 SMT2; SMT2 (STEROL METHYLTRANSFERASE 2); S-adenosylmethionine-dependent
methyltransferase; K08242 24-methylenesterol
C-methyltransferase [EC:2.1.1.143]
Length=361
Score = 52.0 bits (123), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 46/164 (28%), Positives = 79/164 (48%), Gaps = 17/164 (10%)
Query 71 IVLPKGGKAIDVGCGIGGSTAALADKFNANVLGVDLSSNMISIAKERYSSRPDL----KF 126
I + G K +DVGCG+GG A+A ANV+G+ ++ ++ A+ ++ + L +
Sbjct 119 IQVKPGQKILDVGCGVGGPMRAIASHSRANVVGITINEYQVNRAR-LHNKKAGLDALCEV 177
Query 127 LVADALSIPIDSESVDLVYSRDTILHLSVDEKKLLFSKAFDWLKPDGQLVITDYCCGPPE 186
+ + L +P D S D YS + H E+ ++++ + LKP V ++ E
Sbjct 178 VCGNFLQMPFDDNSFDGAYSIEATCHAPKLEE--VYAEIYRVLKPGSMYVSYEWVT--TE 233
Query 187 KW---DDEFKAYLQ--DRNYKLVQLEEYRQLLTEA---GFDVVK 222
K+ DDE +Q +R L L Y + A GF++VK
Sbjct 234 KFKAEDDEHVEVIQGIERGDALPGLRAYVDIAETAKKVGFEIVK 277
> dre:555377 zgc:162780
Length=274
Score = 51.6 bits (122), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 68/246 (27%), Positives = 110/246 (44%), Gaps = 49/246 (19%)
Query 76 GGKAIDVGCGIGGSTAALADKFNANVLGVDLSSNMISIAKERYSSRPDLKFLVADALSIP 135
G A+DVGCG G T LA F V+G D+S + + + ++ + P++ F + A +P
Sbjct 42 NGLAVDVGCGSGQGTLLLAPHF-TRVVGTDISPAQLEMGR-KHVNIPNVSFRESPAEELP 99
Query 136 IDSESVDLVYSRDTILHLSVDEKKLLFSKAFDWLKPDGQLVITDY----------CCGPP 185
+ SVDLV + + H D + L +A LKP G L + +Y C
Sbjct 100 FEDGSVDLVTAM-SAFHW-FDHSRFL-QEADRVLKPHGCLALLNYTLDMELTYGNCSEAL 156
Query 186 EKWDDEFKAYLQD---------------RNYKLVQ--LEEYRQL------LTEAGF-DVV 221
+EF A L R Y +Q ++E+ L + +G+ +V
Sbjct 157 NLICNEFYAALHPLRDPHLGPSSFELYKRTYDSLQYPVKEWHDLFWVKKAVPLSGYIGMV 216
Query 222 KAANHTQRWLKSLDEESQRLEEQKEDFMHLFTLKDFQDLRDGWQSKKERVAKGLQFWGLF 281
K+ + Q LK+ EE++RL + ED LK D+ S + V G++++ F
Sbjct 217 KSFSTFQTLLKTDPEEARRLSQGIED-----RLKRAMDV----TSSETEVIMGVKYF-YF 266
Query 282 IAFKPA 287
+A KPA
Sbjct 267 LAQKPA 272
> dre:100329758 hypothetical protein LOC100329758
Length=274
Score = 50.4 bits (119), Expect = 8e-06, Method: Compositional matrix adjust.
Identities = 67/246 (27%), Positives = 109/246 (44%), Gaps = 49/246 (19%)
Query 76 GGKAIDVGCGIGGSTAALADKFNANVLGVDLSSNMISIAKERYSSRPDLKFLVADALSIP 135
G A+DVGCG G T LA F V+G D+S + + + ++ + P++ F + A +P
Sbjct 42 NGLAVDVGCGSGQGTLLLAPHF-TRVVGTDISPAQLEMGR-KHVNIPNVSFRESPAEELP 99
Query 136 IDSESVDLVYSRDTILHLSVDEKKLLFSKAFDWLKPDGQLVITDY----------CCGPP 185
+ SVDLV + + H D + L +A LKP G L + +Y C
Sbjct 100 FEDGSVDLVTAM-SAFHW-FDHSRFL-QEADRVLKPHGCLALLNYTLDMELTYGNCSEAL 156
Query 186 EKWDDEFKAYLQD---------------RNYKLVQ--LEEYRQL------LTEAGF-DVV 221
+EF A L R Y +Q ++E+ + + +G+ +V
Sbjct 157 NLICNEFYAALHPLRDPHLGPSSFELYKRTYDSLQYPVKEWHDMFGVKKAVPLSGYIGMV 216
Query 222 KAANHTQRWLKSLDEESQRLEEQKEDFMHLFTLKDFQDLRDGWQSKKERVAKGLQFWGLF 281
K+ + Q LK EE++RL + ED LK D+ S + V G++++ F
Sbjct 217 KSFSTFQTLLKKDPEEARRLSQDIED-----RLKRAMDV----TSSETEVIMGVKYF-YF 266
Query 282 IAFKPA 287
+A KPA
Sbjct 267 LAQKPA 272
> xla:100217319 hypothetical protein LOC100217319
Length=252
Score = 50.1 bits (118), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 24/66 (36%), Positives = 43/66 (65%), Gaps = 2/66 (3%)
Query 79 AIDVGCGIGGSTAALADKFNANVLGVDLSSNMISIAKERYSSRPDLKFLVADALSIPIDS 138
A+D GCG G ST LA F V+G+D+S + +S+A+ + +S ++ + ++ A +P++
Sbjct 27 AVDAGCGTGRSTRTLAPYFQ-KVIGIDVSESQLSVAR-KCTSHENISYQISPAEELPLED 84
Query 139 ESVDLV 144
SVDL+
Sbjct 85 ASVDLI 90
> cel:H14E04.1 hypothetical protein
Length=334
Score = 49.7 bits (117), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 43/195 (22%), Positives = 79/195 (40%), Gaps = 29/195 (14%)
Query 65 AEILREIVLPKGGKAIDVGCGIGGSTAALADKFNANVLGVDLSSNMISIAKERYSS---R 121
I ++ L + +D+GCGIGG +AD F A + GV ++ N I E++++
Sbjct 85 CHIAEKLELSENVHCLDIGCGIGGVMLDIAD-FGAKLTGVTIAPNEAEIGNEKFANMGIS 143
Query 122 PDLKFLVADALSIPIDSESVDLVYSRDTILHLSVDEKKLLFSKAFDWLKPDGQLVITD-- 179
K + AD +P + + D+ Y+ ++ ++ +K + + LKP G+ ++ D
Sbjct 144 DRCKIVAADCQKMPFEDSTFDVAYAIYSLKYIPNLDK--VMKEIQRVLKPGGKFIVYDLI 201
Query 180 --------------------YCCGPPE-KWDDEFKAYLQDRNYKLVQLEEYRQLLTEAGF 218
Y CG P E +A + +V+ E + F
Sbjct 202 KTNDYDKDNKEHYKTLHHLEYACGMPSLHTQSEVEAAAEKWEMPVVERENLEETYGNRAF 261
Query 219 DVVKAANHTQRWLKS 233
+A+ WL S
Sbjct 262 HYCFSASPMFMWLVS 276
> ath:AT1G76090 SMT3; SMT3 (STEROL METHYLTRANSFERASE 3); S-adenosylmethionine-dependent
methyltransferase/ sterol 24-C-methyltransferase;
K08242 24-methylenesterol C-methyltransferase
[EC:2.1.1.143]
Length=359
Score = 49.7 bits (117), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 46/164 (28%), Positives = 75/164 (45%), Gaps = 17/164 (10%)
Query 71 IVLPKGGKAIDVGCGIGGSTAALADKFNANVLGVDLSSNMISIAKERYSSRPDL----KF 126
I + G K +D GCG+GG A+A A V G+ ++ + AK ++ + L
Sbjct 119 IKVKPGQKILDAGCGVGGPMRAIAAHSKAQVTGITINEYQVQRAK-LHNKKAGLDSLCNV 177
Query 127 LVADALSIPIDSESVDLVYSRDTILHLSVDEKKLLFSKAFDWLKPDGQLVITDYCCGPPE 186
+ + L +P D + D YS + H E+ ++S+ F +KP V ++ E
Sbjct 178 VCGNFLKMPFDENTFDGAYSIEATCHAPKLEE--VYSEIFRVMKPGSLFVSYEWVT--TE 233
Query 187 KW---DDEFKAYLQ--DRNYKLVQLEEYRQLLTEA---GFDVVK 222
K+ D+E K +Q +R L L Y + A GF+VVK
Sbjct 234 KYRDDDEEHKDVIQGIERGDALPGLRSYADIAVTAKKVGFEVVK 277
> xla:100127257 hypothetical protein LOC100127257
Length=251
Score = 49.7 bits (117), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 24/66 (36%), Positives = 43/66 (65%), Gaps = 2/66 (3%)
Query 79 AIDVGCGIGGSTAALADKFNANVLGVDLSSNMISIAKERYSSRPDLKFLVADALSIPIDS 138
A+D GCG G ST LA F V+G+D+S + +S+A+ + +S ++ + ++ A +P++
Sbjct 27 AVDAGCGTGRSTRTLAPYFQ-KVVGIDVSESQLSVAR-KCTSHENISYQISPAEELPLED 84
Query 139 ESVDLV 144
SVDL+
Sbjct 85 ASVDLI 90
> sce:YML008C ERG6, ISE1, LIS1, SED6, VID1; Delta(24)-sterol C-methyltransferase,
converts zymosterol to fecosterol in the
ergosterol biosynthetic pathway by methylating position C-24;
localized to both lipid particles and mitochondrial outer
membrane (EC:2.1.1.41); K00559 sterol 24-C-methyltransferase
[EC:2.1.1.41]
Length=383
Score = 47.8 bits (112), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 41/158 (25%), Positives = 72/158 (45%), Gaps = 11/158 (6%)
Query 73 LPKGGKAIDVGCGIGGSTAALADKFNANVLGVDLSSNMISIAKERYSS---RPDLKFLVA 129
+ +G +DVGCG+GG +A NV+G++ + I+ AK + F+
Sbjct 117 IQRGDLVLDVGCGVGGPAREIARFTGCNVIGLNNNDYQIAKAKYYAKKYNLSDQMDFVKG 176
Query 130 DALSIPIDSESVDLVYSRDTILHLSVDEKKLLFSKAFDWLKPDGQL-----VITD-YCCG 183
D + + + + D VY+ + H E ++S+ + LKP G V+TD Y
Sbjct 177 DFMKMDFEENTFDKVYAIEATCHAPKLEG--VYSEIYKVLKPGGTFAVYEWVMTDKYDEN 234
Query 184 PPEKWDDEFKAYLQDRNYKLVQLEEYRQLLTEAGFDVV 221
PE ++ L D K+ ++ R+ L GF+V+
Sbjct 235 NPEHRKIAYEIELGDGIPKMFHVDVARKALKNCGFEVL 272
> cel:C35D10.12 hypothetical protein
Length=365
Score = 45.8 bits (107), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 30/103 (29%), Positives = 54/103 (52%), Gaps = 11/103 (10%)
Query 76 GGKAIDVGCGIGGSTAALADKFNANVLGVDLSSNMISIAKERYSSRPDLKFLVADALSIP 135
G +DVGCG T+ ++V+G D S ++S +K + D+ +ADA++IP
Sbjct 49 GSIILDVGCGEAKYTSQ-----KSHVIGFDTCSEVLSSSK-----KDDIDLCLADAINIP 98
Query 136 IDSESVDLVYSRDTILHLSVD-EKKLLFSKAFDWLKPDGQLVI 177
I +SVD + + I HL+ ++ + + L+ GQ++I
Sbjct 99 IRDDSVDAILNVSVIHHLATTARRRQVLQECSRCLRIGGQMLI 141
> ath:AT2G43940 thiol methyltransferase, putative
Length=226
Score = 45.8 bits (107), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 40/170 (23%), Positives = 74/170 (43%), Gaps = 27/170 (15%)
Query 65 AEILREIVLPKGGKAIDVGCGIGGSTAALADKFNANVLGVDLSSNMISIAKERYSSRPDL 124
A ++ LP G +A+ GCG G A+A + +V+G+D+S + + +++S+ P+
Sbjct 50 AHLVETGSLPNG-RALVPGCGTGYDVVAMASP-DRHVVGLDISKTAVERSTKKFSTLPNA 107
Query 125 K---FLVADALSIPIDSESVDLVYSRDTILHLSVDEKKLLFSKAFDWLKPDGQLV----- 176
K FL D + +E DL++ + L + LKP G+L+
Sbjct 108 KYFSFLSEDFFTWE-PAEKFDLIFDYTFFCAFEPGVRPLWAQRMEKLLKPGGELITLMFP 166
Query 177 ITDYCCGPPEKWDDEFKAYLQDRNYKLVQLEEYRQLLTEAGFDVVKAANH 226
I + GPP + V + EY ++L GF+ + ++
Sbjct 167 IDERSGGPPYE----------------VSVSEYEKVLIPLGFEAISIVDN 200
> ath:AT5G13710 SMT1; SMT1 (STEROL METHYLTRANSFERASE 1); sterol
24-C-methyltransferase; K00559 sterol 24-C-methyltransferase
[EC:2.1.1.41]
Length=336
Score = 45.4 bits (106), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 42/174 (24%), Positives = 75/174 (43%), Gaps = 16/174 (9%)
Query 22 HGPEDIKKRQQALDS-HQYSKNGILRYEFIFGRGF--VSSGGGETTAEILR--------E 70
HG + +++ D ++Y YE+ +G F GE+ E ++ +
Sbjct 30 HGGNEEERKANYTDMVNKYYDLATSFYEYGWGESFHFAQRWKGESLRESIKRHEHFLALQ 89
Query 71 IVLPKGGKAIDVGCGIGGSTAALADKFNANVLGVDLSSNMISIAKE--RYSS-RPDLKFL 127
+ + G K +DVGCGIGG +A N+ V G++ + I+ KE R + F+
Sbjct 90 LGIQPGQKVLDVGCGIGGPLREIARFSNSVVTGLNNNEYQITRGKELNRLAGVDKTCNFV 149
Query 128 VADALSIPIDSESVDLVYSRDTILHLSVDEKKLLFSKAFDWLKPDGQLVITDYC 181
AD + +P S D VY+ + H + + + + LKP ++C
Sbjct 150 KADFMKMPFPENSFDAVYAIEATCH--APDAYGCYKEIYRVLKPGQCFAAYEWC 201
> eco:b1822 rlmA, ECK1820, JW1811, rrmA, yebH; 23S rRNA m(1)G745
methyltransferase (EC:2.1.1.51); K00563 23S rRNA (guanine745-N1)-methyltransferase
[EC:2.1.1.187]
Length=269
Score = 45.4 bits (106), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 30/97 (30%), Positives = 45/97 (46%), Gaps = 8/97 (8%)
Query 53 RGFVSSGGGETTAEI----LREIVLPKGGKAIDVGCGIGGSTAALADKF-NANVLGVDLS 107
R F+ +G + + LRE + K +D+GCG G T A AD G+D+S
Sbjct 59 RAFLDAGHYQPLRDAIVAQLRERLDDKATAVLDIGCGEGYYTHAFADALPEITTFGLDVS 118
Query 108 SNMISIAKERYSSRPDLKFLVADALSIPIDSESVDLV 144
I A +RY P + F VA + +P S+D +
Sbjct 119 KVAIKAAAKRY---PQVTFCVASSHRLPFSDTSMDAI 152
> tpv:TP01_0568 hypothetical protein
Length=244
Score = 45.1 bits (105), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 22/81 (27%), Positives = 45/81 (55%), Gaps = 4/81 (4%)
Query 80 IDVGCGIGGSTAALADKFNANVLGVDLSSNMISIAKERYSSRPDLKFLVADALSIPIDSE 139
+DVGCG G + D + +GVD+ S ++ +A+E++ + + ++++AL +P
Sbjct 69 LDVGCGNGKYLSTRTDCY---FIGVDICSELLHLAREKHVN-SNFSLVISNALKLPFKDN 124
Query 140 SVDLVYSRDTILHLSVDEKKL 160
+L + I HLS +++L
Sbjct 125 FANLTLAIAIIHHLSTTQRRL 145
> sce:YHR209W CRG1; Crg1p
Length=291
Score = 44.3 bits (103), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 33/105 (31%), Positives = 54/105 (51%), Gaps = 8/105 (7%)
Query 80 IDVGCGIGGSTAALADKFNANVLGVDLSSNMISIAKERYSSR---PDLKFLVADALSI-P 135
+D+GCG G +T + F V+G+D SS M+SIA++ + R ++F+ A +
Sbjct 43 VDIGCGTGKATFVVEPYFK-EVIGIDPSSAMLSIAEKETNERRLDKKIRFINAPGEDLSS 101
Query 136 IDSESVDLVYSRDTILHLSVDEKKLLFSKAFDWLKPDGQLVITDY 180
I ESVD+V S + I +++ LF + L+ DG Y
Sbjct 102 IRPESVDMVISAEAIHWCNLER---LFQQVSSILRSDGTFAFWFY 143
> pfa:PF10_0274 methyltransferase, putative
Length=593
Score = 43.9 bits (102), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 27/85 (31%), Positives = 48/85 (56%), Gaps = 7/85 (8%)
Query 75 KGGKAIDVGCGIGGSTAALADKFNANVLGVDLSSNMISIAKERYSSRPDLKFLVADALSI 134
+G +DVGCG G + + + F +G+D S ++ +A+++ ++ DL L+A+ ++I
Sbjct 382 EGNIILDVGCGNGKNLSESSKYF---YIGLDFSLYLLMLARKKMNT--DL--LLANCINI 434
Query 135 PIDSESVDLVYSRDTILHLSVDEKK 159
P+ S DL S I HL EK+
Sbjct 435 PLRSNLADLCISIAVIHHLGTHEKR 459
> xla:495315 hypothetical LOC495315
Length=207
Score = 43.5 bits (101), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 27/103 (26%), Positives = 48/103 (46%), Gaps = 9/103 (8%)
Query 76 GGKAIDVGCGIGGSTAALADKFNANVLGVDLSSNMISIAKERYSSRPDLKFLVADALSIP 135
G +D+GCG G ++ +N LG D ++ IAK + +V D L++P
Sbjct 60 GSLIVDIGCGTGKYLRVNSEIYN---LGCDYCKPLVEIAKNNKH-----EVMVCDNLNLP 111
Query 136 IDSESVDLVYSRDTILHLSVDEKKLLFSKAF-DWLKPDGQLVI 177
+ +D V S I H S ++++ K L P G++++
Sbjct 112 FRDQCIDTVISIGVIHHFSTKQRRIQAIKEMARTLVPGGRIML 154
> dre:556362 K1456 protein-like; K10770 alkylated DNA repair protein
alkB homolog 8
Length=693
Score = 43.5 bits (101), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 29/89 (32%), Positives = 39/89 (43%), Gaps = 8/89 (8%)
Query 71 IVLPKGGKAIDVGCGIGGSTAALADKFNANVLGVDLSSNMISIAKERYSSRPDLKFLVAD 130
+ LP G DVGCG G L +G D S N++ I ER V+D
Sbjct 426 LSLPPGSFLADVGCGNG---KYLGINPAVRAVGCDRSVNLVQICIER-----GYDAFVSD 477
Query 131 ALSIPIDSESVDLVYSRDTILHLSVDEKK 159
ALS+P+ S D S I H + E++
Sbjct 478 ALSVPLRRGSCDACISIAVIHHFATQERR 506
> dre:100329679 hexaprenyldihydroxybenzoate methyltransferase,
mitochondrial-like
Length=271
Score = 43.1 bits (100), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 35/121 (28%), Positives = 56/121 (46%), Gaps = 13/121 (10%)
Query 68 LREIVL----PKGGK----AIDVGCGIGGSTAALADKFNANVLGVDLSSNMISIAKERYS 119
L+E++L K GK A+D+GCG G ++ L F V+G+D+S + + A+
Sbjct 26 LKELILQYLDKKKGKPHQLAVDLGCGTGQTSRPLTPYFQ-QVVGIDVSESQVEEAR-AVQ 83
Query 120 SRPDLKFLVADALSIPIDSESVDLVYSRDTILHLSVDEKKLLFSKAFDWLKPDGQLVITD 179
P+L + V A +P SVDL+ + + +A LKP G L +
Sbjct 84 GFPNLTYRVGTAEELPFPDASVDLLTAASAAHWFDAER---FVKEAQRVLKPHGCLALFG 140
Query 180 Y 180
Y
Sbjct 141 Y 141
> dre:555292 fb62c07, si:ch211-31o18.2, wu:fb62c07; zgc:162396
Length=271
Score = 43.1 bits (100), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 35/121 (28%), Positives = 56/121 (46%), Gaps = 13/121 (10%)
Query 68 LREIVL----PKGGK----AIDVGCGIGGSTAALADKFNANVLGVDLSSNMISIAKERYS 119
L+E++L K GK A+D+GCG G ++ L F V+G+D+S + + A+
Sbjct 26 LKELILQYLDKKKGKPHQLAVDLGCGTGQTSRPLTPYFQ-QVVGIDVSESQVEEAR-AVQ 83
Query 120 SRPDLKFLVADALSIPIDSESVDLVYSRDTILHLSVDEKKLLFSKAFDWLKPDGQLVITD 179
P+L + V A +P SVDL+ + + +A LKP G L +
Sbjct 84 GFPNLTYRVGTAEELPFPDASVDLLTAASAAHWFDAER---FVKEAQRVLKPHGCLALFG 140
Query 180 Y 180
Y
Sbjct 141 Y 141
> cel:C14B1.10 hypothetical protein; K10770 alkylated DNA repair
protein alkB homolog 8
Length=591
Score = 42.7 bits (99), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 29/107 (27%), Positives = 49/107 (45%), Gaps = 9/107 (8%)
Query 73 LPKGGKAIDVGCGIGGSTAALADKFNANVLGVDLSSNMISIAKERYSSRPDLKFLVADAL 132
+P+G DVGCG G L K +G D+ + IA+++ D DAL
Sbjct 388 IPRGSVMYDVGCGNG---KYLIPKDGLLKIGCDMCMGLCDIARKK-----DCHVARCDAL 439
Query 133 SIPIDSESVDLVYSRDTILHLSV-DEKKLLFSKAFDWLKPDGQLVIT 178
++P ES D S + H++ + +K L + +KP ++ +T
Sbjct 440 ALPFRYESADAAISIAVLHHIATFERRKRLIEELLRVVKPGSKICVT 486
> eco:b1661 cfa, cdfA, ECK1657, JW1653; cyclopropane fatty acyl
phospholipid synthase (unsaturated-phospholipid methyltransferase)
(EC:2.1.1.79); K00574 cyclopropane-fatty-acyl-phospholipid
synthase [EC:2.1.1.79]
Length=382
Score = 42.7 bits (99), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 28/111 (25%), Positives = 53/111 (47%), Gaps = 4/111 (3%)
Query 67 ILREIVLPKGGKAIDVGCGIGGSTAALADKFNANVLGVDLSSNMISIAKERYSSRPDLKF 126
I ++ L G + +D+GCG GG +A ++ +V+GV +S+ +A+ER D+
Sbjct 159 ICEKLQLKPGMRVLDIGCGWGGLAHYMASNYDVSVVGVTISAEQQKMAQERCEGL-DVTI 217
Query 127 LVADALSIPIDSESVDLVYSRDTILHLSVDEKKLLFSKAFDWLKPDGQLVI 177
L+ D + ++ D + S H+ F+ LKP+G ++
Sbjct 218 LLQDYRDL---NDQFDRIVSVGMFEHVGPKNYDTYFAVVDRNLKPEGIFLL 265
> ath:AT1G36310 methyltransferase
Length=404
Score = 42.4 bits (98), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 51/195 (26%), Positives = 77/195 (39%), Gaps = 35/195 (17%)
Query 73 LPKGGKAIDVGCGIGGSTAALADKFNANVLGVDLSSNMISIAKERYSSRPDLKFLVADAL 132
LP G +D GCG G F +G D+S +I I ++ + LVADA+
Sbjct 105 LPSGSVILDAGCGNGKYLGLNPSCF---FIGCDISHPLIKICSDK-----GQEVLVADAV 156
Query 133 SIPIDSESVDLVYSRDTILHLSVDEKKLLFSKAFDWL----KPDGQLVITDYCCGPPE-- 186
++P E D S + HLS + ++ KA + L KP G ++IT + +
Sbjct 157 NLPYREEFGDAAISIAVLHHLSTENRR---KKAIEELVRVVKPGGFVLITVWAAEQEDTS 213
Query 187 ---KWDDEFKAYLQD-------------RNYKLVQLEEYRQLLTEAGFDVVKAANHTQRW 230
KW Y+++ RN LE + TE K N
Sbjct 214 LLTKWTPLSAKYVEEWVGPGSPMNSPRVRNNPFFSLESIPE--TEVSTKEQKVENSQFIG 271
Query 231 LKSLDEESQRLEEQK 245
L+S+ E + EQK
Sbjct 272 LESIPESEESTREQK 286
> mmu:230027 Coq3, 4732433J24, C77934; coenzyme Q3 homolog, methyltransferase
(yeast) (EC:2.1.1.114); K00591 hexaprenyldihydroxybenzoate
methyltransferase [EC:2.1.1.114]
Length=370
Score = 42.4 bits (98), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 35/108 (32%), Positives = 52/108 (48%), Gaps = 8/108 (7%)
Query 76 GGKAIDVGCGIGGSTAALADKFNANVLGVDLSSNMISIAKERYSSRPDLKFLV---ADAL 132
G K +DVGCG G T L + A+V+G+D + I IA+ S P L + +L
Sbjct 148 GMKILDVGCGGGLLTEPLG-RLGASVVGIDPVAENIKIAQHHKSFDPVLDKRIQYKVCSL 206
Query 133 SIPID--SESVDLVYSRDTILHLSVDEKKLLFSKAFDWLKPDGQLVIT 178
+D +E D V + + + H+S E + + LKP G L IT
Sbjct 207 EEAVDESAECFDAVVASEVVEHVSHLE--MFIQCCYQVLKPGGSLFIT 252
> cel:R08F11.4 hypothetical protein
Length=354
Score = 41.6 bits (96), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 36/121 (29%), Positives = 63/121 (52%), Gaps = 14/121 (11%)
Query 68 LREIVLPKGGKAIDVGCGIGGSTAALADKF-NANVLGVDLSSNMISIAKERYSSR----P 122
++E + G + +DVGCG G + LA+ + + +G+D++ I A+ + S
Sbjct 158 IKEKLEAGGIRVLDVGCGGGFHSGLLAEHYPKSQFVGLDITEKAIKAARLKKKSDGTDFE 217
Query 123 DLKFLVADALSIPID-SESVDLVYSRDTILHLSVDEK---KLLFSKAFDWLKPDGQLVIT 178
+L+F+VADA +P ++S DLV IL S ++ L + +KPDG + +T
Sbjct 218 NLEFVVADAAIMPSSWTDSFDLV-----ILFGSCHDQMRPDLCLLEVHRVVKPDGLVAVT 272
Query 179 D 179
D
Sbjct 273 D 273
> cel:K12D9.1 hypothetical protein
Length=391
Score = 40.8 bits (94), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 34/110 (30%), Positives = 57/110 (51%), Gaps = 8/110 (7%)
Query 76 GGKAIDVGCGIGGSTAALADKFN-ANVLGVDLSSNMISIAKERYSSR----PDLKFLVAD 130
G + +DVGCG G + LA+ ++ + +G+D+ I AK S +L+F+V D
Sbjct 203 GFRVLDVGCGEGFHSCLLAENYSKSQFVGLDICEKAIKSAKLNKKSDGSDFQNLEFVVGD 262
Query 131 ALSIPID-SESVDLVYSRDTILHLSVDEKKLLFSKAFDWLKPDGQLVITD 179
A+ +P D + DLV ++ L + LL + LKP G +V+T+
Sbjct 263 AMIMPEDWTGCFDLVAFFGSLHDLLRPDLSLL--EVHRVLKPGGMVVLTE 310
> cel:R08E5.1 hypothetical protein
Length=250
Score = 40.8 bits (94), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 30/103 (29%), Positives = 54/103 (52%), Gaps = 21/103 (20%)
Query 76 GGKAIDVGCGIGGSTAALADKF-NANVLGVDLSSNMISIAKERYS----SRPDLKFLVAD 130
G + +DVGCG G ++ LA+++ A+ +G+++ + I AK+R + + +L+F+ D
Sbjct 113 GMRVLDVGCGGGSHSSLLAEQYPKAHFVGLEIGEDAIRQAKQRKTKSGAAFNNLEFIQCD 172
Query 131 ALSIP-IDSESVDLVYSRDTILHLSVDEKKLLFSKAFDWLKPD 172
A +P I ++S DLV L+F D +PD
Sbjct 173 AGKMPEIWTDSFDLV---------------LIFDACHDQCRPD 200
> cpv:cgd7_630 Ym1014wp-like, Ymb4 methylase
Length=315
Score = 40.8 bits (94), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 28/103 (27%), Positives = 48/103 (46%), Gaps = 5/103 (4%)
Query 76 GGKAIDVGCGIGGSTAALADKFNANVLGVDLSSNMISIAKERYSSRPDLKFLVADALSIP 135
G +DVGCG G + D +G D +++ A R PDL+ V D + +
Sbjct 81 GSLLLDVGCGNGRFMDCIKDS-KVCFMGTDRCKSLLGSAIAR---NPDLQVFVDDCMRLN 136
Query 136 IDSESVDLVYSRDTILHLSVDEKKL-LFSKAFDWLKPDGQLVI 177
+ S + D + + HLS E+++ S+ L+ +G L+I
Sbjct 137 VRSGTFDGIICIAVLHHLSTPERRIQAVSELIRCLRRNGTLLI 179
> cel:ZK652.9 coq-5; COenzyme Q (ubiquinone) biosynthesis family
member (coq-5); K06127 ubiquinone biosynthesis methyltransferase
[EC:2.1.1.-]
Length=285
Score = 40.8 bits (94), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 43/181 (23%), Positives = 73/181 (40%), Gaps = 28/181 (15%)
Query 73 LPKGGKAIDVGCGIGG-STAALADKFNANVLGVDLSSNMISIAKERYSSRPDLK-----F 126
+P K +D+ G G + L A V D++ M+ + K+R D++ +
Sbjct 93 VPYNAKCLDMAGGTGDIAFRILRHSPTAKVTVSDINQPMLDVGKKRAEKERDIQPSRAEW 152
Query 127 LVADALSIPIDSESVDLVYSRDTILHLSVDEKKLLFSKAFDWLKPDGQLVITDY------ 180
+ A+A +P +S + DL I + + EK + +AF LKP GQL I ++
Sbjct 153 VCANAEQMPFESNTYDLFTMSFGIRNCTHPEK--VVREAFRVLKPGGQLAILEFSEVNSA 210
Query 181 --------------CCGPPEKWDDEFKAYLQDRNYKLVQLEEYRQLLTEAGFDVVKAANH 226
G D YL + K +E+ +++ E GF V+ N
Sbjct 211 LKPIYDAYSFNVIPVLGEILASDRASYQYLVESIRKFPNQDEFARIIREEGFSNVRYENL 270
Query 227 T 227
T
Sbjct 271 T 271
> ath:AT3G17365 catalytic/ methyltransferase
Length=239
Score = 40.4 bits (93), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 19/67 (28%), Positives = 35/67 (52%), Gaps = 0/67 (0%)
Query 65 AEILREIVLPKGGKAIDVGCGIGGSTAALADKFNANVLGVDLSSNMISIAKERYSSRPDL 124
A ++ V + + + +GCG + + D +V+ +D+SS +I ++YS RP L
Sbjct 37 APLINLYVPQRNQRVLVIGCGNSAFSEGMVDDGYEDVVSIDISSVVIDTMIKKYSDRPQL 96
Query 125 KFLVADA 131
K+L D
Sbjct 97 KYLKMDV 103
> xla:414682 mettl13; methyltransferase like 13
Length=693
Score = 39.7 bits (91), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 31/108 (28%), Positives = 46/108 (42%), Gaps = 2/108 (1%)
Query 53 RGFVSSGGGETTAEILREIVLPKGGKAIDVGCGIGGSTAALADKFNANVLGVDLSSNMIS 112
R F GG +L + + P+ K VGCG + L D N+ +D+S +I
Sbjct 27 RAFEWYGGYLELCGLLHKYIKPRD-KVFVVGCGNSELSEQLYDAGCQNLTNIDVSEVVIR 85
Query 113 IAKERYSS-RPDLKFLVADALSIPIDSESVDLVYSRDTILHLSVDEKK 159
ER S+ RP++ F V DA D V + T+ + D K
Sbjct 86 QMNERNSNRRPNMTFQVMDATQTTFDDSCFQAVLDKGTLDAIMTDTDK 133
> eco:b1870 cmoA, ECK1871, JW1859, yecO; tRNA cmo(5)U34 methyltransferase,
SAM-dependent
Length=247
Score = 39.7 bits (91), Expect = 0.013, Method: Compositional matrix adjust.
Identities = 29/119 (24%), Positives = 54/119 (45%), Gaps = 8/119 (6%)
Query 67 ILREIVLPKGGKAIDVGCGIGGSTAALADKF---NANVLGVDLSSNMISIAKER---YSS 120
+L E + G + D+GC +G +T ++ N ++ +D S MI + Y +
Sbjct 48 MLAERFVQPGTQVYDLGCSLGAATLSVRRNIHHDNCKIIAIDNSPAMIERCRRHIDAYKA 107
Query 121 RPDLKFLVADALSIPIDSESVDLVYSRDTILHLSVDEKKLLFSKAFDWLKPDGQLVITD 179
+ + D I I++ S +V T+ L E++ L K + L P G LV+++
Sbjct 108 PTPVDVIEGDIRDIAIENAS--MVVLNFTLQFLEPSERQALLDKIYQGLNPGGALVLSE 164
> sce:YER175C TMT1, TAM1; Tmt1p
Length=299
Score = 39.7 bits (91), Expect = 0.014, Method: Compositional matrix adjust.
Identities = 27/81 (33%), Positives = 41/81 (50%), Gaps = 20/81 (24%)
Query 80 IDVGCGIGGSTAALADKFNA--NVLGVDLSSNMISIA---KE-----------RYSSRPD 123
+DVGCG G +T +A + ++G DLS+ MI A KE + SS D
Sbjct 41 VDVGCGPGTATLQMAQELKPFEQIIGSDLSATMIKTAEVIKEGSPDTYKNVSFKISSSDD 100
Query 124 LKFLVADALSIPIDSESVDLV 144
KFL AD+ +D + +D++
Sbjct 101 FKFLGADS----VDKQKIDMI 117
> cel:R08E5.3 hypothetical protein
Length=365
Score = 39.3 bits (90), Expect = 0.017, Method: Compositional matrix adjust.
Identities = 32/112 (28%), Positives = 59/112 (52%), Gaps = 12/112 (10%)
Query 76 GGKAIDVGCGIGGSTAALADKF-NANVLGVDLSSNMISIAKERYSSR----PDLKFLVAD 130
G + +DVGCG G ++ LA+++ ++ +G+D+ + I AK+R + +L+F+ D
Sbjct 176 GVRVLDVGCGGGFHSSLLAEQYPKSHFVGLDIGEDAIRQAKQRKTKSGAAFNNLEFIECD 235
Query 131 ALSIP-IDSESVDLVYSRDTILHLSVDEKK--LLFSKAFDWLKPDGQLVITD 179
A +P I ++S DLV I D+++ L + LKP G + +
Sbjct 236 AGKMPEIWTDSFDLVL----IFDACHDQRRPDLCVQEIHRVLKPSGMFAMVE 283
> mmu:71449 Mettl13, 5630401D24Rik; methyltransferase like 13
Length=698
Score = 39.3 bits (90), Expect = 0.018, Method: Compositional matrix adjust.
Identities = 28/113 (24%), Positives = 54/113 (47%), Gaps = 2/113 (1%)
Query 65 AEILREIVLPKGGKAIDVGCGIGGSTAALADKFNANVLGVDLSSNMISIAKERYSS-RPD 123
E+L + + PK K + +GCG + L D +++ +D+S +I KER S RP
Sbjct 39 CEVLHKYIKPKE-KVLVIGCGNSELSEQLYDVGYQDIVNIDISEVVIKQMKERNGSRRPH 97
Query 124 LKFLVADALSIPIDSESVDLVYSRDTILHLSVDEKKLLFSKAFDWLKPDGQLV 176
+ FL D + + +V + T+ + DE+++ + L G+++
Sbjct 98 MSFLKMDMTQLEFPDATFQVVLDKGTLDAVLTDEEEVTLRQVDRMLAEVGRVL 150
> ath:AT2G43920 thiol methyltransferase, putative
Length=200
Score = 38.9 bits (89), Expect = 0.023, Method: Compositional matrix adjust.
Identities = 35/122 (28%), Positives = 64/122 (52%), Gaps = 16/122 (13%)
Query 77 GKAIDVGCGIGGSTAALA--DKFNANVLGVDLSSNMISIAKERYSSRPDLKF--LVADAL 132
G+ + GCG G A+A ++F V+G+D+S ++ A E Y S P ++ V + +
Sbjct 68 GRTLVPGCGGGHDVVAMASPERF---VVGLDISDKALNKANETYGSSPKAEYFSFVKEDV 124
Query 133 SIPIDSESVDLVYSRDTILHLSVD-EKKLLFSKAF-DWLKPDGQLV-----ITDYCCGPP 185
+E DL++ D + +++ E + + K+ + LKPDG+L+ +TD+ G P
Sbjct 125 FTWRPNELFDLIF--DYVFFCAIEPEMRPAWGKSMHELLKPDGELITLMYPMTDHEGGAP 182
Query 186 EK 187
K
Sbjct 183 YK 184
Lambda K H
0.318 0.136 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 11647553988
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40