bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_0067_orf1
Length=269
Score E
Sequences producing significant alignments: (Bits) Value
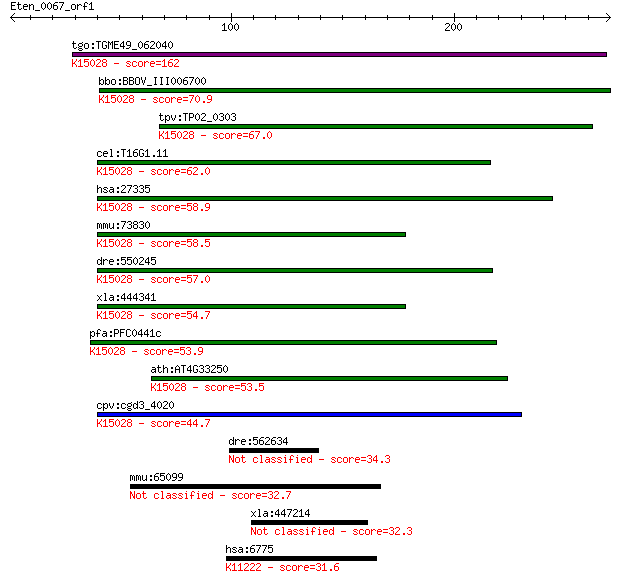
tgo:TGME49_062040 eukaryotic translation initiation factor 3 s... 162 1e-39
bbo:BBOV_III006700 17.m07593; hypothetical protein; K15028 tra... 70.9 4e-12
tpv:TP02_0303 hypothetical protein; K15028 translation initiat... 67.0 7e-11
cel:T16G1.11 eif-3.K; Eukaryotic Initiation Factor family memb... 62.0 2e-09
hsa:27335 EIF3K, EIF3-p28, EIF3S12, HSPC029, M9, MSTP001, PLAC... 58.9 2e-08
mmu:73830 Eif3k, 1200009C21Rik, Eif3s12; eukaryotic translatio... 58.5 2e-08
dre:550245 zgc:110726; K15028 translation initiation factor 3 ... 57.0 8e-08
xla:444341 eif3k, MGC82783, eif3s12; eukaryotic translation in... 54.7 3e-07
pfa:PFC0441c SAC3/GNAP family-related protein, putative; K1502... 53.9 6e-07
ath:AT4G33250 EIF3K; EIF3K (EUKARYOTIC TRANSLATION INITIATION ... 53.5 8e-07
cpv:cgd3_4020 eIF-3 p25/subunit 11 ; K15028 translation initia... 44.7 4e-04
dre:562634 fibrillin 2-like 34.3 0.43
mmu:65099 Irak1bp1, 4921528N06Rik, AI851240, Aabp3, Aip70, Sim... 32.7 1.4
xla:447214 MGC81969 protein 32.3 1.8
hsa:6775 STAT4, SLEB11; signal transducer and activator of tra... 31.6 3.0
> tgo:TGME49_062040 eukaryotic translation initiation factor 3
subunit 11, putative ; K15028 translation initiation factor
3 subunit K
Length=269
Score = 162 bits (410), Expect = 1e-39, Method: Compositional matrix adjust.
Identities = 99/265 (37%), Positives = 142/265 (53%), Gaps = 39/265 (14%)
Query 29 VEDLLNIHQMIYLPSTLDLLVSHLEKGIAAGGASPDLCSAVLRLYLLYPQCANAEVVRKI 88
V+ LL ++ Y PS+L LL + E+ +A + C AVL+LYLLYPQC + ++ RKI
Sbjct 16 VQALLQAPELRYDPSSLTLLCQYTEEQVAKNTYDQEACLAVLKLYLLYPQCFSVDLARKI 75
Query 89 LVQTIVALPENQFGYYACIAPSHFSNPEDEKKVQDVLTLQELLESCDFKAAWAMLREPEM 148
LV+ I+ LP + + +D KKVQ+ + L ELLE C FK W L+EP M
Sbjct 76 LVKGIMNLPNEDCAVFVGMLQQQ-PGKKDWKKVQEAIHLWELLEKCRFKQVWEFLKEPAM 134
Query 149 ADVLHTPGVVEALRRYVCEVLQQTFSTISLSDFCQLLNLNAAAAAAAAGAAAAADEKVEK 208
ADV+ TPG+V+++RR+VC+V+ T+S +SL+D C LN+ + A E+
Sbjct 135 ADVVSTPGLVDSIRRFVCDVVSLTYSALSLADLCAFLNVVPGTSEA------------ER 182
Query 209 LVKARGWTIE-------DAAAGQ---KIVRVV----------------PRETDAPKALAT 242
L+K GWT+E +AA + K+VRVV R D A
Sbjct 183 LLKNLGWTVEEVFVPAKEAARSRETVKVVRVVGSASLAAAALKEGEERNRNRDGKPGAAG 242
Query 243 RASVAHKTVEKYLSVEGLRQCLALL 267
+A KTVEKY+ E L+ C+A L
Sbjct 243 QALAVRKTVEKYMMPENLKVCMATL 267
> bbo:BBOV_III006700 17.m07593; hypothetical protein; K15028 translation
initiation factor 3 subunit K
Length=247
Score = 70.9 bits (172), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 62/239 (25%), Positives = 106/239 (44%), Gaps = 33/239 (13%)
Query 41 LPSTLDLLVSHLEKGIAAGGASPDLCSAVLRLYLLYPQCANAEVVRKILVQTIVALPENQ 100
+P+ L +EK A S D A+L++Y L+P A+ V++KILVQ + LP N
Sbjct 29 IPALTAYLDEQMEK---ANAYSLDNNVAILKIYTLFPHIADPLVIQKILVQCLTQLPAND 85
Query 101 FGYYACIAPSHFSNPEDEKKVQDVLTLQELLESCDFKAAWAMLREPEMAD----VLHTPG 156
F C A ++ + V+ L LL++C F W R M + L PG
Sbjct 86 FN--ICFAQVPLPT-QEHPVIAKVVALHNLLQNCLFHKFWEEARHNMMDNDDRTFLDVPG 142
Query 157 VVEALRRYVCEVLQQTFSTISLSDFCQLLNLNAAAAAAAAGAAAAADEKVEKLVKARGWT 216
+ E++RR++ +V+ + +S+ + +L+ + E+ E+L+K+ WT
Sbjct 143 LRESIRRFILDVVPLVYLQMSVPELRSMLDYPSNC------------EEFEQLLKSYKWT 190
Query 217 IE------DAAAGQKIVRVVPRETDAPKALATRASVAHKTVEKYLSVEGLRQCLALLES 269
+E D +G V P + K A VEKY ++ +R + L+
Sbjct 191 LEGGYNRDDPNSG---TCVPPTRDEVIK--QHMAQDKEPGVEKYFKLDTMRTYYSTLKG 244
> tpv:TP02_0303 hypothetical protein; K15028 translation initiation
factor 3 subunit K
Length=244
Score = 67.0 bits (162), Expect = 7e-11, Method: Compositional matrix adjust.
Identities = 53/204 (25%), Positives = 95/204 (46%), Gaps = 30/204 (14%)
Query 68 AVLRLYLLYPQCANAEVVRKILVQTIVALPENQFGYYACIAPSHFSNPEDEKKVQDVLTL 127
+L++YLLYP ++ V++KILV ++ +P F C P H N E V V+ L
Sbjct 53 GILKIYLLYPDVSDVLVIQKILVLSLTQVPAPDFTVCLCQIPLHLHNQE---PVSKVINL 109
Query 128 QELLESCDFKAAW--AMLREPEM--ADVLHTPGVVEALRRYVCEVLQQTFSTISLSDFCQ 183
L++C F W A L M VL G+ +++R++V +V ++ +++ + +
Sbjct 110 HNFLQNCMFLRFWESANLELESMPGKTVLDVSGLSDSVRKFVLDVSPLVYNQMTVPELRK 169
Query 184 LLNLNAAAAAAAAGAAAAADEKVEKLVKARGWTI------EDAAAGQKIVRVVPRETDAP 237
LLN + ++ E+L+ GW+ ED +G + RE D+
Sbjct 170 LLNFQTNS------------QEFEQLLSVCGWSFDSSYNREDENSG--LCYSTSRE-DSS 214
Query 238 KALATRASVAHKTVEKYLSVEGLR 261
K + A+ +EKYL + ++
Sbjct 215 KVHKSNAT--QNPLEKYLKSDPMK 236
> cel:T16G1.11 eif-3.K; Eukaryotic Initiation Factor family member
(eif-3.K); K15028 translation initiation factor 3 subunit
K
Length=240
Score = 62.0 bits (149), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 45/192 (23%), Positives = 87/192 (45%), Gaps = 32/192 (16%)
Query 40 YLPSTLDLLVSHLEKGIAAGGASPDLCSAVLRLYLLYPQCANAEVVRKILVQTIVALPEN 99
Y P + L + ++ + D+ +L+LY L P+ + VVR++L++T++ LP +
Sbjct 20 YNPENVADLAACVQAMVNENKYDKDIVLTILKLYQLNPEKYDEAVVRQVLLKTLMVLPSS 79
Query 100 QFGYYACIAPSHFSNPEDEKKVQDVLTLQELLESCDFKAAWAMLR---EPEM--ADVLHT 154
F C+ ++ ++ +++ D L +LESC+F W +++ +P +
Sbjct 80 DFALAKCLIDTNRLGSQELRRIFD---LGAVLESCNFAVFWKLVKGAYKPTTNPNEPFKV 136
Query 155 PGVV-----------EALRRYVCEVLQQTFSTISLSDFCQLLNLNAAAAAAAAGAAAAAD 203
PG V +A++ Y C V+ TF I +LL A+D
Sbjct 137 PGEVPKMIKPMVGFEDAVKHYACRVISVTFQKIEKKMLSRLL-------------GGASD 183
Query 204 EKVEKLVKARGW 215
++V L ++ GW
Sbjct 184 KEVTALAQSFGW 195
> hsa:27335 EIF3K, EIF3-p28, EIF3S12, HSPC029, M9, MSTP001, PLAC-24,
PLAC24, PRO1474, PTD001; eukaryotic translation initiation
factor 3, subunit K; K15028 translation initiation factor
3 subunit K
Length=218
Score = 58.9 bits (141), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 48/206 (23%), Positives = 90/206 (43%), Gaps = 24/206 (11%)
Query 40 YLPSTLDLLVSHLEKGIAAGGASPDLCSAVLRLYLLYPQCANAEVVRKILVQTIVALPEN 99
Y P L L ++E + AVL+LY P V +IL++ + LP
Sbjct 21 YNPENLATLERYVETQAKENAYDLEANLAVLKLYQFNPAFFQTTVTAQILLKALTNLPHT 80
Query 100 QFGYYAC-IAPSHFSNPEDEKKVQDVLTLQELLESCDFKAAWAMLREPEMADVLH-TPGV 157
F C I +H ++E+ ++ +L L +LLE+C F+A W L E D+L G
Sbjct 81 DFTLCKCMIDQAH----QEERPIRQILYLGDLLETCHFQAFWQALDEN--MDLLEGITGF 134
Query 158 VEALRRYVCEVLQQTFSTISLSDFCQLLNLNAAAAAAAAGAAAAADEKVEKLVKARGWTI 217
+++R+++C V+ T+ I ++L +D +++ + GW+
Sbjct 135 EDSVRKFICHVVGITYQHIDRWLLAEML-------------GDLSDSQLKVWMSKYGWSA 181
Query 218 EDAAAGQKIVRVVPRETDAPKALATR 243
+++ +I E+ PK + +
Sbjct 182 DESG---QIFICSQEESIKPKNIVEK 204
> mmu:73830 Eif3k, 1200009C21Rik, Eif3s12; eukaryotic translation
initiation factor 3, subunit K; K15028 translation initiation
factor 3 subunit K
Length=218
Score = 58.5 bits (140), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 40/140 (28%), Positives = 67/140 (47%), Gaps = 8/140 (5%)
Query 40 YLPSTLDLLVSHLEKGIAAGGASPDLCSAVLRLYLLYPQCANAEVVRKILVQTIVALPEN 99
Y P L L ++E + AVL+LY P V +IL++ + LP
Sbjct 21 YNPENLATLERYVETQAKENAYDLEANLAVLKLYQFNPAFFQTTVTAQILLKALTNLPHT 80
Query 100 QFGYYAC-IAPSHFSNPEDEKKVQDVLTLQELLESCDFKAAWAMLREPEMADVLH-TPGV 157
F C I +H ++E+ ++ +L L +LLE+C F+A W L E D+L G
Sbjct 81 DFTLCKCMIDQAH----QEERPIRQILYLGDLLETCHFQAFWQALDEN--MDLLEGITGF 134
Query 158 VEALRRYVCEVLQQTFSTIS 177
+++R+++C V+ T+ I
Sbjct 135 EDSVRKFICHVVGITYQHID 154
> dre:550245 zgc:110726; K15028 translation initiation factor
3 subunit K
Length=219
Score = 57.0 bits (136), Expect = 8e-08, Method: Compositional matrix adjust.
Identities = 44/179 (24%), Positives = 78/179 (43%), Gaps = 21/179 (11%)
Query 40 YLPSTLDLLVSHLEKGIAAGGASPDLCSAVLRLYLLYPQCANAEVVRKILVQTIVALPEN 99
Y P L L +++ + AVL+LY V +IL++ + LP
Sbjct 22 YNPENLATLERYVDTQARENAYDLEANLAVLKLYQFNLAYFQTTVTAQILLKALTNLPHT 81
Query 100 QFGYYAC-IAPSHFSNPEDEKKVQDVLTLQELLESCDFKAAWAMLREP-EMADVLHTPGV 157
F C I +H ++E+ ++ +L L LLE+C F++ WA L E ++ D + G
Sbjct 82 DFTLCKCMIDQTH----QEERPIRQILYLGNLLETCHFQSFWASLEENRDLIDGI--TGF 135
Query 158 VEALRRYVCEVLQQTFSTISLSDFCQLLNLNAAAAAAAAGAAAAADEKVEKLVKARGWT 216
E++R+++C V+ T+ I ++L D +V+ + GWT
Sbjct 136 EESVRKFICHVVGITYQNIEYRLLAEML-------------GDPLDTQVKVWMNKYGWT 181
> xla:444341 eif3k, MGC82783, eif3s12; eukaryotic translation
initiation factor 3, subunit K; K15028 translation initiation
factor 3 subunit K
Length=218
Score = 54.7 bits (130), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 35/139 (25%), Positives = 66/139 (47%), Gaps = 6/139 (4%)
Query 40 YLPSTLDLLVSHLEKGIAAGGASPDLCSAVLRLYLLYPQCANAEVVRKILVQTIVALPEN 99
Y P L L ++E + AVL+LY P V ++L++ + LP
Sbjct 21 YNPENLATLERYVETQAKENAYDLEANLAVLKLYQFNPAFFQTTVTAQVLLKALTNLPHT 80
Query 100 QFGYYAC-IAPSHFSNPEDEKKVQDVLTLQELLESCDFKAAWAMLREPEMADVLHTPGVV 158
F C I +H ++++ ++ +L L +LLE+C F++ W L E + + G
Sbjct 81 DFTLCKCMIDQAH----QEDRPIRQILYLGDLLETCHFQSFWQALDE-NLDLIDGVTGFE 135
Query 159 EALRRYVCEVLQQTFSTIS 177
+++R+++C V+ T+ I
Sbjct 136 DSVRKFICHVVGITYQHID 154
> pfa:PFC0441c SAC3/GNAP family-related protein, putative; K15028
translation initiation factor 3 subunit K
Length=235
Score = 53.9 bits (128), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 34/191 (17%), Positives = 81/191 (42%), Gaps = 26/191 (13%)
Query 37 QMIYLPSTLDLLVSHLEKGIAAG-GASPDLCSAVLRLYLLYPQCANAEVVRKILVQTIVA 95
M++ S L +L ++ D+ +LRL+ LYP C + ++++KIL+ +
Sbjct 18 HMMFNASKLKVLSDYVNVAFENNEYYDNDVMLTLLRLFCLYPHCYDKDIIKKILICVLYN 77
Query 96 LPENQFGYYACIAPSHFSNPEDEKKVQDVLTLQELLESCDFKAAWAMLREP--------E 147
+ Y + S + ++ V+ + EL+++C +K W + +
Sbjct 78 INNVDMNMYMSLINSSLY----DDNIKSVIYIYELIKNCQYKKLWNCINNNIGNEYNNCD 133
Query 148 MADVLHTPGVVEALRRYVCEVLQQTFSTISLSDFCQLLNLNAAAAAAAAGAAAAADEKVE 207
+ + + + +R+Y+ + +F +I L + + LN+ + ++E
Sbjct 134 YSYLKNDKNFICNIRKYILNTISISFESIYLKNMSEYLNI-------------QDNTQLE 180
Query 208 KLVKARGWTIE 218
+ + WTI+
Sbjct 181 QFLNENKWTIK 191
> ath:AT4G33250 EIF3K; EIF3K (EUKARYOTIC TRANSLATION INITIATION
FACTOR 3K); translation initiation factor; K15028 translation
initiation factor 3 subunit K
Length=226
Score = 53.5 bits (127), Expect = 8e-07, Method: Compositional matrix adjust.
Identities = 39/164 (23%), Positives = 73/164 (44%), Gaps = 24/164 (14%)
Query 64 DLCSAVLRLYLLYPQCANAEVVRKILVQTIVALPENQFGYYACIAPSHFSNPEDEKKVQD 123
+LC +LRLY P+ N +V +ILV+ ++A+P F + P + E++ +
Sbjct 49 NLC--LLRLYQFEPERMNTHIVARILVKALMAMPTPDFSLCLFLIPERV---QMEEQFKS 103
Query 124 VLTLQELLESCDFKAAWAMLREPEMADVLH----TPGVVEALRRYVCEVLQQTFSTISLS 179
++ L LE+ F+ W E A H PG +A++ Y +L ++ + S
Sbjct 104 LIVLSHYLETGRFQQFWD-----EAAKNRHILEAVPGFEQAIQAYASHLLSLSYQKVPRS 158
Query 180 DFCQLLNLNAAAAAAAAGAAAAADEKVEKLVKARGWTIEDAAAG 223
+ +N++ A+ D+ +E+ V GW +E
Sbjct 159 VLAEAVNMD----------GASLDKFIEQQVTNSGWIVEKEGGS 192
> cpv:cgd3_4020 eIF-3 p25/subunit 11 ; K15028 translation initiation
factor 3 subunit K
Length=262
Score = 44.7 bits (104), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 40/192 (20%), Positives = 87/192 (45%), Gaps = 19/192 (9%)
Query 40 YLPSTLDLLVSHLEKGIAAGGASPDLCSAVLRLYLLYPQCANAEVVRKILVQTIVALPEN 99
Y S LD+ + + I G S VL Y +YP+ N E+V+ IL+ +I+ P +
Sbjct 51 YDVSNLDIFENCVRDQIIKGQYSILNNLVVLLQYSIYPKRTNLEIVQDILLLSIIRGPLS 110
Query 100 QFGYYACIAPSHFSNPEDEKKVQDVLTLQELLESCDFKAAWAMLREPEM--ADVLHTPGV 157
+ +C S +++ ++ ++ L +LL SC + W +L+ + + V G
Sbjct 111 S-DFLSCTYQIPLS-VQNDPNIKQIIQLNDLLTSCRYANMWNLLKTNDQLRSKVEKIQGF 168
Query 158 VEALRRYVCEVLQQTFSTISLSDFCQLLNLNAAAAAAAAGAAAAADEKVEKLVKARGWTI 217
+++R + + + S IS + +LL+ + +++ +++ W +
Sbjct 169 YDSIRDIIIYSVNCSHSCISTAVLSELLDFPKDST------------ELKNIIEKNKWIL 216
Query 218 EDAAAGQKIVRV 229
+ A +I+R+
Sbjct 217 DSAC---EIIRI 225
> dre:562634 fibrillin 2-like
Length=732
Score = 34.3 bits (77), Expect = 0.43, Method: Compositional matrix adjust.
Identities = 16/40 (40%), Positives = 21/40 (52%), Gaps = 0/40 (0%)
Query 99 NQFGYYACIAPSHFSNPEDEKKVQDVLTLQELLESCDFKA 138
N FG Y C PS ++ ED + QDV E L +CD +
Sbjct 271 NTFGSYECSCPSGYTLREDGRMCQDVDECAEDLHNCDSRG 310
> mmu:65099 Irak1bp1, 4921528N06Rik, AI851240, Aabp3, Aip70, Simpl;
interleukin-1 receptor-associated kinase 1 binding protein
1
Length=237
Score = 32.7 bits (73), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 34/116 (29%), Positives = 51/116 (43%), Gaps = 23/116 (19%)
Query 55 GIAAGGASPDLCSAVLRLYLLYPQCANAE--VVRKI--LVQTIVALPENQFGYYACIAPS 110
G A ASPD +R+ A A+ V R++ + Q++ Q G+ CI +
Sbjct 56 GAAEVSASPDRALVTVRVSSTKEVSAEAKKSVCRRLDYITQSL-----QQQGFQVCITFT 110
Query 111 HFSNPEDEKKVQDVLTLQELLESCDFKAAWAMLREPEMADVLHTPGVVEALRRYVC 166
F K+Q++ L+E D + ++ PE HTPG VE LRR C
Sbjct 111 EFG------KMQNICNF--LVEKLD---SSVVISPPEF---YHTPGSVENLRRQAC 152
> xla:447214 MGC81969 protein
Length=841
Score = 32.3 bits (72), Expect = 1.8, Method: Composition-based stats.
Identities = 18/52 (34%), Positives = 29/52 (55%), Gaps = 3/52 (5%)
Query 109 PSHFSNPEDEKKVQDVLTLQELLESCDFKAAWAMLREPEMADVLHTPGVVEA 160
P S+P+D+K+V+ +LT +E E +K+ W RE +V+ GV E
Sbjct 750 PEEGSDPDDKKEVKSILT-KEFKEDPGYKSVW--FREDATPEVVVIQGVEEG 798
> hsa:6775 STAT4, SLEB11; signal transducer and activator of transcription
4; K11222 signal transducer and activator of transcription
4
Length=748
Score = 31.6 bits (70), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 22/71 (30%), Positives = 37/71 (52%), Gaps = 4/71 (5%)
Query 98 ENQFGY-YACIAPSHFSNPEDEKKVQDVLTLQELLESCDFKAAWAMLREPEM---ADVLH 153
+++F Y Y I S+ Q+VLTLQE+L S DFK A+ + ++ D+L
Sbjct 168 QDEFDYRYKTIQTMDQSDKNSAMVNQEVLTLQEMLNSLDFKRKEALSKMTQIIHETDLLM 227
Query 154 TPGVVEALRRY 164
++E L+ +
Sbjct 228 NTMLIEELQDW 238
Lambda K H
0.320 0.133 0.380
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 10082348456
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40