bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_0111_orf5
Length=197
Score E
Sequences producing significant alignments: (Bits) Value
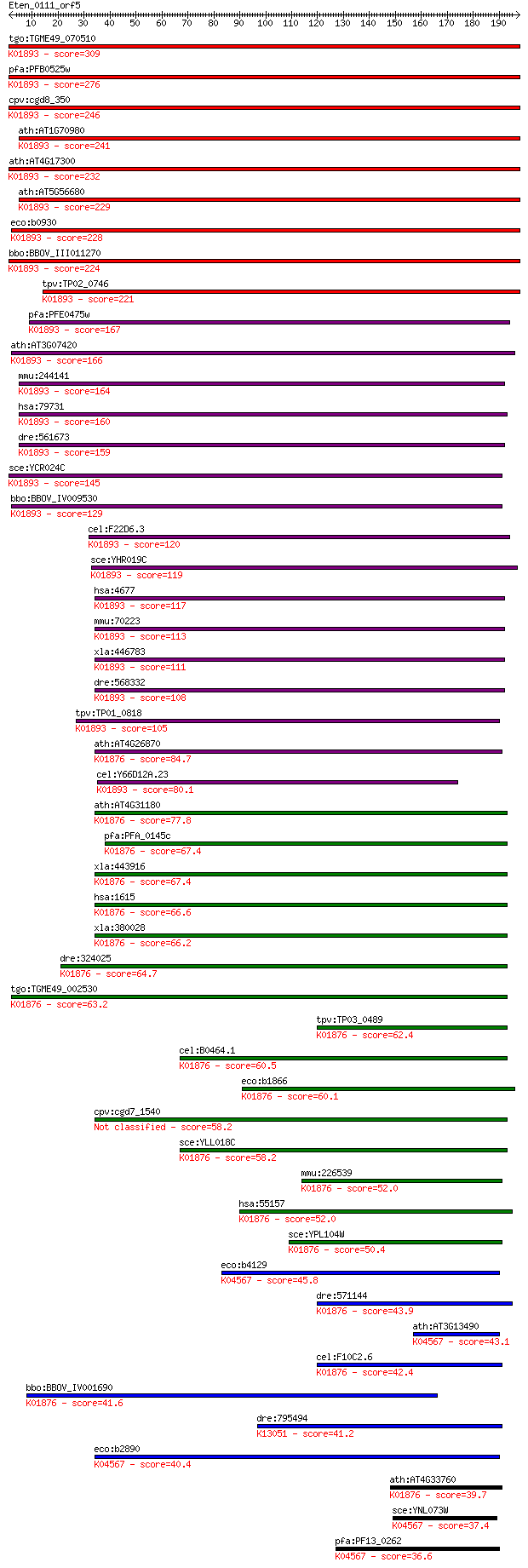
tgo:TGME49_070510 asparaginyl-tRNA synthetase, putative (EC:6.... 309 4e-84
pfa:PFB0525w asparagine-tRNA ligase, putative; K01893 asparagi... 276 4e-74
cpv:cgd8_350 asparaginyl-tRNA synthetase (NOB+tRNA synthase) ;... 246 3e-65
ath:AT1G70980 SYNC3; SYNC3; ATP binding / aminoacyl-tRNA ligas... 241 2e-63
ath:AT4G17300 NS1; NS1; asparagine-tRNA ligase (EC:6.1.1.22); ... 232 5e-61
ath:AT5G56680 SYNC1; SYNC1; ATP binding / aminoacyl-tRNA ligas... 229 5e-60
eco:b0930 asnS, ECK0921, JW0913, lcs, tss; asparaginyl tRNA sy... 228 1e-59
bbo:BBOV_III011270 17.m07970; asparaginyl-tRNA synthetase fami... 224 1e-58
tpv:TP02_0746 asparaginyl-tRNA synthetase; K01893 asparaginyl-... 221 1e-57
pfa:PFE0475w asparagine-tRNA ligase, putative (EC:6.1.1.22); K... 167 3e-41
ath:AT3G07420 NS2; NS2; asparagine-tRNA ligase (EC:6.1.1.22); ... 166 6e-41
mmu:244141 Nars2, AI875199, MGC41336; asparaginyl-tRNA synthet... 164 2e-40
hsa:79731 NARS2, FLJ23441, SLM5; asparaginyl-tRNA synthetase 2... 160 2e-39
dre:561673 MGC153218; zgc:153218 (EC:6.1.1.22); K01893 asparag... 159 8e-39
sce:YCR024C SLM5; Mitochondrial asparaginyl-tRNA synthetase (E... 145 1e-34
bbo:BBOV_IV009530 23.m06478; asparaginyl-tRNA synthetase (EC:6... 129 8e-30
cel:F22D6.3 nrs-1; asparagiNyl tRNA Synthetase family member (... 120 3e-27
sce:YHR019C DED81; Ded81p (EC:6.1.1.22); K01893 asparaginyl-tR... 119 8e-27
hsa:4677 NARS, ASNRS, NARS1; asparaginyl-tRNA synthetase (EC:6... 117 4e-26
mmu:70223 Nars, 3010001M15Rik, AA960128, ASNRS, C78150; aspara... 113 3e-25
xla:446783 nars, MGC80438; asparaginyl-tRNA synthetase (EC:6.1... 111 2e-24
dre:568332 nars, fb57a10, si:dkey-274m14.2, wu:fb57a10; aspara... 108 1e-23
tpv:TP01_0818 asparaginyl-tRNA synthetase; K01893 asparaginyl-... 105 9e-23
ath:AT4G26870 aspartyl-tRNA synthetase, putative / aspartate--... 84.7 2e-16
cel:Y66D12A.23 hypothetical protein; K01893 asparaginyl-tRNA s... 80.1 4e-15
ath:AT4G31180 aspartyl-tRNA synthetase, putative / aspartate--... 77.8 2e-14
pfa:PFA_0145c aspartyl-tRNA synthetase, putative (EC:6.1.1.12)... 67.4 3e-11
xla:443916 dars, MGC80207; aspartyl-tRNA synthetase (EC:6.1.1.... 67.4 3e-11
hsa:1615 DARS, DKFZp781B11202, MGC111579; aspartyl-tRNA synthe... 66.6 5e-11
xla:380028 dars, MGC53970; aspartyl-tRNA synthetase (EC:6.1.1.... 66.2 7e-11
dre:324025 dars, MGC154056, wu:fc17a11, zgc:154056; aspartyl-t... 64.7 2e-10
tgo:TGME49_002530 aspartyl-tRNA synthetase, putative (EC:6.1.1... 63.2 5e-10
tpv:TP03_0489 aspartyl-tRNA synthetase (EC:6.1.1.12); K01876 a... 62.4 9e-10
cel:B0464.1 drs-1; aspartyl(D) tRNA Synthetase family member (... 60.5 4e-09
eco:b1866 aspS, ECK1867, JW1855, tls; aspartyl-tRNA synthetase... 60.1 5e-09
cpv:cgd7_1540 aspartate--tRNA ligase 58.2 2e-08
sce:YLL018C DPS1; AspRS; aspartyl-tRNA synthetase (EC:6.1.1.12... 58.2 2e-08
mmu:226539 Dars2, 5830468K18Rik, MGC99963; aspartyl-tRNA synth... 52.0 1e-06
hsa:55157 DARS2, ASPRS, FLJ10514, LBSL, MT-ASPRS, RP3-383J4.2;... 52.0 1e-06
sce:YPL104W MSD1, LPG5; Msd1p (EC:6.1.1.12); K01876 aspartyl-t... 50.4 4e-06
eco:b4129 lysU, ECK4123, JW4090; lysine tRNA synthetase, induc... 45.8 9e-05
dre:571144 dars2, si:dkey-21n10.1; aspartyl-tRNA synthetase 2,... 43.9 4e-04
ath:AT3G13490 OVA5; OVA5 (OVULE ABORTION 5); ATP binding / ami... 43.1 6e-04
cel:F10C2.6 drs-2; aspartyl(D) tRNA Synthetase family member (... 42.4 0.001
bbo:BBOV_IV001690 21.m02784; aspartyl-tRNA synthetase (EC:6.1.... 41.6 0.002
dre:795494 L-asparaginase-like; K13051 beta-aspartyl-peptidase... 41.2 0.003
eco:b2890 lysS, asuD, ECK2885, herC, JW2858; lysine tRNA synth... 40.4 0.004
ath:AT4G33760 tRNA synthetase class II (D, K and N) family pro... 39.7 0.007
sce:YNL073W MSK1; Mitochondrial lysine-tRNA synthetase, requir... 37.4 0.035
pfa:PF13_0262 lysine-tRNA ligase (EC:6.1.1.6); K04567 lysyl-tR... 36.6 0.058
> tgo:TGME49_070510 asparaginyl-tRNA synthetase, putative (EC:6.1.1.22);
K01893 asparaginyl-tRNA synthetase [EC:6.1.1.22]
Length=676
Score = 309 bits (791), Expect = 4e-84, Method: Compositional matrix adjust.
Identities = 147/201 (73%), Positives = 169/201 (84%), Gaps = 6/201 (2%)
Query 1 VKSC----LDNCSGEIDWFEKNQEEGLRARLENILAAPFERLTYTDAVIRLQREEAEGRV 56
VK C LDNC +I+WF+KNQEEGL ARLENILA PF R++YT+A+ L+ EE +
Sbjct 478 VKFCVQWVLDNCRADIEWFQKNQEEGLIARLENILAEPFARVSYTEAIEVLKAEEP--KA 535
Query 57 KFQEKVEWGMDLGSEHERYLTEQLAKKPVIVHDYPKDIKAFYMKQNEDGKTVRAMDVLVP 116
+F+EKVEWGMD+GSEHERYLTE + KKP IV++YPKDIKAFYMK NEDGKTVRAMDVLVP
Sbjct 536 QFKEKVEWGMDMGSEHERYLTENVYKKPCIVYNYPKDIKAFYMKLNEDGKTVRAMDVLVP 595
Query 117 KIGELIGGSQREEDLEKLDRMIKEKGLNLDSYWWYRELRSYGTVPHAGFGLGFERLIMLV 176
KIGEL+GGSQRE+D ++L MI K L+ YWWY ELR YGT+PHAGFGLGFERL+MLV
Sbjct 596 KIGELVGGSQREDDRDRLAAMIAAKDLDPKPYWWYMELREYGTIPHAGFGLGFERLVMLV 655
Query 177 TGIENIRDTIPYPRYPGHAEF 197
TGIENIRDTIPYPRYPGHAEF
Sbjct 656 TGIENIRDTIPYPRYPGHAEF 676
> pfa:PFB0525w asparagine-tRNA ligase, putative; K01893 asparaginyl-tRNA
synthetase [EC:6.1.1.22]
Length=610
Score = 276 bits (705), Expect = 4e-74, Method: Compositional matrix adjust.
Identities = 132/201 (65%), Positives = 162/201 (80%), Gaps = 8/201 (3%)
Query 1 VKSCLD----NCSGEIDWFEKNQEEGLRARLENILAAPFERLTYTDAVIRLQREEAEGRV 56
+K C+D N +I +FE+N E L RL+NIL F ++TYT+A+ LQ
Sbjct 414 IKYCIDYVLNNNFHDIYYFEENVETNLIKRLKNILNEDFAKITYTNAIEILQNYSDS--- 470
Query 57 KFQEKVEWGMDLGSEHERYLTEQLAKKPVIVHDYPKDIKAFYMKQNEDGKTVRAMDVLVP 116
F+ KVEWGMDL SEHER++ E++ KKPVIV++YPKD+KAFYMK NED KTV AMDVLVP
Sbjct 471 -FEVKVEWGMDLQSEHERFIAEKIFKKPVIVYNYPKDLKAFYMKLNEDNKTVAAMDVLVP 529
Query 117 KIGELIGGSQREEDLEKLDRMIKEKGLNLDSYWWYRELRSYGTVPHAGFGLGFERLIMLV 176
KIGE+IGGSQRE++LE+LD+MIKEK LN+DSYWWYR+LR YG+ PHAGFGLGFERLIMLV
Sbjct 530 KIGEVIGGSQREDNLERLDKMIKEKKLNIDSYWWYRQLRQYGSHPHAGFGLGFERLIMLV 589
Query 177 TGIENIRDTIPYPRYPGHAEF 197
TG++NI+DTIP+PRYPGHAEF
Sbjct 590 TGVDNIKDTIPFPRYPGHAEF 610
> cpv:cgd8_350 asparaginyl-tRNA synthetase (NOB+tRNA synthase)
; K01893 asparaginyl-tRNA synthetase [EC:6.1.1.22]
Length=499
Score = 246 bits (628), Expect = 3e-65, Method: Compositional matrix adjust.
Identities = 114/197 (57%), Positives = 148/197 (75%), Gaps = 4/197 (2%)
Query 1 VKSCLDNCSGEIDWFEKNQEEGLRARLENILAAPFERLTYTDAVIRLQREEAEGRVKFQE 60
V+ L N ++ + +KN E GL RL+ I F R++YT+A+ L+ + E F
Sbjct 307 VEYVLINNMPDLLYLDKNIENGLVERLKVICKEEFARISYTEAIEMLKPHDKE----FTV 362
Query 61 KVEWGMDLGSEHERYLTEQLAKKPVIVHDYPKDIKAFYMKQNEDGKTVRAMDVLVPKIGE 120
V WGMDLGSEHE+Y+T+ L K+P I+++YPKDIK+FYMK NEDG TV AMD+LVPKIGE
Sbjct 363 PVSWGMDLGSEHEKYITDVLEKRPCIIYNYPKDIKSFYMKLNEDGNTVAAMDILVPKIGE 422
Query 121 LIGGSQREEDLEKLDRMIKEKGLNLDSYWWYRELRSYGTVPHAGFGLGFERLIMLVTGIE 180
+IGGSQRE+D+EKL+ IK + ++ YWWY ELR YG+VPH+GFGLGFERLIM+VTG+E
Sbjct 423 VIGGSQREDDIEKLENAIKSRNMDPAPYWWYNELRKYGSVPHSGFGLGFERLIMMVTGVE 482
Query 181 NIRDTIPYPRYPGHAEF 197
N+RD IP+PRYP H EF
Sbjct 483 NVRDVIPFPRYPNHCEF 499
> ath:AT1G70980 SYNC3; SYNC3; ATP binding / aminoacyl-tRNA ligase/
asparagine-tRNA ligase/ aspartate-tRNA ligase/ nucleic
acid binding / nucleotide binding (EC:6.1.1.22); K01893 asparaginyl-tRNA
synthetase [EC:6.1.1.22]
Length=571
Score = 241 bits (614), Expect = 2e-63, Method: Compositional matrix adjust.
Identities = 112/194 (57%), Positives = 147/194 (75%), Gaps = 1/194 (0%)
Query 5 LDNCSGEIDWFEKNQEEGLRARLENILAAPFERLTYTDAVIRLQREEAEGRVKFQEKVEW 64
+D C +++ +KN +EG RL + A F+R+TYT+A+ RL++ A+G+V F KVEW
Sbjct 378 MDKCGDDMELMDKNVDEGCTKRLNMVAKASFKRVTYTEAIERLEKAVAQGKVVFDNKVEW 437
Query 65 GMDLGSEHERYLTE-QLAKKPVIVHDYPKDIKAFYMKQNEDGKTVRAMDVLVPKIGELIG 123
G+DL SEHERYLTE + +KP+IV++YPK IKAFYM+ N+D KTV AMDVLVPK+GELIG
Sbjct 438 GIDLASEHERYLTEVEFDQKPIIVYNYPKGIKAFYMRLNDDEKTVAAMDVLVPKVGELIG 497
Query 124 GSQREEDLEKLDRMIKEKGLNLDSYWWYRELRSYGTVPHAGFGLGFERLIMLVTGIENIR 183
GSQREE + + + I+E GL ++ Y WY +LR YGTV H GFGLGFER+I TGI+NIR
Sbjct 498 GSQREERYDVIKQRIEEMGLPMEPYEWYLDLRRYGTVKHCGFGLGFERMIQFATGIDNIR 557
Query 184 DTIPYPRYPGHAEF 197
D IP+PRYPG A+
Sbjct 558 DVIPFPRYPGKADL 571
> ath:AT4G17300 NS1; NS1; asparagine-tRNA ligase (EC:6.1.1.22);
K01893 asparaginyl-tRNA synthetase [EC:6.1.1.22]
Length=567
Score = 232 bits (592), Expect = 5e-61, Method: Compositional matrix adjust.
Identities = 114/198 (57%), Positives = 146/198 (73%), Gaps = 5/198 (2%)
Query 1 VKSCLDNCSGEIDWFEKNQEEGLRARLENILAAPFERLTYTDAVIRLQREEAEGRVKFQE 60
VK LDNC ++++F+ E+G+ RL ++ F +L YTDA+ L + KF
Sbjct 374 VKYVLDNCKEDMEFFDTWIEKGIIRRLSDVAEKEFLQLGYTDAIEILLKANK----KFDF 429
Query 61 KVEWGMDLGSEHERYLTEQ-LAKKPVIVHDYPKDIKAFYMKQNEDGKTVRAMDVLVPKIG 119
V+WG+DL SEHERY+TE+ +PVI+ DYPK+IKAFYM++N+DGKTV AMD+LVP+IG
Sbjct 430 PVKWGLDLQSEHERYITEEAFGGRPVIIRDYPKEIKAFYMRENDDGKTVAAMDMLVPRIG 489
Query 120 ELIGGSQREEDLEKLDRMIKEKGLNLDSYWWYRELRSYGTVPHAGFGLGFERLIMLVTGI 179
ELIGGSQREE LE L+ + E LN +SYWWY +LR YG+VPHAGFGLGFERL+ VTGI
Sbjct 490 ELIGGSQREERLEVLEARLDELKLNKESYWWYLDLRRYGSVPHAGFGLGFERLVQFVTGI 549
Query 180 ENIRDTIPYPRYPGHAEF 197
+NIRD IP+PR P AEF
Sbjct 550 DNIRDVIPFPRTPASAEF 567
> ath:AT5G56680 SYNC1; SYNC1; ATP binding / aminoacyl-tRNA ligase/
asparagine-tRNA ligase/ aspartate-tRNA ligase/ nucleic
acid binding / nucleotide binding (EC:6.1.1.22); K01893 asparaginyl-tRNA
synthetase [EC:6.1.1.22]
Length=572
Score = 229 bits (583), Expect = 5e-60, Method: Compositional matrix adjust.
Identities = 108/193 (55%), Positives = 143/193 (74%), Gaps = 1/193 (0%)
Query 5 LDNCSGEIDWFEKNQEEGLRARLENILAAPFERLTYTDAVIRLQREEAEGRVKFQEKVEW 64
L+ C +++ KN + G RL+ + + PF R+TYT A+ L+ A+G+ +F VEW
Sbjct 381 LEKCYADMELMAKNFDSGCIDRLKLVASTPFGRITYTKAIELLEEAVAKGK-EFDNNVEW 439
Query 65 GMDLGSEHERYLTEQLAKKPVIVHDYPKDIKAFYMKQNEDGKTVRAMDVLVPKIGELIGG 124
G+DL SEHERYLTE L +KP+IV++YPK IKAFYM+ N+D KTV AMDVLVPK+GELIGG
Sbjct 440 GIDLASEHERYLTEVLFQKPLIVYNYPKGIKAFYMRLNDDEKTVAAMDVLVPKVGELIGG 499
Query 125 SQREEDLEKLDRMIKEKGLNLDSYWWYRELRSYGTVPHAGFGLGFERLIMLVTGIENIRD 184
SQREE + + + I+E GL ++ Y WY +LR YGTV H GFGLGFER+I+ TG++NIRD
Sbjct 500 SQREERYDVIKKRIEEMGLPIEPYEWYLDLRRYGTVKHCGFGLGFERMILFATGLDNIRD 559
Query 185 TIPYPRYPGHAEF 197
IP+PRYPG A+
Sbjct 560 VIPFPRYPGKADL 572
> eco:b0930 asnS, ECK0921, JW0913, lcs, tss; asparaginyl tRNA
synthetase (EC:6.1.1.22); K01893 asparaginyl-tRNA synthetase
[EC:6.1.1.22]
Length=466
Score = 228 bits (581), Expect = 1e-59, Method: Compositional matrix adjust.
Identities = 113/196 (57%), Positives = 143/196 (72%), Gaps = 4/196 (2%)
Query 2 KSCLDNCSGEIDWFEKNQEEGLRARLENILAAPFERLTYTDAVIRLQREEAEGRVKFQEK 61
K+ L+ + ++ +F + ++ +RLE + A F ++ YTDAV L E GR KF+
Sbjct 275 KAVLEERADDMKFFAERVDKDAVSRLERFIEADFAQVDYTDAVTIL---ENCGR-KFENP 330
Query 62 VEWGMDLGSEHERYLTEQLAKKPVIVHDYPKDIKAFYMKQNEDGKTVRAMDVLVPKIGEL 121
V WG+DL SEHERYL E+ K PV+V +YPKDIKAFYM+ NEDGKTV AMDVL P IGE+
Sbjct 331 VYWGVDLSSEHERYLAEEHFKAPVVVKNYPKDIKAFYMRLNEDGKTVAAMDVLAPGIGEI 390
Query 122 IGGSQREEDLEKLDRMIKEKGLNLDSYWWYRELRSYGTVPHAGFGLGFERLIMLVTGIEN 181
IGGSQREE L+ LD + E GLN + YWWYR+LR YGTVPH+GFGLGFERLI VTG++N
Sbjct 391 IGGSQREERLDVLDERMLEMGLNKEDYWWYRDLRRYGTVPHSGFGLGFERLIAYVTGVQN 450
Query 182 IRDTIPYPRYPGHAEF 197
+RD IP+PR P +A F
Sbjct 451 VRDVIPFPRTPRNASF 466
> bbo:BBOV_III011270 17.m07970; asparaginyl-tRNA synthetase family
protein; K01893 asparaginyl-tRNA synthetase [EC:6.1.1.22]
Length=605
Score = 224 bits (571), Expect = 1e-58, Method: Compositional matrix adjust.
Identities = 107/218 (49%), Positives = 147/218 (67%), Gaps = 21/218 (9%)
Query 1 VKSCLDNCSGEIDWFEKNQEEGLRARLENILAAPFERLTYTDAVIRLQREEAE---GRVK 57
+K LD+C ++ + E ++GL L + F R+TYTD + L + EA+ GRV+
Sbjct 388 IKYALDHCLDDLLYLESIGDKGLVDSLRSFATEKFVRITYTDVIDILMQHEADFQAGRVE 447
Query 58 ------------------FQEKVEWGMDLGSEHERYLTEQLAKKPVIVHDYPKDIKAFYM 99
F+ VEWG+DL SEHER++ E + KKP I+++YP IKAFYM
Sbjct 448 IDSAQCKREDGSEPRKTAFENHVEWGIDLSSEHERFIAEAVFKKPTIIYNYPAKIKAFYM 507
Query 100 KQNEDGKTVRAMDVLVPKIGELIGGSQREEDLEKLDRMIKEKGLNLDSYWWYRELRSYGT 159
++N+DGKT AMDV+VP+ GEL+GGSQREE + L+ IK+ GL +D Y WY +LR+YG+
Sbjct 508 RRNDDGKTCAAMDVIVPRFGELVGGSQREERRDVLEDSIKQNGLCMDDYTWYMDLRTYGS 567
Query 160 VPHAGFGLGFERLIMLVTGIENIRDTIPYPRYPGHAEF 197
+PH+GFGLGFERL+MLVTG+ NIRD IP+PRY G A+F
Sbjct 568 IPHSGFGLGFERLVMLVTGVSNIRDVIPFPRYVGKADF 605
> tpv:TP02_0746 asparaginyl-tRNA synthetase; K01893 asparaginyl-tRNA
synthetase [EC:6.1.1.22]
Length=710
Score = 221 bits (563), Expect = 1e-57, Method: Composition-based stats.
Identities = 100/187 (53%), Positives = 140/187 (74%), Gaps = 4/187 (2%)
Query 14 WFEKNQEEGLRARLENILAAPFERLTYTDAVIRLQ---REEAEGRVKFQEKVEWGMDLGS 70
+F ++ L +RL NI+ F L+YT + LQ ++ E ++ + V WG+DL S
Sbjct 525 FFNNTVDKELLSRLYNIVDNEFVHLSYTSVIDILQEYIKQNPEQPFEYND-VHWGIDLQS 583
Query 71 EHERYLTEQLAKKPVIVHDYPKDIKAFYMKQNEDGKTVRAMDVLVPKIGELIGGSQREED 130
EHER+++E++ PVI+++YPK IKAFYM++N+DGKTV AMD+++PKIGELIGGSQREE
Sbjct 584 EHERFISEKVFNGPVIIYNYPKQIKAFYMRRNDDGKTVAAMDLIIPKIGELIGGSQREER 643
Query 131 LEKLDRMIKEKGLNLDSYWWYRELRSYGTVPHAGFGLGFERLIMLVTGIENIRDTIPYPR 190
+ L++ IKE LN+ YWWY +LR YGT+ H+GFGLGFERLIM+VTG++NI+D IP+PR
Sbjct 644 FDYLEKSIKENKLNMQDYWWYMDLRRYGTIVHSGFGLGFERLIMMVTGVQNIKDVIPFPR 703
Query 191 YPGHAEF 197
Y GH+ F
Sbjct 704 YSGHSLF 710
> pfa:PFE0475w asparagine-tRNA ligase, putative (EC:6.1.1.22);
K01893 asparaginyl-tRNA synthetase [EC:6.1.1.22]
Length=722
Score = 167 bits (422), Expect = 3e-41, Method: Composition-based stats.
Identities = 78/185 (42%), Positives = 120/185 (64%), Gaps = 6/185 (3%)
Query 9 SGEIDWFEKNQEEGLRARLENILAAPFERLTYTDAVIRLQREEAEGRVKFQEKVEWGMDL 68
S ++++ + ++ L+ +LENIL F +TY +A+ L++E F + +WG D
Sbjct 540 SEDVNYLNEYHDKDLKNKLENILNKKFVLITYDEAINILKKENV-----FYD-FQWGKDF 593
Query 69 GSEHERYLTEQLAKKPVIVHDYPKDIKAFYMKQNEDGKTVRAMDVLVPKIGELIGGSQRE 128
E ++YL K PV++ +YP IK FYMK NED KTV MD++ P +GE++GGS+RE
Sbjct 594 SFEEQKYLATNYFKAPVVIINYPTHIKPFYMKLNEDEKTVSCMDIIFPDVGEVVGGSERE 653
Query 129 EDLEKLDRMIKEKGLNLDSYWWYRELRSYGTVPHAGFGLGFERLIMLVTGIENIRDTIPY 188
+L+ L + ++ K LN+ Y Y +LR +G +PHAGFG+G +RLIM ++ I NIRD + +
Sbjct 654 INLKNLKKQMENKKLNMKLYDPYIQLRKHGNIPHAGFGIGMDRLIMFLSSINNIRDVVTF 713
Query 189 PRYPG 193
PRYPG
Sbjct 714 PRYPG 718
> ath:AT3G07420 NS2; NS2; asparagine-tRNA ligase (EC:6.1.1.22);
K01893 asparaginyl-tRNA synthetase [EC:6.1.1.22]
Length=638
Score = 166 bits (419), Expect = 6e-41, Method: Compositional matrix adjust.
Identities = 81/194 (41%), Positives = 120/194 (61%), Gaps = 3/194 (1%)
Query 2 KSCLDNCSGEIDWFEKNQEEGLRARLENILAAPFERLTYTDAVIRLQREEAEGRVKFQEK 61
K L+N ++ + K ++ + RLE ++ R +YT+ + LQ+ KF+ K
Sbjct 446 KYVLENRDEDMKFISKRVDKTITTRLEATASSSLLRFSYTEVISLLQKATT---TKFETK 502
Query 62 VEWGMDLGSEHERYLTEQLAKKPVIVHDYPKDIKAFYMKQNEDGKTVRAMDVLVPKIGEL 121
EWG+ L +EH YLT+++ K PVIVH YPK IK FY++ N+D KTV A D++VPK+G +
Sbjct 503 PEWGVALTTEHLSYLTDEIYKGPVIVHTYPKAIKQFYVRLNDDKKTVAAFDLVVPKVGVV 562
Query 122 IGGSQREEDLEKLDRMIKEKGLNLDSYWWYRELRSYGTVPHAGFGLGFERLIMLVTGIEN 181
I GSQ EE E LD I E G + + WY +LR +GTV H+G L E++++ TG+ +
Sbjct 563 ITGSQNEERFEILDARIGESGFTREKFEWYLDLRRHGTVKHSGISLSMEQMLLYATGLPD 622
Query 182 IRDTIPYPRYPGHA 195
I+D IP+PR G A
Sbjct 623 IKDAIPFPRSWGKA 636
> mmu:244141 Nars2, AI875199, MGC41336; asparaginyl-tRNA synthetase
2 (mitochondrial)(putative) (EC:6.1.1.22); K01893 asparaginyl-tRNA
synthetase [EC:6.1.1.22]
Length=477
Score = 164 bits (414), Expect = 2e-40, Method: Compositional matrix adjust.
Identities = 80/189 (42%), Positives = 120/189 (63%), Gaps = 7/189 (3%)
Query 5 LDNCSGEIDWFEKNQEEGLRARLENILAAPFERLTYTDAVIRLQREEAEGRVKFQEKVEW 64
L +C +++ + G + RLE++L F ++YT+A+ L+ +A F K W
Sbjct 288 LSHCPEDVELCHQFIAAGQKGRLEHMLKNNFLIISYTEAIEILK--QASQNFAFTPK--W 343
Query 65 GMDLGSEHERYLTEQLAKKPVIVHDYPKDIKAFYMKQNEDG--KTVRAMDVLVPKIGELI 122
G+DL +EHE+YL PV V +YP ++K FYM++NEDG TV A+D+LVP +GEL
Sbjct 344 GVDLQTEHEKYLVRHCGNIPVFVINYPSELKPFYMRENEDGPQNTVAAVDLLVPGVGELF 403
Query 123 GGSQREEDLEKLDRMIKEKGLNLDSYWWYRELRSYGTVPHAGFGLGFERLIMLVTGIENI 182
GGS REE L++ + GL +Y WY +LR +G+VPH GFG+GFER + + G++NI
Sbjct 404 GGSLREERYHVLEQRLARSGLT-KAYQWYLDLRKFGSVPHGGFGMGFERYLQCILGVDNI 462
Query 183 RDTIPYPRY 191
+D IP+PR+
Sbjct 463 KDVIPFPRF 471
> hsa:79731 NARS2, FLJ23441, SLM5; asparaginyl-tRNA synthetase
2, mitochondrial (putative) (EC:6.1.1.22); K01893 asparaginyl-tRNA
synthetase [EC:6.1.1.22]
Length=477
Score = 160 bits (406), Expect = 2e-39, Method: Compositional matrix adjust.
Identities = 81/190 (42%), Positives = 115/190 (60%), Gaps = 7/190 (3%)
Query 5 LDNCSGEIDWFEKNQEEGLRARLENILAAPFERLTYTDAVIRLQREEAEGRVKFQEKVEW 64
L C +++ K G + RLE++L F ++YT+AV L++ F EW
Sbjct 288 LSKCPEDVELCHKFIAPGQKDRLEHMLKNNFLIISYTEAVEILKQASQ----NFTFTPEW 343
Query 65 GMDLGSEHERYLTEQLAKKPVIVHDYPKDIKAFYMKQNEDG--KTVRAMDVLVPKIGELI 122
G DL +EHE+YL + PV V +YP +K FYM+ NEDG TV A+D+LVP +GEL
Sbjct 344 GADLRTEHEKYLVKHCGNIPVFVINYPLTLKPFYMRDNEDGPQHTVAAVDLLVPGVGELF 403
Query 123 GGSQREEDLEKLDRMIKEKGLNLDSYWWYRELRSYGTVPHAGFGLGFERLIMLVTGIENI 182
GG REE L+ + GL + Y WY +LR +G+VPH GFG+GFER + + G++NI
Sbjct 404 GGGLREERYHFLEERLARSGLT-EVYQWYLDLRRFGSVPHGGFGMGFERYLQCILGVDNI 462
Query 183 RDTIPYPRYP 192
+D IP+PR+P
Sbjct 463 KDVIPFPRFP 472
> dre:561673 MGC153218; zgc:153218 (EC:6.1.1.22); K01893 asparaginyl-tRNA
synthetase [EC:6.1.1.22]
Length=485
Score = 159 bits (401), Expect = 8e-39, Method: Compositional matrix adjust.
Identities = 75/189 (39%), Positives = 116/189 (61%), Gaps = 7/189 (3%)
Query 5 LDNCSGEIDWFEKNQEEGLRARLENILAAPFERLTYTDAVIRLQREEAEGRVKFQEKVEW 64
NC+ +++ F K+ G R +++ +L F ++YT+A+ L+ F EW
Sbjct 296 FSNCTEDVELFHKHVAPGHREQVDPMLNRKFNVISYTEAIDILKSSSQ----NFTFSPEW 351
Query 65 GMDLGSEHERYLTEQLAKKPVIVHDYPKDIKAFYMKQNEDG--KTVRAMDVLVPKIGELI 122
G DL +EHE++L + PV V +YP ++K FY + N+D T A+D+LVP +GEL
Sbjct 352 GRDLQTEHEKFLVKHCGNTPVFVVNYPYELKPFYARDNQDHPRHTAAAVDLLVPGVGELC 411
Query 123 GGSQREEDLEKLDRMIKEKGLNLDSYWWYRELRSYGTVPHAGFGLGFERLIMLVTGIENI 182
GGS REE L+ L + + + GL D+Y WY +LR +G+VPH GFG+GFER + + G+ NI
Sbjct 412 GGSLREERLQLLTQRLSQAGLE-DTYAWYLQLREFGSVPHGGFGMGFERYLQCILGLNNI 470
Query 183 RDTIPYPRY 191
+D IP+ R+
Sbjct 471 KDAIPFARF 479
> sce:YCR024C SLM5; Mitochondrial asparaginyl-tRNA synthetase
(EC:6.1.1.22); K01893 asparaginyl-tRNA synthetase [EC:6.1.1.22]
Length=492
Score = 145 bits (365), Expect = 1e-34, Method: Compositional matrix adjust.
Identities = 81/195 (41%), Positives = 118/195 (60%), Gaps = 11/195 (5%)
Query 1 VKSCLDNCSGEIDWFEKNQEEGLRARLENILAAPFERLTYTDAVIRLQREEAEGRVKFQE 60
+ S +N S E+ ++ Q+ ++ R E+++ + +TYT+A+ L++ E F+
Sbjct 296 ISSQENNASSELSINQETQQ--IKTRWEDLINEKWHNITYTNAIEILKKRHNEVS-HFKY 352
Query 61 KVEWGMDLGSEHERYLTEQLAKKPVIVHDYPKDIKAFYMKQNED-GKTVRAMDVLVPKIG 119
+ +WG L +EHE++L + K PV V DYP+ K FYMKQN TV D+LVP +G
Sbjct 353 EPKWGQPLQTEHEKFLAGEYFKSPVFVTDYPRLCKPFYMKQNSTPDDTVGCFDLLVPGMG 412
Query 120 ELIGGSQREEDLEKLDRMIKEKGLN----LDSYWWYRELRSYGTVPHAGFGLGFERLIML 175
E+IGGS RE+D +KL R +K +G+N LD WY LR G+ PH GFGLGFER I
Sbjct 413 EIIGGSLREDDYDKLCREMKARGMNRSGELD---WYVSLRKEGSAPHGGFGLGFERFISY 469
Query 176 VTGIENIRDTIPYPR 190
+ G NI+D IP+ R
Sbjct 470 LYGNHNIKDAIPFYR 484
> bbo:BBOV_IV009530 23.m06478; asparaginyl-tRNA synthetase (EC:6.1.1.22);
K01893 asparaginyl-tRNA synthetase [EC:6.1.1.22]
Length=557
Score = 129 bits (323), Expect = 8e-30, Method: Compositional matrix adjust.
Identities = 73/211 (34%), Positives = 107/211 (50%), Gaps = 23/211 (10%)
Query 2 KSCLDNCSGEIDWFEKNQEEGLRARLENILAA----------------------PFERLT 39
++ LDNC +I E++ + + RLE ++ P L
Sbjct 340 RTILDNCRDDILLLERHFKTSIIKRLETLVGTGSSGQDSLPRHLTDDPNYCEPQPIHTLE 399
Query 40 YTDAVIRLQREEAEGRVKFQEKVEWGMDLGSEHERYLTEQLAKKPVIVHDYPKDIKAFYM 99
Y +A+ L R K+EWG L + HER L L V V YP+ I FYM
Sbjct 400 YKEALSILNRAIETDESYEHSKLEWGDQLSAAHERSLLSILESPLVAVVQYPRAIAPFYM 459
Query 100 KQNEDGKTVRAMDVLVPKIGELIGGSQREEDLEKLDRMIKEKGLNLDSYWWYRELRSYGT 159
+Q DG V +D+ +GE+ GGS RE + +L++ ++E G+ Y Y ELR +G
Sbjct 460 QQ-LDGGVVNNVDIFAEGVGEIAGGSIREVNYPELEKSMQEHGVYGKDYAGYLELRKFGN 518
Query 160 VPHAGFGLGFERLIMLVTGIENIRDTIPYPR 190
VPH GFG+GFER+IM + GI+NI+D I +P+
Sbjct 519 VPHGGFGIGFERMIMFLLGIQNIKDVIHFPK 549
> cel:F22D6.3 nrs-1; asparagiNyl tRNA Synthetase family member
(nrs-1); K01893 asparaginyl-tRNA synthetase [EC:6.1.1.22]
Length=545
Score = 120 bits (300), Expect = 3e-27, Method: Compositional matrix adjust.
Identities = 61/163 (37%), Positives = 101/163 (61%), Gaps = 4/163 (2%)
Query 32 AAPFERLTYTDAVIRLQREEAEGRVKFQEKVEWGMDLGSEHERYLTEQLAKKPVIVHDYP 91
A PF+R+ Y +A+ LQ+ + R + EK +G D+ ER +T+ + P++++ +P
Sbjct 382 ARPFKRMPYKEAIEWLQKNDV--RNEMGEKFVYGEDIAEAAERRMTDTIGV-PILLNRFP 438
Query 92 KDIKAFYM-KQNEDGKTVRAMDVLVPKIGELIGGSQREEDLEKLDRMIKEKGLNLDSYWW 150
IKAFYM + +D + ++D+L+P +GE++GGS R ++L ++ GL+ +Y+W
Sbjct 439 HGIKAFYMPRCADDNELTESVDLLMPGVGEIVGGSMRIWKEDQLLAAFEKGGLDSKNYYW 498
Query 151 YRELRSYGTVPHAGFGLGFERLIMLVTGIENIRDTIPYPRYPG 193
Y + R YG+VPH G+GLG ER I +T +IRD YPR+ G
Sbjct 499 YMDQRKYGSVPHGGYGLGLERFICWLTDTNHIRDVCLYPRFVG 541
> sce:YHR019C DED81; Ded81p (EC:6.1.1.22); K01893 asparaginyl-tRNA
synthetase [EC:6.1.1.22]
Length=554
Score = 119 bits (297), Expect = 8e-27, Method: Compositional matrix adjust.
Identities = 58/165 (35%), Positives = 100/165 (60%), Gaps = 4/165 (2%)
Query 33 APFERLTYTDAVIRLQREEAEGRVKFQEKVEWGMDLGSEHERYLTEQLAKKPVIVHDYPK 92
APF RL Y DA+ L + + E ++G D+ ER +T+ + P+ + +P
Sbjct 392 APFMRLQYKDAITWLNEHDIKNEEG--EDFKFGDDIAEAAERKMTDTIGV-PIFLTRFPV 448
Query 93 DIKAFYMKQ-NEDGKTVRAMDVLVPKIGELIGGSQREEDLEKLDRMIKEKGLNLDSYWWY 151
+IK+FYMK+ ++D + ++DVL+P +GE+ GGS R +D+++L K +G++ D+Y+W+
Sbjct 449 EIKSFYMKRCSDDPRVTESVDVLMPNVGEITGGSMRIDDMDELMAGFKREGIDTDAYYWF 508
Query 152 RELRSYGTVPHAGFGLGFERLIMLVTGIENIRDTIPYPRYPGHAE 196
+ R YGT PH G+G+G ER++ + +RD YPR+ G +
Sbjct 509 IDQRKYGTCPHGGYGIGTERILAWLCDRFTVRDCSLYPRFSGRCK 553
> hsa:4677 NARS, ASNRS, NARS1; asparaginyl-tRNA synthetase (EC:6.1.1.22);
K01893 asparaginyl-tRNA synthetase [EC:6.1.1.22]
Length=548
Score = 117 bits (292), Expect = 4e-26, Method: Compositional matrix adjust.
Identities = 60/161 (37%), Positives = 101/161 (62%), Gaps = 8/161 (4%)
Query 34 PFERLTYTDAVIRLQREEAEGRVKFQEKV--EWGMDLGSEHERYLTEQLAKKPVIVHDYP 91
PF+R+ Y+DA++ L+ E VK ++ E+G D+ ER +T+ + +P+++ +P
Sbjct 387 PFKRMNYSDAIVWLK----EHDVKKEDGTFYEFGEDIPEAPERLMTDTI-NEPILLCRFP 441
Query 92 KDIKAFYMKQN-EDGKTVRAMDVLVPKIGELIGGSQREEDLEKLDRMIKEKGLNLDSYWW 150
+IK+FYM++ ED + ++DVL+P +GE++GGS R D E++ K +G++ Y+W
Sbjct 442 VEIKSFYMQRCPEDSRLTESVDVLMPNVGEIVGGSMRIFDSEEILAGYKREGIDPTPYYW 501
Query 151 YRELRSYGTVPHAGFGLGFERLIMLVTGIENIRDTIPYPRY 191
Y + R YGT PH G+GLG ER + + +IRD YPR+
Sbjct 502 YTDQRKYGTCPHGGYGLGLERFLTWILNRYHIRDVCLYPRF 542
> mmu:70223 Nars, 3010001M15Rik, AA960128, ASNRS, C78150; asparaginyl-tRNA
synthetase (EC:6.1.1.22); K01893 asparaginyl-tRNA
synthetase [EC:6.1.1.22]
Length=559
Score = 113 bits (283), Expect = 3e-25, Method: Compositional matrix adjust.
Identities = 60/161 (37%), Positives = 99/161 (61%), Gaps = 8/161 (4%)
Query 34 PFERLTYTDAVIRLQREEAEGRVKFQEKV--EWGMDLGSEHERYLTEQLAKKPVIVHDYP 91
PF R+ Y+DA+ L+ E VK ++ E+G D+ ER +T+ + +P+++ +P
Sbjct 398 PFRRMNYSDAIEWLK----EHDVKKEDGTFYEFGDDIPEAPERLMTDTI-NEPILLCRFP 452
Query 92 KDIKAFYMKQN-EDGKTVRAMDVLVPKIGELIGGSQREEDLEKLDRMIKEKGLNLDSYWW 150
+IK+FYM++ ED + ++DVL+P +GE++GGS R D E++ K +G++ Y+W
Sbjct 453 VEIKSFYMQRCPEDPRLTESVDVLMPNVGEIVGGSMRSWDSEEILEGYKREGIDPAPYYW 512
Query 151 YRELRSYGTVPHAGFGLGFERLIMLVTGIENIRDTIPYPRY 191
Y + R YGT PH G+GLG ER + + +IRD YPR+
Sbjct 513 YTDQRKYGTCPHGGYGLGLERFLSWILNRYHIRDVCLYPRF 553
> xla:446783 nars, MGC80438; asparaginyl-tRNA synthetase (EC:6.1.1.22);
K01893 asparaginyl-tRNA synthetase [EC:6.1.1.22]
Length=559
Score = 111 bits (277), Expect = 2e-24, Method: Compositional matrix adjust.
Identities = 59/161 (36%), Positives = 97/161 (60%), Gaps = 8/161 (4%)
Query 34 PFERLTYTDAVIRLQREEAEGRVKFQEKV--EWGMDLGSEHERYLTEQLAKKPVIVHDYP 91
PF R+ Y++A+ L+ E VK + E+G D+ ER +T+ + +P+++ +P
Sbjct 398 PFRRMNYSEAITWLK----EHDVKKDDGTYYEFGEDIPEAPERLMTDAI-NEPILLCRFP 452
Query 92 KDIKAFYMKQ-NEDGKTVRAMDVLVPKIGELIGGSQREEDLEKLDRMIKEKGLNLDSYWW 150
+IK+FYM++ ED + ++DVL+P +GE++GGS R D E+L K + ++ Y+W
Sbjct 453 AEIKSFYMQRCAEDKRLTESVDVLMPNVGEIVGGSMRIWDSEELLEGYKREEIDPTPYYW 512
Query 151 YRELRSYGTVPHAGFGLGFERLIMLVTGIENIRDTIPYPRY 191
Y + R YGT PH G+GLG ER + + +IRD YPR+
Sbjct 513 YTDQRKYGTCPHGGYGLGLERFLTWILNRHHIRDVCLYPRF 553
> dre:568332 nars, fb57a10, si:dkey-274m14.2, wu:fb57a10; asparaginyl-tRNA
synthetase; K01893 asparaginyl-tRNA synthetase
[EC:6.1.1.22]
Length=556
Score = 108 bits (270), Expect = 1e-23, Method: Compositional matrix adjust.
Identities = 57/161 (35%), Positives = 98/161 (60%), Gaps = 8/161 (4%)
Query 34 PFERLTYTDAVIRLQREEAEGRVKFQEKV--EWGMDLGSEHERYLTEQLAKKPVIVHDYP 91
PF+R+ YT+A+ L+ E +K + E+G D+ ER +T+ + + +++ +P
Sbjct 395 PFKRMNYTEAIDWLK----EHNIKKDDGTFYEFGEDIPEAPERLMTDSI-NETILLCRFP 449
Query 92 KDIKAFYMKQN-EDGKTVRAMDVLVPKIGELIGGSQREEDLEKLDRMIKEKGLNLDSYWW 150
+IK+FYM++ ED + ++DVL+P +GE++GGS R D ++L K +G++ Y+W
Sbjct 450 AEIKSFYMQRCPEDRRLTESVDVLMPNVGEIVGGSMRIWDSDELLEGYKREGIDPTPYYW 509
Query 151 YRELRSYGTVPHAGFGLGFERLIMLVTGIENIRDTIPYPRY 191
Y + R +GT PH G+GLG ER + + +IRD YPR+
Sbjct 510 YTDQRKFGTCPHGGYGLGLERFLTWLLNRHHIRDVCLYPRF 550
> tpv:TP01_0818 asparaginyl-tRNA synthetase; K01893 asparaginyl-tRNA
synthetase [EC:6.1.1.22]
Length=558
Score = 105 bits (262), Expect = 9e-23, Method: Compositional matrix adjust.
Identities = 64/173 (36%), Positives = 91/173 (52%), Gaps = 11/173 (6%)
Query 27 LENILAAPFERLTYTDAVIRLQREEAEGRVKFQEKVEWGMDLGSEHERYLTEQLAKKPVI 86
L ++ A + +TY DAV R+ E G K + EWG L ER L + +
Sbjct 383 LVHLSNANIKMITYEDAV-RILNETNSGLDKQDKVFEWGKRLSEPQERRLLKYFPNDIIA 441
Query 87 VHDYPKDIKAFYMKQN-------EDGK---TVRAMDVLVPKIGELIGGSQREEDLEKLDR 136
+ ++P+ I FYMK + E G TV D+L P E+ GGS RE + E L R
Sbjct 442 IANFPESITPFYMKSSSGTDKSVESGNLKNTVENFDILAPFGYEIAGGSIREHEYEVLKR 501
Query 137 MIKEKGLNLDSYWWYRELRSYGTVPHAGFGLGFERLIMLVTGIENIRDTIPYP 189
+ E+ ++ + Y Y LR +PH GFG+GF+RL+ML+T ENIRD P+P
Sbjct 502 KVNERKISGEDYEKYLNLRKNVYLPHGGFGIGFDRLVMLMTMKENIRDVTPFP 554
> ath:AT4G26870 aspartyl-tRNA synthetase, putative / aspartate--tRNA
ligase, putative (EC:6.1.1.12); K01876 aspartyl-tRNA
synthetase [EC:6.1.1.12]
Length=532
Score = 84.7 bits (208), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 53/163 (32%), Positives = 87/163 (53%), Gaps = 7/163 (4%)
Query 34 PFERLTYTDAVIRLQREEAEGRVK-FQEKVEWGMDLGSEHERYLTEQLAKKP----VIVH 88
PF+ L + +RL E +K E+V+ DL +E ER L + + +K ++H
Sbjct 364 PFQSLKFLPQTLRLTFAEGIQMLKEAGEEVDPLGDLNTESERKLGQLVLEKYKTEFYMLH 423
Query 89 DYPKDIKAFY-MKQNEDGKTVRAMDVLVPKIGELIGGSQREEDLEKLDRMIKEKGLNLDS 147
YP ++ FY M D + DV + + E++ G+QR D E L++ +E G+++ +
Sbjct 424 RYPSAVRPFYTMPYENDSNYSNSFDVFI-RGEEIMSGAQRIHDPELLEKRARECGIDVKT 482
Query 148 YWWYRELRSYGTVPHAGFGLGFERLIMLVTGIENIRDTIPYPR 190
Y + YG PH GFG+G ER++ML+ + NIR T +PR
Sbjct 483 ISTYIDAFRYGAPPHGGFGVGLERVVMLLCALNNIRKTSLFPR 525
> cel:Y66D12A.23 hypothetical protein; K01893 asparaginyl-tRNA
synthetase [EC:6.1.1.22]
Length=448
Score = 80.1 bits (196), Expect = 4e-15, Method: Compositional matrix adjust.
Identities = 53/141 (37%), Positives = 76/141 (53%), Gaps = 21/141 (14%)
Query 35 FERLTYTDAV--IRLQREEAEGRVKFQEKVEWGMDLGSEHERYLTEQLAKKPVIVHDYPK 92
F R+ Y +AV + L+ + + F +K E +DL H+ + P+ V YP
Sbjct 273 FPRIRYDEAVHLLALKGSKVTAKSGFSKKNE--LDLVKLHDDH--------PIFVTHYPV 322
Query 93 DIKAFYMKQNEDGKTVRAMDVLVPKIGELIGGSQREEDLEKLDRMIKEKGLNLDSYWWYR 152
K FYM + DG+ + D+LVP IGEL GGS RE+ E+L R +G +D WY
Sbjct 323 GQKPFYMAR--DGQLTLSFDLLVPGIGELAGGSLREQSAEELQR----RGCGID---WYL 373
Query 153 ELRSYGTVPHAGFGLGFERLI 173
E+R G P GFG+GFER++
Sbjct 374 EMRRRGQPPTGGFGIGFERML 394
> ath:AT4G31180 aspartyl-tRNA synthetase, putative / aspartate--tRNA
ligase, putative (EC:6.1.1.12); K01876 aspartyl-tRNA
synthetase [EC:6.1.1.12]
Length=558
Score = 77.8 bits (190), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 56/170 (32%), Positives = 84/170 (49%), Gaps = 17/170 (10%)
Query 34 PFERLTYTDAVIRLQREEAEGRVKFQEKVEWGM------DLGSEHERYLTEQLAKKP--- 84
PFE L + +RL EE +K E G+ DL +E ER L + + +K
Sbjct 390 PFEPLKFLPKTLRLTFEEGVQMLK-----EAGVEVDPLGDLNTESERKLGQLVLEKYNTE 444
Query 85 -VIVHDYPKDIKAFYMKQNEDGKT-VRAMDVLVPKIGELIGGSQREEDLEKLDRMIKEKG 142
I+H YPK ++ FY D + DV + + E+I G+QR E L++ E G
Sbjct 445 FYILHRYPKAVRPFYTMTCADNPLYSNSFDVFI-RGEEIISGAQRVHIPEVLEQRAGECG 503
Query 143 LNLDSYWWYRELRSYGTVPHAGFGLGFERLIMLVTGIENIRDTIPYPRYP 192
+++ + Y + YG H GFG+G ER++ML + NIR T +PR P
Sbjct 504 IDVKTISTYIDSFRYGAPLHGGFGVGLERVVMLFCALNNIRKTSLFPRDP 553
> pfa:PFA_0145c aspartyl-tRNA synthetase, putative (EC:6.1.1.12);
K01876 aspartyl-tRNA synthetase [EC:6.1.1.12]
Length=626
Score = 67.4 bits (163), Expect = 3e-11, Method: Composition-based stats.
Identities = 47/160 (29%), Positives = 85/160 (53%), Gaps = 9/160 (5%)
Query 38 LTYTDAVIRLQREEAEGRVKFQEKVEWGMDLGSEHERYLTEQLA----KKPVIVHDYPKD 93
TY +A+ L + G++ +E+ DL ++ E+ L + + I+ ++P
Sbjct 466 FTYEEAIKILIKN---GKLFLKEEDILTYDLTTDLEKELGKLIKLSHNTDYYIIINFPSS 522
Query 94 IKAFY-MKQNEDGKTVRAMDVLVPKIGELIGGSQREEDLEKLDRMIKEKGLNLDSYWWYR 152
++ FY M + +D K + D + + E++ GSQR D++ L IK L+ + +Y
Sbjct 523 LRPFYTMYKEDDPKISNSYDFFM-RGEEILSGSQRISDMKLLLENIKLFNLDPNKLNFYI 581
Query 153 ELRSYGTVPHAGFGLGFERLIMLVTGIENIRDTIPYPRYP 192
+ +Y + PH+G G+G ER++ML G+ NIR T +PR P
Sbjct 582 DSFAYSSYPHSGCGIGLERVLMLFLGLNNIRKTSLFPRDP 621
> xla:443916 dars, MGC80207; aspartyl-tRNA synthetase (EC:6.1.1.12);
K01876 aspartyl-tRNA synthetase [EC:6.1.1.12]
Length=530
Score = 67.4 bits (163), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 46/170 (27%), Positives = 81/170 (47%), Gaps = 17/170 (10%)
Query 34 PFERLTYTDAVIRLQREEAEGRVKFQEKVEWGMDLGSEHE----------RYLTEQLAKK 83
P E + + +RL+ E ++ E G+++G E + R + E+
Sbjct 362 PCEPFKFLEPTLRLEYTEGVAMLR-----EAGVEMGDEEDLSTPNEKLLGRLVKEKYDTD 416
Query 84 PVIVHDYPKDIKAFY-MKQNEDGKTVRAMDVLVPKIGELIGGSQREEDLEKLDRMIKEKG 142
I+ YP ++ FY M ++ K + D+ + + E++ G+QR D + L G
Sbjct 417 FYILDKYPLAVRPFYTMPDPQNPKYSNSYDMFM-RGEEILSGAQRVHDPQLLTERATHHG 475
Query 143 LNLDSYWWYRELRSYGTVPHAGFGLGFERLIMLVTGIENIRDTIPYPRYP 192
++L+ Y + +G PHAG G+G ER+ ML G+ N+R T +PR P
Sbjct 476 IDLEKIKAYIDSFRFGAPPHAGGGIGLERVTMLYLGLHNVRQTSMFPRDP 525
> hsa:1615 DARS, DKFZp781B11202, MGC111579; aspartyl-tRNA synthetase
(EC:6.1.1.12); K01876 aspartyl-tRNA synthetase [EC:6.1.1.12]
Length=501
Score = 66.6 bits (161), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 49/166 (29%), Positives = 83/166 (50%), Gaps = 9/166 (5%)
Query 34 PFERLTYTDAVIRLQREEAEGRVKFQEKVEWGM--DLGSEHERYLTEQLAKKP----VIV 87
P E + + +RL+ EA ++ + VE G DL + +E+ L + +K I+
Sbjct 333 PCEPFKFLEPTLRLEYCEALAMLR-EAGVEMGDEDDLSTPNEKLLGHLVKEKYDTDFYIL 391
Query 88 HDYPKDIKAFY-MKQNEDGKTVRAMDVLVPKIGELIGGSQREEDLEKLDRMIKEKGLNLD 146
YP ++ FY M + K + D+ + + E++ G+QR D + L G++L+
Sbjct 392 DKYPLAVRPFYTMPDPRNPKQSNSYDMFM-RGEEILSGAQRIHDPQLLTERALHHGIDLE 450
Query 147 SYWWYRELRSYGTVPHAGFGLGFERLIMLVTGIENIRDTIPYPRYP 192
Y + +G PHAG G+G ER+ ML G+ N+R T +PR P
Sbjct 451 KIKAYIDSFRFGAPPHAGGGIGLERVTMLFLGLHNVRQTSMFPRDP 496
> xla:380028 dars, MGC53970; aspartyl-tRNA synthetase (EC:6.1.1.12);
K01876 aspartyl-tRNA synthetase [EC:6.1.1.12]
Length=530
Score = 66.2 bits (160), Expect = 7e-11, Method: Compositional matrix adjust.
Identities = 46/170 (27%), Positives = 80/170 (47%), Gaps = 17/170 (10%)
Query 34 PFERLTYTDAVIRLQREEAEGRVKFQEKVEWGMDLGSEHE----------RYLTEQLAKK 83
P E + + +RL+ E ++ E G+++G E + R + E+
Sbjct 362 PCEPFKFLEPTLRLEYTEGVAMLR-----EAGVEMGDEEDLSTPNEKLLGRLVKEKYDTD 416
Query 84 PVIVHDYPKDIKAFY-MKQNEDGKTVRAMDVLVPKIGELIGGSQREEDLEKLDRMIKEKG 142
I+ YP ++ FY M + K + D+ + + E++ G+QR D + L G
Sbjct 417 FYILDKYPLAVRPFYTMPDPNNPKYSNSYDMFM-RGEEILSGAQRVHDPQLLTDRATHHG 475
Query 143 LNLDSYWWYRELRSYGTVPHAGFGLGFERLIMLVTGIENIRDTIPYPRYP 192
++L+ Y + +G PHAG G+G ER+ ML G+ N+R T +PR P
Sbjct 476 IDLEKIKAYIDSFRFGAPPHAGGGIGLERVTMLYLGLHNVRQTSMFPRDP 525
> dre:324025 dars, MGC154056, wu:fc17a11, zgc:154056; aspartyl-tRNA
synthetase (EC:6.1.1.12); K01876 aspartyl-tRNA synthetase
[EC:6.1.1.12]
Length=531
Score = 64.7 bits (156), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 51/184 (27%), Positives = 88/184 (47%), Gaps = 14/184 (7%)
Query 21 EGLRARLENILAA-----PFERLTYTDAVIRLQREEAEGRVKFQEKVEWG--MDLGSEHE 73
+GLR R + + P E + + +RL+ +E ++ VE G DL + +E
Sbjct 345 KGLRDRFQTEIQTVNKQYPSEPFKFLEPTLRLEYKEGLAMLR-AAGVEMGDEEDLSTPNE 403
Query 74 RYLTEQLAKKP----VIVHDYPKDIKAFY-MKQNEDGKTVRAMDVLVPKIGELIGGSQRE 128
+ L + +K ++ YP ++ FY M + K + D+ + + E++ G+QR
Sbjct 404 KLLGRLVKEKYDTDFYVLDKYPLAVRPFYTMPDPNNPKYSNSYDMFM-RGEEILSGAQRI 462
Query 129 EDLEKLDRMIKEKGLNLDSYWWYRELRSYGTVPHAGFGLGFERLIMLVTGIENIRDTIPY 188
D + L ++L+ Y + YG PHAG G+G ER+ ML G+ N+R T +
Sbjct 463 HDAQLLTERAMHHNIDLEKIKAYIDSFRYGAPPHAGGGIGLERVTMLYLGLHNVRQTSMF 522
Query 189 PRYP 192
PR P
Sbjct 523 PRDP 526
> tgo:TGME49_002530 aspartyl-tRNA synthetase, putative (EC:6.1.1.12);
K01876 aspartyl-tRNA synthetase [EC:6.1.1.12]
Length=773
Score = 63.2 bits (152), Expect = 5e-10, Method: Composition-based stats.
Identities = 54/201 (26%), Positives = 80/201 (39%), Gaps = 25/201 (12%)
Query 2 KSCLDNCSGEIDWFEKNQEEGLRARLENILAAPFERLTYTDAVIRLQREEA------EGR 55
K + C EID F + P E T+ + RL EE G
Sbjct 583 KGLSERCKKEIDLFHQQH--------------PAEPFTWIEETPRLSFEEGVQMLREAGC 628
Query 56 VKFQEKVEWGMDLGSEHERYLTEQLAKKP----VIVHDYPKDIKAFYMKQNEDGKTVRAM 111
E + DL +E E+ L + +K ++ YP ++ FY + K
Sbjct 629 PNIPEDLS-EFDLSTEQEKMLGRLVKEKYHTDFYMLLQYPLKVRPFYTMPDPHNKMYSNS 687
Query 112 DVLVPKIGELIGGSQREEDLEKLDRMIKEKGLNLDSYWWYRELRSYGTVPHAGFGLGFER 171
+ E+ G+QR D + L + E G+ S Y + GT PH G G+G ER
Sbjct 688 YDFFMRNEEITSGAQRVHDADLLTQRCLECGVPPSSIQTYIDSFRLGTAPHGGAGIGLER 747
Query 172 LIMLVTGIENIRDTIPYPRYP 192
++ML G +NIR +PR P
Sbjct 748 VVMLFLGAKNIRQVSLFPRDP 768
> tpv:TP03_0489 aspartyl-tRNA synthetase (EC:6.1.1.12); K01876
aspartyl-tRNA synthetase [EC:6.1.1.12]
Length=604
Score = 62.4 bits (150), Expect = 9e-10, Method: Composition-based stats.
Identities = 29/73 (39%), Positives = 47/73 (64%), Gaps = 0/73 (0%)
Query 120 ELIGGSQREEDLEKLDRMIKEKGLNLDSYWWYRELRSYGTVPHAGFGLGFERLIMLVTGI 179
E++ G+QR D +L+ + G+++++ Y E+ YG+ PHAG G+G ER++ML G+
Sbjct 527 EILSGAQRIHDSVELEARARACGIDVNTIKDYIEVFRYGSFPHAGSGIGLERVVMLFLGL 586
Query 180 ENIRDTIPYPRYP 192
NIR T +PR P
Sbjct 587 TNIRRTSMFPRDP 599
> cel:B0464.1 drs-1; aspartyl(D) tRNA Synthetase family member
(drs-1); K01876 aspartyl-tRNA synthetase [EC:6.1.1.12]
Length=531
Score = 60.5 bits (145), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 39/131 (29%), Positives = 65/131 (49%), Gaps = 6/131 (4%)
Query 67 DLGSEHERYL----TEQLAKKPVIVHDYPKDIKAFY-MKQNEDGKTVRAMDVLVPKIGEL 121
DL + E++L E+ + ++ +P ++ FY M D + + D+ + + E+
Sbjct 397 DLSTPVEKFLGKLVKEKYSTDFYVLDKFPLSVRPFYTMPDAHDERYSNSYDMFM-RGEEI 455
Query 122 IGGSQREEDLEKLDRMIKEKGLNLDSYWWYRELRSYGTVPHAGFGLGFERLIMLVTGIEN 181
+ G+QR D + L K ++L Y + YG PHAG G+G ER+ ML G+ N
Sbjct 456 LSGAQRIHDADMLVERAKHHQVDLAKIQSYIDSFKYGCPPHAGGGIGLERVTMLFLGLHN 515
Query 182 IRDTIPYPRYP 192
IR +PR P
Sbjct 516 IRLASLFPRDP 526
> eco:b1866 aspS, ECK1867, JW1855, tls; aspartyl-tRNA synthetase
(EC:6.1.1.12); K01876 aspartyl-tRNA synthetase [EC:6.1.1.12]
Length=590
Score = 60.1 bits (144), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 35/109 (32%), Positives = 55/109 (50%), Gaps = 5/109 (4%)
Query 91 PKDIKAFYMKQNEDGKTVRAMDVLVPKIGELIGGSQREEDLEKLDRMIKEKGLN----LD 146
PKD+ A +K + A D+++ E+ GGS R + + + G+N +
Sbjct 454 PKDMTAAELKAAPENAVANAYDMVINGY-EVGGGSVRIHNGDMQQTVFGILGINEEEQRE 512
Query 147 SYWWYRELRSYGTVPHAGFGLGFERLIMLVTGIENIRDTIPYPRYPGHA 195
+ + + YGT PHAG G +RL ML+TG +NIRD I +P+ A
Sbjct 513 KFGFLLDALKYGTPPHAGLAFGLDRLTMLLTGTDNIRDVIAFPKTTAAA 561
> cpv:cgd7_1540 aspartate--tRNA ligase
Length=532
Score = 58.2 bits (139), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 48/170 (28%), Positives = 76/170 (44%), Gaps = 13/170 (7%)
Query 34 PFERLTYTDAVIRLQREEA-----EGRVKFQEKVEWGMDLGSEHERYL----TEQLAKKP 84
PF ++ RL EE E + E + D+ +E ER L E+
Sbjct 360 PFTPFVISEKTPRLTFEEGCNLLKEAGAEIPEDLS-DFDISTEQERLLGSIVKEKYNSDF 418
Query 85 VIVHDYPKDIKAFYMKQN--EDGKTVRAMDVLVPKIGELIGGSQREEDLEKLDRMIKEKG 142
++ YP ++ FY + E K + D + + E++ G+QR D + L + +E G
Sbjct 419 YMMLKYPLKVRPFYTMPDFDEPEKWSNSFDFFM-RGEEILSGAQRVHDYDLLVKRCQECG 477
Query 143 LNLDSYWWYRELRSYGTVPHAGFGLGFERLIMLVTGIENIRDTIPYPRYP 192
++ S Y G PH G G+G ER+IML + NIR + +PR P
Sbjct 478 VSEHSLRDYLNSFKLGAPPHGGCGIGLERVIMLFLNLGNIRKSSMFPRDP 527
> sce:YLL018C DPS1; AspRS; aspartyl-tRNA synthetase (EC:6.1.1.12);
K01876 aspartyl-tRNA synthetase [EC:6.1.1.12]
Length=557
Score = 58.2 bits (139), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 42/133 (31%), Positives = 69/133 (51%), Gaps = 8/133 (6%)
Query 67 DLGSEHERYLTEQLAKKP----VIVHDYPKDIKAFY-MKQNEDGKTVRAMDVLVPKIGEL 121
DL +E+E++L + + K I+ +P +I+ FY M + K + D + + E+
Sbjct 421 DLSTENEKFLGKLVRDKYDTDFYILDKFPLEIRPFYTMPDPANPKYSNSYDFFM-RGEEI 479
Query 122 IGGSQREEDLEKLDRMIKEKGLNLDSYWW--YRELRSYGTVPHAGFGLGFERLIMLVTGI 179
+ G+QR D L +K GL+ + Y + SYG PHAG G+G ER++M +
Sbjct 480 LSGAQRIHDHALLQERMKAHGLSPEDPGLKDYCDGFSYGCPPHAGGGIGLERVVMFYLDL 539
Query 180 ENIRDTIPYPRYP 192
+NIR +PR P
Sbjct 540 KNIRRASLFPRDP 552
> mmu:226539 Dars2, 5830468K18Rik, MGC99963; aspartyl-tRNA synthetase
2 (mitochondrial) (EC:6.1.1.12); K01876 aspartyl-tRNA
synthetase [EC:6.1.1.12]
Length=653
Score = 52.0 bits (123), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 33/79 (41%), Positives = 42/79 (53%), Gaps = 3/79 (3%)
Query 114 LVPKIGELIGGSQREEDLEKLDRMIKEKGLNLDSYWWYRELRS--YGTVPHAGFGLGFER 171
LV E+ GGS R D +L R I E L D L++ YG PH G LG +R
Sbjct 528 LVLNGNEIGGGSVRIHD-AQLQRYILETLLKEDVKLLSHLLQALDYGAPPHGGIALGLDR 586
Query 172 LIMLVTGIENIRDTIPYPR 190
L+ LVTG +IRD I +P+
Sbjct 587 LVCLVTGAPSIRDVIAFPK 605
> hsa:55157 DARS2, ASPRS, FLJ10514, LBSL, MT-ASPRS, RP3-383J4.2;
aspartyl-tRNA synthetase 2, mitochondrial (EC:6.1.1.12);
K01876 aspartyl-tRNA synthetase [EC:6.1.1.12]
Length=645
Score = 52.0 bits (123), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 41/108 (37%), Positives = 51/108 (47%), Gaps = 7/108 (6%)
Query 90 YPKDIKAFYMKQNEDGKTVRAMDVLVPKIGELIGGSQREEDLEKLDRMIKEKGLNLDSYW 149
+P DI Y E K LV E+ GGS R + E L R I L D
Sbjct 508 HPSDIHLLY---TEPKKARSQHYDLVLNGNEIGGGSIRIHNAE-LQRYILATLLKEDVKM 563
Query 150 WYRELRS--YGTVPHAGFGLGFERLIMLVTGIENIRDTIPYPR-YPGH 194
L++ YG PH G LG +RLI LVTG +IRD I +P+ + GH
Sbjct 564 LSHLLQALDYGAPPHGGIALGLDRLICLVTGSPSIRDVIAFPKSFRGH 611
> sce:YPL104W MSD1, LPG5; Msd1p (EC:6.1.1.12); K01876 aspartyl-tRNA
synthetase [EC:6.1.1.12]
Length=658
Score = 50.4 bits (119), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 33/86 (38%), Positives = 49/86 (56%), Gaps = 6/86 (6%)
Query 109 RAMDVLVPKIGELIGGSQREEDLEKLDRMIKEKGLNLD-SYWWYRELRS---YGTVPHAG 164
R D++V + EL GGS R D +L I E L +D +Y + L + GT PHAG
Sbjct 543 RHYDLVVNGV-ELGGGSTRIHD-PRLQDYIFEDILKIDNAYELFGHLLNAFDMGTPPHAG 600
Query 165 FGLGFERLIMLVTGIENIRDTIPYPR 190
F +GF+R+ ++ E+IRD I +P+
Sbjct 601 FAIGFDRMCAMICETESIRDVIAFPK 626
> eco:b4129 lysU, ECK4123, JW4090; lysine tRNA synthetase, inducible
(EC:6.1.1.6); K04567 lysyl-tRNA synthetase, class II
[EC:6.1.1.6]
Length=505
Score = 45.8 bits (107), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 29/116 (25%), Positives = 49/116 (42%), Gaps = 11/116 (9%)
Query 83 KPVIVHDYPKDIKAFYMKQNEDGKTVRAMDVLVPKIGELIGG-----SQREEDLEKLDRM 137
+P + +YP ++ + + + + + + G IG + E+ E+
Sbjct 386 QPTFITEYPAEVSPLARRNDVNPEITDRFEFFIG--GREIGNGFSELNDAEDQAERFQEQ 443
Query 138 IKEKGLNLDSYWWYRE----LRSYGTVPHAGFGLGFERLIMLVTGIENIRDTIPYP 189
+ K D +Y E YG P AG G+G +R+IML T IRD I +P
Sbjct 444 VNAKAAGDDEAMFYDEDYVTALEYGLPPTAGLGIGIDRMIMLFTNSHTIRDVILFP 499
> dre:571144 dars2, si:dkey-21n10.1; aspartyl-tRNA synthetase
2, mitochondrial; K01876 aspartyl-tRNA synthetase [EC:6.1.1.12]
Length=660
Score = 43.9 bits (102), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 29/80 (36%), Positives = 43/80 (53%), Gaps = 8/80 (10%)
Query 120 ELIGGSQR----EEDLEKLDRMIKEKGLNLDSYWWYRELRSYGTVPHAGFGLGFERLIML 175
E+ GGS R + L LD ++KE +L S+ E G PH G LG +RL+ +
Sbjct 556 EIGGGSIRIHNASQQLYILDTILKEDP-SLLSHLL--EALDSGAPPHGGIALGLDRLVSI 612
Query 176 VTGIENIRDTIPYPR-YPGH 194
+ G +IRD I +P+ + GH
Sbjct 613 IVGAPSIRDVIAFPKSFRGH 632
> ath:AT3G13490 OVA5; OVA5 (OVULE ABORTION 5); ATP binding / aminoacyl-tRNA
ligase/ lysine-tRNA ligase/ nucleic acid binding
/ nucleotide binding (EC:6.1.1.6); K04567 lysyl-tRNA synthetase,
class II [EC:6.1.1.6]
Length=602
Score = 43.1 bits (100), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 17/33 (51%), Positives = 23/33 (69%), Gaps = 0/33 (0%)
Query 157 YGTVPHAGFGLGFERLIMLVTGIENIRDTIPYP 189
YG P +G GLG +RL+ML+T +IRD I +P
Sbjct 564 YGMPPASGMGLGIDRLVMLLTNSASIRDVIAFP 596
> cel:F10C2.6 drs-2; aspartyl(D) tRNA Synthetase family member
(drs-2); K01876 aspartyl-tRNA synthetase [EC:6.1.1.12]
Length=593
Score = 42.4 bits (98), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 25/75 (33%), Positives = 37/75 (49%), Gaps = 4/75 (5%)
Query 120 ELIGGSQREEDLEKLDRMIKEKGLNLDSYWWYRELRSYGTVPHAGFGLGFERLIMLVTGI 179
E+ GGS R E+ E ++K G D S+G PH GF LG +R + ++T
Sbjct 486 EMGGGSIRIENSEIQRHVLKVLGEPTDEMEHLLNALSHGAPPHGGFALGLDRFVAMLTSD 545
Query 180 EN----IRDTIPYPR 190
N +RD I +P+
Sbjct 546 GNPLTPVRDVIAFPK 560
> bbo:BBOV_IV001690 21.m02784; aspartyl-tRNA synthetase (EC:6.1.1.12);
K01876 aspartyl-tRNA synthetase [EC:6.1.1.12]
Length=557
Score = 41.6 bits (96), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 42/169 (24%), Positives = 73/169 (43%), Gaps = 24/169 (14%)
Query 8 CSGEIDWFEKNQEEGLRARLENILAAPFE------RLTYTDAVIRLQREEAEGRVKFQEK 61
C E++ F+K+Q PF+ R+ +T+AV L+ E + +
Sbjct 401 CKTEVEIFQKHQPS----------VEPFKWLDQTPRIRFTEAVEMLRGTELAESIP--DD 448
Query 62 VEWGMDLGSEHE----RYLTEQLAKKPVIVHDYPKDIKAFY-MKQNEDGKTVRAMDVLVP 116
+ D +E E R + E+ IV++YP + + FY M + DG
Sbjct 449 IS-SYDFSTEQEKRLGRLVKEKYNTDYYIVYEYPLNARPFYTMPKERDGIMFTHSYDFFM 507
Query 117 KIGELIGGSQREEDLEKLDRMIKEKGLNLDSYWWYRELRSYGTVPHAGF 165
+ E++ G+QR + + L + KE G++ + Y E G PHAG+
Sbjct 508 RGEEILSGAQRIHNPDLLLQRAKECGIDPKTISCYIEAFKMGASPHAGY 556
> dre:795494 L-asparaginase-like; K13051 beta-aspartyl-peptidase
(threonine type) [EC:3.4.19.5]
Length=1428
Score = 41.2 bits (95), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 26/96 (27%), Positives = 46/96 (47%), Gaps = 3/96 (3%)
Query 97 FYMKQNEDG--KTVRAMDVLVPKIGELIGGSQREEDLEKLDRMIKEKGLNLDSYWWYREL 154
FY ED +A D+L+ +GE +G +R + + ++ + DSY WY +
Sbjct 1326 FYQAYIEDSGQSKAKAADLLL-GLGETVGLGERHSTPDMVREALQHHAVPEDSYKWYINM 1384
Query 155 RSYGTVPHAGFGLGFERLIMLVTGIENIRDTIPYPR 190
R + +G+G+G ER + + ++RD PR
Sbjct 1385 RQVMPLLTSGWGMGTERYLCWLLQHNDVRDMQIIPR 1420
> eco:b2890 lysS, asuD, ECK2885, herC, JW2858; lysine tRNA synthetase,
constitutive (EC:6.1.1.6); K04567 lysyl-tRNA synthetase,
class II [EC:6.1.1.6]
Length=505
Score = 40.4 bits (93), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 39/180 (21%), Positives = 73/180 (40%), Gaps = 27/180 (15%)
Query 34 PFERLTYTDAVIRLQREE--------------AEGRVKFQEKVEWGMD-LGSEHERYLTE 78
PFE+LT +A+ + + E AE + + WG+ + +E + E
Sbjct 323 PFEKLTMREAIKKYRPETDMADLDNFDSAKAIAES-IGIHVEKSWGLGRIVTEIFEEVAE 381
Query 79 QLAKKPVIVHDYPKDIKAFYMKQNEDGKTVRAMDVLVPKIGELIGG-----SQREEDLEK 133
+P + +YP ++ + + + + + + G IG + E+ ++
Sbjct 382 AHLIQPTFITEYPAEVSPLARRNDVNPEITDRFEFFIG--GREIGNGFSELNDAEDQAQR 439
Query 134 LDRMIKEKGLNLDSYWWYRE----LRSYGTVPHAGFGLGFERLIMLVTGIENIRDTIPYP 189
+ K D +Y E +G P AG G+G +R++ML T IRD I +P
Sbjct 440 FLDQVAAKDAGDDEAMFYDEDYVTALEHGLPPTAGLGIGIDRMVMLFTNSHTIRDVILFP 499
> ath:AT4G33760 tRNA synthetase class II (D, K and N) family protein
(EC:6.1.1.12); K01876 aspartyl-tRNA synthetase [EC:6.1.1.12]
Length=664
Score = 39.7 bits (91), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 14/43 (32%), Positives = 24/43 (55%), Gaps = 0/43 (0%)
Query 148 YWWYRELRSYGTVPHAGFGLGFERLIMLVTGIENIRDTIPYPR 190
+ + E G PH G G +R++M++ G +IRD I +P+
Sbjct 594 FGYLLEALDMGAPPHGGIAYGLDRMVMMLGGASSIRDVIAFPK 636
> sce:YNL073W MSK1; Mitochondrial lysine-tRNA synthetase, required
for import of both aminoacylated and deacylated forms of
tRNA(Lys) into mitochondria and for aminoacylation of mitochondrially
encoded tRNA(Lys) (EC:6.1.1.6); K04567 lysyl-tRNA
synthetase, class II [EC:6.1.1.6]
Length=576
Score = 37.4 bits (85), Expect = 0.035, Method: Compositional matrix adjust.
Identities = 16/40 (40%), Positives = 22/40 (55%), Gaps = 0/40 (0%)
Query 149 WWYRELRSYGTVPHAGFGLGFERLIMLVTGIENIRDTIPY 188
+ Y E YG P GFGLG +RL ML + I + +P+
Sbjct 528 YQYVETMKYGMPPVGGFGLGIDRLCMLFCDKKRIEEVLPF 567
> pfa:PF13_0262 lysine-tRNA ligase (EC:6.1.1.6); K04567 lysyl-tRNA
synthetase, class II [EC:6.1.1.6]
Length=583
Score = 36.6 bits (83), Expect = 0.058, Method: Compositional matrix adjust.
Identities = 22/67 (32%), Positives = 35/67 (52%), Gaps = 6/67 (8%)
Query 127 REEDLEKLDRMIKEKG----LNLDSYWWYRELRSYGTVPHAGFGLGFERLIMLVTGIENI 182
++++ KL + +EKG LDS + YG P G GLG +R+ M +T +I
Sbjct 513 KQKECFKLQQKDREKGDTEAAQLDSAFCTS--LEYGLPPTGGLGLGIDRITMFLTNKNSI 570
Query 183 RDTIPYP 189
+D I +P
Sbjct 571 KDVILFP 577
Lambda K H
0.318 0.139 0.420
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5802328440
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40