bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_0063_orf9
Length=425
Score E
Sequences producing significant alignments: (Bits) Value
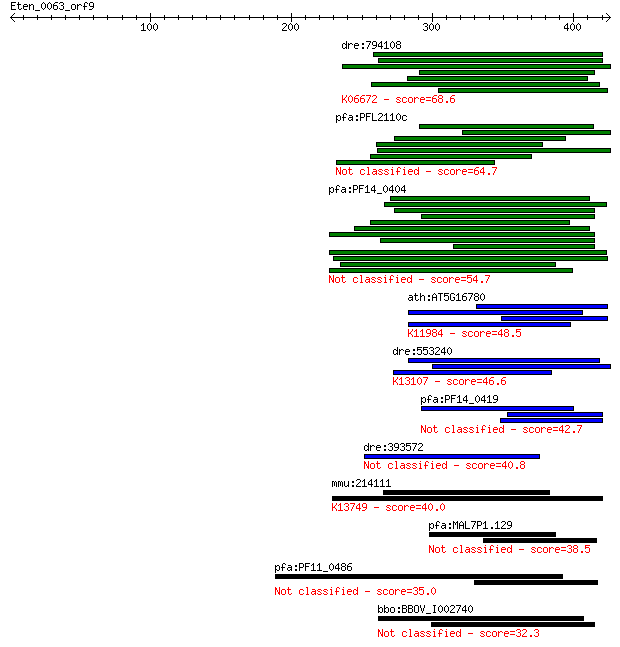
dre:794108 nipblb, nipbl, wu:fi33d08; nipped-b homolog b (Dros... 68.6 4e-11
pfa:PFL2110c hypothetical protein 64.7 6e-10
pfa:PF14_0404 TREP; TRAP-related protein 54.7 7e-07
ath:AT5G16780 DOT2; DOT2 (DEFECTIVELY ORGANIZED TRIBUTARIES 2)... 48.5 5e-05
dre:553240 rbmx2, im:7141316, zgc:113884; RNA binding motif pr... 46.6 2e-04
pfa:PF14_0419 conserved Plasmodium protein, unknown function 42.7 0.002
dre:393572 traf3ip1, Elipsa, MGC63522, zgc:63522; TNF receptor... 40.8 0.011
mmu:214111 Slc24a1, MGC27617; solute carrier family 24 (sodium... 40.0 0.016
pfa:MAL7P1.129 conserved Plasmodium protein, unknown function 38.5 0.045
pfa:PF11_0486 MAEBL, putative 35.0 0.49
bbo:BBOV_I002740 19.m02242; 200 kDa antigen p200 32.3 3.7
> dre:794108 nipblb, nipbl, wu:fi33d08; nipped-b homolog b (Drosophila);
K06672 cohesin loading factor subunit SCC2
Length=2856
Score = 68.6 bits (166), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 70/188 (37%), Positives = 108/188 (57%), Gaps = 29/188 (15%)
Query 258 EDAKAEKSEAEDKGE-----REGESEEAGD--------KDREEGAGREGEAE--TEVERE 302
D KAEK AE KG EG SE + D KD + GR + + VE
Sbjct 630 HDGKAEKIRAEGKGHETSRKHEGRSELSRDCKEERHREKDSDSSKGRRSDTSKSSRVEHN 689
Query 303 REKEREE-------GEKQREKEREGGEDERERGGEREKEKEKERESEKE----GEKEKEG 351
R+KE+E+ EK REKE E G D +ER ++EK++EK R+ E E E+ K+
Sbjct 690 RDKEQEQEKVGDKGLEKGREKELEKGRD-KERVKDQEKDQEKGRDKEVEKGRYKERVKDR 748
Query 352 EKEEEEGREREKEEEREKEKERESEEEREKGKERESEKEREEGNEGEGEGEREEEKERES 411
KE+E+ R++E+ + R+K++ ++ E+ REK +++E EK+RE+ + E E +R +K R+
Sbjct 749 VKEQEKVRDKEQVKGRDKKRSKDLEKCREKDQDKELEKDREKNQDKELEKDR--DKVRDK 806
Query 412 DRERVGEK 419
DR++V EK
Sbjct 807 DRDKVREK 814
Score = 65.1 bits (157), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 57/161 (35%), Positives = 105/161 (65%), Gaps = 6/161 (3%)
Query 262 AEKSEAEDKGEREGESEEAGDKDREEGAGREGEAETEVEREREKERE-EGEKQREKEREG 320
++ S E ++E E E+ GDK E+G RE E E ++ER K++E + EK R+KE E
Sbjct 681 SKSSRVEHNRDKEQEQEKVGDKGLEKG--REKELEKGRDKERVKDQEKDQEKGRDKEVEK 738
Query 321 GEDERERGGEREKEKEKERESEKEGEKEKEGEKEEEEGREREKEEEREKEKERESEEERE 380
G +ER +R KE+EK R+ E+ ++K+ K+ E+ RE+++++E EK++E+ ++E E
Sbjct 739 GR-YKERVKDRVKEQEKVRDKEQVKGRDKKRSKDLEKCREKDQDKELEKDREKNQDKELE 797
Query 381 KGKERESEKEREEGNEGEGEGEREEEKE--RESDRERVGEK 419
K +++ +K+R++ E + + RE++++ RE DRE++ E+
Sbjct 798 KDRDKVRDKDRDKVREKDRDKVREKDRDKLREKDREKIRER 838
Score = 61.2 bits (147), Expect = 7e-09, Method: Compositional matrix adjust.
Identities = 59/207 (28%), Positives = 122/207 (58%), Gaps = 17/207 (8%)
Query 236 KDPQKRPGEKSEGDRDGDGVGDEDAKAEKSEAEDKGEREG---------ESEEAGDKDRE 286
KD G +S+ + + D + E+ + DKG +G + E D++++
Sbjct 668 KDSDSSKGRRSDTSKSSRVEHNRDKEQEQEKVGDKGLEKGREKELEKGRDKERVKDQEKD 727
Query 287 EGAGREGEAETEVEREREKER-EEGEKQREKEREGGED-------ERERGGEREKEKEKE 338
+ GR+ E E +ER K+R +E EK R+KE+ G D E+ R +++KE EK+
Sbjct 728 QEKGRDKEVEKGRYKERVKDRVKEQEKVRDKEQVKGRDKKRSKDLEKCREKDQDKELEKD 787
Query 339 RESEKEGEKEKEGEKEEEEGREREKEEEREKEKERESEEEREKGKERESEKEREEGNEGE 398
RE ++ E EK+ +K ++ R++ +E++R+K +E++ ++ REK +E+ E++R++G E +
Sbjct 788 REKNQDKELEKDRDKVRDKDRDKVREKDRDKVREKDRDKLREKDREKIRERDRDKGREKD 847
Query 399 GEGEREEEKERESDRERVGEKIKKKKK 425
+ E+ + +E++ ++ER+ ++ K+++K
Sbjct 848 RDKEQVKTREKDQEKERLKDRDKEREK 874
Score = 55.1 bits (131), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 43/125 (34%), Positives = 87/125 (69%), Gaps = 2/125 (1%)
Query 291 REGEAETEVEREREKERE-EGEKQREKEREGGEDERERGGEREKEKEKERESEKEGEKEK 349
RE + + E+E++REK ++ E EK R+K R+ D + R +R+K +EK+R+ +E ++EK
Sbjct 776 REKDQDKELEKDREKNQDKELEKDRDKVRDKDRD-KVREKDRDKVREKDRDKLREKDREK 834
Query 350 EGEKEEEEGREREKEEEREKEKERESEEEREKGKERESEKEREEGNEGEGEGEREEEKER 409
E++ ++GRE+++++E+ K +E++ E+ER K +++E EK R++G + + + E++ KE
Sbjct 835 IRERDRDKGREKDRDKEQVKTREKDQEKERLKDRDKEREKVRDKGRDRDRDQEKKRNKEL 894
Query 410 ESDRE 414
D++
Sbjct 895 TEDKQ 899
Score = 50.1 bits (118), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 44/129 (34%), Positives = 89/129 (68%), Gaps = 2/129 (1%)
Query 282 DKDREEGAGREGEAETEVEREREKEREEG-EKQREKEREGGEDERERGGEREKEKEKERE 340
D+D+E RE + E+E++R+K R++ +K REK+R+ E++R REK++EK RE
Sbjct 779 DQDKELEKDREKNQDKELEKDRDKVRDKDRDKVREKDRDKVR-EKDRDKLREKDREKIRE 837
Query 341 SEKEGEKEKEGEKEEEEGREREKEEEREKEKERESEEEREKGKERESEKEREEGNEGEGE 400
+++ +EK+ +KE+ + RE+++E+ER K++++E E+ R+KG++R+ ++E++ E +
Sbjct 838 RDRDKGREKDRDKEQVKTREKDQEKERLKDRDKEREKVRDKGRDRDRDQEKKRNKELTED 897
Query 401 GEREEEKER 409
+ E++ R
Sbjct 898 KQAPEQRSR 906
Score = 42.7 bits (99), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 50/166 (30%), Positives = 95/166 (57%), Gaps = 7/166 (4%)
Query 257 DEDAKAEKS-EAEDKGEREGESEEAGDKDREEGAGREGEAETEVEREREKEREEG-EKQR 314
D+D + EK E E E + ++ DKDR++ RE + + E++R+K RE+ EK R
Sbjct 779 DQDKELEKDREKNQDKELEKDRDKVRDKDRDKV--REKDRDKVREKDRDKLREKDREKIR 836
Query 315 EKEREGGED---ERERGGEREKEKEKERESEKEGEKEKEGEKEEEEGREREKEEEREKEK 371
E++R+ G + ++E+ REK++EKER +++ E+EK +K + R++EK+ +E +
Sbjct 837 ERDRDKGREKDRDKEQVKTREKDQEKERLKDRDKEREKVRDKGRDRDRDQEKKRNKELTE 896
Query 372 ERESEEEREKGKERESEKEREEGNEGEGEGEREEEKERESDRERVG 417
++++ E+R + ++E G E + + ER K + E+ G
Sbjct 897 DKQAPEQRSRPNSPRVKQEPRNGEESKIKPERSVHKNSNNKDEKRG 942
Score = 40.4 bits (93), Expect = 0.014, Method: Compositional matrix adjust.
Identities = 40/126 (31%), Positives = 67/126 (53%), Gaps = 8/126 (6%)
Query 304 EKEREEGEKQREKEREGGEDERERGGEREKEKEKERESEKEGEKEKEGEKEEEEGREREK 363
EK R EG+ + G E R + E+ +EK+ +S K + E R++E+
Sbjct 635 EKIRAEGKGHETSRKHEGRSELSRDCKEERHREKDSDSSKGRRSDTSKSSRVEHNRDKEQ 694
Query 364 EEER------EKEKERESEEEREKGKERESEKEREEGNEGEGEGEREEEKERESDRERVG 417
E+E+ EK +E+E E+ R+K + ++ EK++E+G + E E R KER DR +
Sbjct 695 EQEKVGDKGLEKGREKELEKGRDKERVKDQEKDQEKGRDKEVEKGR--YKERVKDRVKEQ 752
Query 418 EKIKKK 423
EK++ K
Sbjct 753 EKVRDK 758
> pfa:PFL2110c hypothetical protein
Length=1846
Score = 64.7 bits (156), Expect = 6e-10, Method: Composition-based stats.
Identities = 42/123 (34%), Positives = 72/123 (58%), Gaps = 2/123 (1%)
Query 291 REGEAETEVEREREKEREEGEKQREKEREGGEDERERGGEREKEKEKERESEKEGEKEKE 350
R GE E E KE ++ ++++ E E ++ GE +K +E+++ E++ K +E
Sbjct 1487 RRGEEEKMSADENMKEEQKMREEQKVGEEQKVGEEQKVGEEQKLREEQKMREEQ--KMRE 1544
Query 351 GEKEEEEGREREKEEEREKEKERESEEEREKGKERESEKEREEGNEGEGEGEREEEKERE 410
+K EE + RE+++ RE++K RE ++ RE+ K RE +K REE E + REE+K RE
Sbjct 1545 EQKMREEQKMREEQKVREEQKMREEQKMREEQKMREEQKVREEQKLREEQKMREEQKMRE 1604
Query 411 SDR 413
+
Sbjct 1605 EQK 1607
Score = 61.2 bits (147), Expect = 8e-09, Method: Composition-based stats.
Identities = 44/119 (36%), Positives = 71/119 (59%), Gaps = 14/119 (11%)
Query 321 GEDERERGGEREKEKEKERESEKEGEKEKEGE----------KEEEEGREREK-EEEREK 369
GE+E+ E KE++K RE +K GE++K GE +EE++ RE +K EE++
Sbjct 1489 GEEEKMSADENMKEEQKMREEQKVGEEQKVGEEQKVGEEQKLREEQKMREEQKMREEQKM 1548
Query 370 EKE---RESEEEREKGKERESEKEREEGNEGEGEGEREEEKERESDRERVGEKIKKKKK 425
+E RE ++ RE+ K RE +K REE E + REE+K RE + R +K+++++K
Sbjct 1549 REEQKMREEQKVREEQKMREEQKMREEQKMREEQKVREEQKLREEQKMREEQKMREEQK 1607
Score = 55.8 bits (133), Expect = 3e-07, Method: Composition-based stats.
Identities = 41/123 (33%), Positives = 76/123 (61%), Gaps = 6/123 (4%)
Query 273 REGESEE--AGDKDREEGAGREGEAETEVEREREKEREEGEKQREKEREGGEDERERGGE 330
R GE E+ A + +EE RE + E ++ E+++ E++ +E++ E+++ R +
Sbjct 1487 RRGEEEKMSADENMKEEQKMREEQKVGEEQKVGEEQKVGEEQKLREEQKMREEQKMREEQ 1546
Query 331 REKEKEKERESEKEGEKEKEGEKEEEEGREREKEEEREKEKERESEEEREKGKERESEKE 390
+ +E++K RE +K E++K +EE++ RE +K RE++K RE ++ RE+ K RE +K
Sbjct 1547 KMREEQKMREEQKVREEQKM--REEQKMREEQK--MREEQKVREEQKLREEQKMREEQKM 1602
Query 391 REE 393
REE
Sbjct 1603 REE 1605
Score = 53.1 bits (126), Expect = 2e-06, Method: Composition-based stats.
Identities = 35/119 (29%), Positives = 65/119 (54%), Gaps = 1/119 (0%)
Query 260 AKAEKSEAEDKGEREGESEEAGDKDREEGAGREGE-AETEVEREREKEREEGEKQREKER 318
+ EK A++ + E + E E+ G E + E + RE +K REE + + E++
Sbjct 1489 GEEEKMSADENMKEEQKMREEQKVGEEQKVGEEQKVGEEQKLREEQKMREEQKMREEQKM 1548
Query 319 EGGEDERERGGEREKEKEKERESEKEGEKEKEGEKEEEEGREREKEEEREKEKERESEE 377
+ RE RE++K +E + +E +K +E +K EE + RE+++ RE++K RE ++
Sbjct 1549 REEQKMREEQKVREEQKMREEQKMREEQKMREEQKVREEQKLREEQKMREEQKMREEQK 1607
Score = 52.8 bits (125), Expect = 2e-06, Method: Composition-based stats.
Identities = 56/192 (29%), Positives = 98/192 (51%), Gaps = 27/192 (14%)
Query 261 KAEKSEAEDKGEREGESEE-AGDKDREEGAGREGEAETEVEREREKEREEGEKQREKER- 318
+ +K ED + E + E+ +K R E ++ E +++ ++EKE E EK K
Sbjct 1386 QEKKKLQEDIKKLEKDKEQFQKEKIRREEKEKQLLLEEKIKLQKEKELFENEKLERKMSY 1445
Query 319 --EGGEDERERGGEREKEKEKERESE---------------KEGEKEK----EGEKEEEE 357
+ E E+++ + EK +R + + GE+EK E KEE++
Sbjct 1446 MLKINELEKKKNERNKMEKSYKRMIQKDKEKKKKKESRDKIRRGEEEKMSADENMKEEQK 1505
Query 358 GREREK--EEEREKEKERESEEE--REKGKERESEKEREEGNEGEGEGEREEEKERESDR 413
RE +K EE++ E+++ EE+ RE+ K RE +K REE E + REE+K RE +
Sbjct 1506 MREEQKVGEEQKVGEEQKVGEEQKLREEQKMREEQKMREEQKMREEQKMREEQKVREEQK 1565
Query 414 ERVGEKIKKKKK 425
R +K+++++K
Sbjct 1566 MREEQKMREEQK 1577
Score = 47.8 bits (112), Expect = 8e-05, Method: Composition-based stats.
Identities = 33/116 (28%), Positives = 70/116 (60%), Gaps = 6/116 (5%)
Query 256 GDEDAKAEKSEAEDKGEREGESEEAGDKDR--EEGAGREGEAETEVEREREKEREEGEKQ 313
DE+ K E+ E+ ++ GE ++ G++ + EE RE + E ++ RE+++ E++
Sbjct 1496 ADENMKEEQKMREE--QKVGEEQKVGEEQKVGEEQKLREEQKMREEQKMREEQKMREEQK 1553
Query 314 REKEREGGEDERERGGEREKEKEKERESEKEGEKEKEGEKEEEEGREREKEEEREK 369
+E++ E+++ R ++ +E++K RE +K E++K +EE++ RE +K E +K
Sbjct 1554 MREEQKVREEQKMREEQKMREEQKMREEQKVREEQK--LREEQKMREEQKMREEQK 1607
Score = 36.2 bits (82), Expect = 0.25, Method: Composition-based stats.
Identities = 34/114 (29%), Positives = 61/114 (53%), Gaps = 5/114 (4%)
Query 232 DSPGKDPQKRPGEKSEGDRDGDGVGDEDAKAEKSE-AEDKGEREGESEEAGDKDREEGAG 290
D K+ QK E+ G+ VG+E E+ + E++ RE + K REE
Sbjct 1497 DENMKEEQKMREEQKVGEEQK--VGEEQKVGEEQKLREEQKMREEQKMREEQKMREEQKM 1554
Query 291 REGEAETEVEREREKER-EEGEKQREKEREGGEDERERGGEREKEKEKERESEK 343
RE + E ++ RE+++ E +K RE E++ E+++ R ++ +E++K RE +K
Sbjct 1555 REEQKVREEQKMREEQKMREEQKMRE-EQKVREEQKLREEQKMREEQKMREEQK 1607
> pfa:PF14_0404 TREP; TRAP-related protein
Length=3543
Score = 54.7 bits (130), Expect = 7e-07, Method: Composition-based stats.
Identities = 48/141 (34%), Positives = 74/141 (52%), Gaps = 0/141 (0%)
Query 270 KGEREGESEEAGDKDREEGAGREGEAETEVEREREKEREEGEKQREKEREGGEDERERGG 329
KGE + +++ + ++ + E E+E ER+ E +E ++Q KE + E E R
Sbjct 2529 KGEIKEDTKHEKENAKQMNEEIKNVKEQEIEHERKNETKEEKEQSIKEGKNKEFEEGRNK 2588
Query 330 EREKEKEKERESEKEGEKEKEGEKEEEEGREREKEEEREKEKERESEEEREKGKERESEK 389
E E+ + KE E + E E+ KE EEGR +E EE R KE E +E E+G+ +E E+
Sbjct 2589 EFEEGRNKEFEEGRNKEFEEGRNKEFEEGRNKEFEEGRNKEFEDGRNKEFEEGRNKEFEE 2648
Query 390 EREEGNEGEGEGEREEEKERE 410
R + E E EE K +E
Sbjct 2649 RRNKEFEEGRNKEFEEGKNKE 2669
Score = 53.1 bits (126), Expect = 2e-06, Method: Composition-based stats.
Identities = 56/161 (34%), Positives = 85/161 (52%), Gaps = 11/161 (6%)
Query 266 EAEDKGEREGESEEAGDKDREEGAGREGEAETEVEREREKEREEGEKQREKEREGGEDER 325
E E + ER+ E++E ++ +EG + E E R KE EEG + +E E E
Sbjct 2555 EQEIEHERKNETKEEKEQSIKEGKNK------EFEEGRNKEFEEGRNKEFEEGRNKEFEE 2608
Query 326 ERGGEREKEKEKERESEKEGEKEKEGEKEEEEGREREKEEEREKEKERESEEEREKGKER 385
R E E+ + KE E + E E KE EEGR +E EE R KE E +E E+GK +
Sbjct 2609 GRNKEFEEGRNKEFEEGRNKEFEDGRNKEFEEGRNKEFEERRNKEFEEGRNKEFEEGKNK 2668
Query 386 E----SEKEREEGNEGEGEGEREEEKERESDRERVGEKIKK 422
E ++K+ EEG E + E ++K +E + ++ E+I+K
Sbjct 2669 EIKEVNDKKFEEGKNKEIK-EGNKKKVQEQNDKKFNEEIEK 2708
Score = 50.1 bits (118), Expect = 2e-05, Method: Composition-based stats.
Identities = 51/156 (32%), Positives = 80/156 (51%), Gaps = 14/156 (8%)
Query 273 REGESEEAGDKDREEGAGREGEA--ETEVEREREKEREEGEKQREKEREGGEDERERGGE 330
R E EE +K+ EEG +E E E E R KE EEG + +ER E E R E
Sbjct 2602 RNKEFEEGRNKEFEEGRNKEFEEGRNKEFEDGRNKEFEEGRNKEFEERRNKEFEEGRNKE 2661
Query 331 REKEKEKERESEKEGEKEKEGEKEEEEGREREKEEEREKEKERESEEEREKGKERE---- 386
E+ K KE + + + E+ KE +EG +++ +E+ +K+ E E+ E+G E+E
Sbjct 2662 FEEGKNKEIKEVNDKKFEEGKNKEIKEGNKKKVQEQNDKKFNEEIEKLIEEGNEKEIKEG 2721
Query 387 --------SEKEREEGNEGEGEGEREEEKERESDRE 414
++KE EGN+ E E E++ E +++E
Sbjct 2722 RNKEIRERNDKEINEGNDKNFEKEIEKQSEEANEKE 2757
Score = 47.0 bits (110), Expect = 2e-04, Method: Composition-based stats.
Identities = 46/133 (34%), Positives = 69/133 (51%), Gaps = 10/133 (7%)
Query 292 EGEAETEVEREREKEREEGEKQREKEREGGEDERERGGEREKE------KEKERESEKEG 345
+GE + + + E+E ++ E+ + + + E ER+ + EKE K KE E +
Sbjct 2529 KGEIKEDTKHEKENAKQMNEEIKNVKEQEIEHERKNETKEEKEQSIKEGKNKEFEEGRNK 2588
Query 346 EKEKEGEKEEEEGREREKEEEREKEKERESEEEREKGKERESE----KEREEGNEGEGEG 401
E E+ KE EEGR +E EE R KE E +E E+G+ +E E KE EEG E E
Sbjct 2589 EFEEGRNKEFEEGRNKEFEEGRNKEFEEGRNKEFEEGRNKEFEDGRNKEFEEGRNKEFEE 2648
Query 402 EREEEKERESDRE 414
R +E E ++E
Sbjct 2649 RRNKEFEEGRNKE 2661
Score = 46.6 bits (109), Expect = 2e-04, Method: Composition-based stats.
Identities = 50/147 (34%), Positives = 80/147 (54%), Gaps = 6/147 (4%)
Query 256 GDEDAKAEKSEAEDKGEREGESEEAGDKDREEGAGREGEAETEVEREREKEREEGEKQRE 315
G++ E+ E + + E + +E DK EEG +E + E + E ++ ++E E +
Sbjct 2953 GNDKEIEEEIEKKIEEANEKKIKEENDKKFEEGNDKEVKEENDKEVKKGNDKEFEEGNDK 3012
Query 316 KEREGGEDERERGGEREKEKEKERESEKEGEKE-KEG-EKEEEEGREREKEEER----EK 369
K EG + E + G E+E ++ E+E ++ EKE KEG EKE +EG ++ EE+ E+
Sbjct 3013 KFEEGNDKEIKEGNEKEIKEGNEKEIKEGNEKEIKEGNEKEIKEGNDKNFEEDDYKKFEE 3072
Query 370 EKERESEEEREKGKERESEKEREEGNE 396
K +E EE K E KE EEGN+
Sbjct 3073 GKNKEFEERMNKEFEERMNKEFEEGNK 3099
Score = 41.6 bits (96), Expect = 0.005, Method: Composition-based stats.
Identities = 62/181 (34%), Positives = 89/181 (49%), Gaps = 16/181 (8%)
Query 245 KSEGDRDGDGVGDEDAKAEKSEAEDKGEREGESEEAGDKDREEGAGREGEA--ETEVERE 302
K E D+ + D++ K E + KG + E EE DK EEG +E + E E++
Sbjct 2975 KEENDKKFEEGNDKEVKEENDKEVKKGN-DKEFEEGNDKKFEEGNDKEIKEGNEKEIKEG 3033
Query 303 REKEREEGEKQR-----EKEREGGED---ERERGGEREKEKEKERESEKEGEKEKEGEKE 354
EKE +EG ++ EKE + G D E + + E+ K KE E E E+ KE
Sbjct 3034 NEKEIKEGNEKEIKEGNEKEIKEGNDKNFEEDDYKKFEEGKNKEFEERMNKEFEERMNKE 3093
Query 355 EEEGREREKEEEREKEKERESEEEREKGKERESE----KEREEGNEGE-GEGEREEEKER 409
EEG ++ EE +K E +E E+GK ++ E K +EGNE E EG +E E
Sbjct 3094 FEEGNKKVVEEGNKKVVEGGRNKEFEEGKNKQFEEVMNKVIKEGNEKEINEGNDKEINEG 3153
Query 410 E 410
E
Sbjct 3154 E 3154
Score = 41.2 bits (95), Expect = 0.007, Method: Composition-based stats.
Identities = 63/213 (29%), Positives = 102/213 (47%), Gaps = 25/213 (11%)
Query 227 GDPQEDSPGKDPQKRPGEKSEGDRDGDGVGDEDAKAEKSEAEDKGEREG---ESEEAGDK 283
G +E G++ + G E + + +E E E +K EG E E+ +K
Sbjct 2577 GKNKEFEEGRNKEFEEGRNKEFEEGRNKEFEEGRNKEFEEGRNKEFEEGRNKEFEDGRNK 2636
Query 284 DREEGAGREGEAE--TEVEREREKEREEGEKQREKE------REGGEDERERGGEREKEK 335
+ EEG +E E E E R KE EEG+ + KE EG E + G +++ ++
Sbjct 2637 EFEEGRNKEFEERRNKEFEEGRNKEFEEGKNKEIKEVNDKKFEEGKNKEIKEGNKKKVQE 2696
Query 336 EKERESEKEGEK--EKEGEKEEEEGR-----EREKEE-------EREKEKERESEEEREK 381
+ +++ +E EK E+ EKE +EGR ER +E EKE E++SEE EK
Sbjct 2697 QNDKKFNEEIEKLIEEGNEKEIKEGRNKEIRERNDKEINEGNDKNFEKEIEKQSEEANEK 2756
Query 382 GKERESEKEREEGNEGEGEGEREEEKERESDRE 414
+ ++K EEGN E + +E E ++++
Sbjct 2757 EIKEGNDKIFEEGNYKEINEGKNKEFEEGNNKD 2789
Score = 39.7 bits (91), Expect = 0.020, Method: Composition-based stats.
Identities = 53/158 (33%), Positives = 95/158 (60%), Gaps = 6/158 (3%)
Query 263 EKSEAEDKGEREGESEEAGDKDREEGAGREGEAETEVEREREKEREEGEKQREKEREGGE 322
E ++ E K E E E DK+ +EG ++ E ETE + E ++E E + +K E E
Sbjct 2888 EGNDKEIKEGNEKEINEGNDKEIKEGEYKKFEEETEKQIEEGNDKEIKEGEYKKFEEETE 2947
Query 323 DERERGGEREKEKEKERESEKEGEKE--KEGEKEEEEGREREKEEEREKEKERESEEERE 380
+ E G ++E E+E E++ E+ EK+ +E +K+ EEG ++E +EE +KE ++ +++E E
Sbjct 2948 KQIEEGNDKEIEEEIEKKIEEANEKKIKEENDKKFEEGNDKEVKEENDKEVKKGNDKEFE 3007
Query 381 KGKERESE----KEREEGNEGEGEGEREEEKERESDRE 414
+G +++ E KE +EGNE E + E+E + +++E
Sbjct 3008 EGNDKKFEEGNDKEIKEGNEKEIKEGNEKEIKEGNEKE 3045
Score = 38.1 bits (87), Expect = 0.071, Method: Composition-based stats.
Identities = 34/106 (32%), Positives = 65/106 (61%), Gaps = 6/106 (5%)
Query 315 EKEREGGEDERERGGEREKEKEKERESEKEGEKEKE--GEKEEEEGREREKEEEREKEKE 372
+K +E + + E G ++E ++E ++E +K +KE E +K+ EEG ++E +E EKE +
Sbjct 2972 KKIKEENDKKFEEGNDKEVKEENDKEVKKGNDKEFEEGNDKKFEEGNDKEIKEGNEKEIK 3031
Query 373 RESEEEREKGKERE----SEKEREEGNEGEGEGEREEEKERESDRE 414
+E+E ++G E+E +EKE +EGN+ E + ++ E ++E
Sbjct 3032 EGNEKEIKEGNEKEIKEGNEKEIKEGNDKNFEEDDYKKFEEGKNKE 3077
Score = 37.0 bits (84), Expect = 0.16, Method: Composition-based stats.
Identities = 55/201 (27%), Positives = 92/201 (45%), Gaps = 6/201 (2%)
Query 227 GDPQEDSPGKDPQKRPG---EKSEGDRDGDGVGDEDAKAEKSEAEDKGEREGESEEAGDK 283
G+ +E G D + G E EG+ G+E E +E E K E E +E DK
Sbjct 3001 GNDKEFEEGNDKKFEEGNDKEIKEGNEKEIKEGNEKEIKEGNEKEIKEGNEKEIKEGNDK 3060
Query 284 DREEGAGREGEAETEVEREREKEREEGEKQREKEREGGEDERERGGER--EKEKEKERES 341
+ EE ++ E E E +E E+ ++ EG + E G ++ E + KE E
Sbjct 3061 NFEEDDYKKFEEGKNKEFEERMNKEFEERMNKEFEEGNKKVVEEGNKKVVEGGRNKEFEE 3120
Query 342 EKEGEKEKEGEKEEEEGREREKEEEREKEKERESEEEREKGKERESEKEREEGNEGEGEG 401
K + E+ K +EG E+E E +KE E E ++ + + + +E + EG+
Sbjct 3121 GKNKQFEEVMNKVIKEGNEKEINEGNDKEIN-EGEYKKFEEETEKQIEEGNDKEIKEGDH 3179
Query 402 EREEEKERESDRERVGEKIKK 422
++ EEK + E + + IK+
Sbjct 3180 KKFEEKIEKQVEEGMNKVIKE 3200
Score = 35.4 bits (80), Expect = 0.37, Method: Composition-based stats.
Identities = 51/197 (25%), Positives = 90/197 (45%), Gaps = 6/197 (3%)
Query 230 QEDSPGKDPQKRPGEKSEGDRDGDGVGDEDAKAEKSEAEDKGEREGESEEAGDKDREEGA 289
+E G D + G + + D E + E E +K +EG +E K+ E
Sbjct 2996 KEVKKGNDKEFEEGNDKKFEEGNDKEIKEGNEKEIKEGNEKEIKEGNEKEI--KEGNEKE 3053
Query 290 GREGEAETEVEREREKEREEGEKQREKEREGGEDERERGGEREKEKEKERESEKEGEKEK 349
+EG + E + K+ EEG+ + +ER E E E E+ +K E + E
Sbjct 3054 IKEGN-DKNFEEDDYKKFEEGKNKEFEERMNKEFEERMNKEFEEGNKKVVEEGNKKVVEG 3112
Query 350 EGEKEEEEGREREKEEEREKEKERESEEEREKGKER---ESEKEREEGNEGEGEGEREEE 406
KE EEG+ ++ EE K + +E+E +G ++ E E ++ E + E ++
Sbjct 3113 GRNKEFEEGKNKQFEEVMNKVIKEGNEKEINEGNDKEINEGEYKKFEEETEKQIEEGNDK 3172
Query 407 KERESDRERVGEKIKKK 423
+ +E D ++ EKI+K+
Sbjct 3173 EIKEGDHKKFEEKIEKQ 3189
Score = 33.9 bits (76), Expect = 1.3, Method: Composition-based stats.
Identities = 46/165 (27%), Positives = 77/165 (46%), Gaps = 13/165 (7%)
Query 235 GKDPQKRPGEKSEGDRDGDGVGDEDAKA---EKSEAEDKGEREGESEEAGDKDREEGAGR 291
GK+ + + G K + D +E+ + E +E E K R E E DK+ EG +
Sbjct 2681 GKNKEIKEGNKKKVQEQNDKKFNEEIEKLIEEGNEKEIKEGRNKEIRERNDKEINEGNDK 2740
Query 292 EGEAETEVEREREKEREEGEKQREKEREGGEDERERGGEREKEKEKERESEKEGEKEKEG 351
E E E + E E+E E + EG E G +E E+ ++ ++E EKE
Sbjct 2741 NFEKEIEKQSEEANEKEIKEGNDKIFEEGNYKEINEGKNKEFEEGNNKDVKEENEKEINE 2800
Query 352 EKEE----------EEGREREKEEEREKEKERESEEEREKGKERE 386
EE +EG E+E +E +K+ E +++E ++G ++E
Sbjct 2801 GNEEEINEGNEKEIKEGNEKEIKEGNDKKFEEGNDKEIKEGNDKE 2845
Score = 33.1 bits (74), Expect = 2.0, Method: Composition-based stats.
Identities = 56/186 (30%), Positives = 94/186 (50%), Gaps = 14/186 (7%)
Query 227 GDPQEDSPGKDPQKRPGEKSEGDRDGDGVGDEDAKAEKSEAEDKGERE---GESEEAGDK 283
G+ +E G + + + G + E D +ED + E ++K E E EE +K
Sbjct 3033 GNEKEIKEGNEKEIKEGNEKEIKEGNDKNFEEDDYKKFEEGKNKEFEERMNKEFEERMNK 3092
Query 284 DREEGAGREGEAETE--VEREREKEREEGEKQREKE------REGGEDERERGGERE--K 333
+ EEG + E + VE R KE EEG+ ++ +E +EG E E G ++E +
Sbjct 3093 EFEEGNKKVVEEGNKKVVEGGRNKEFEEGKNKQFEEVMNKVIKEGNEKEINEGNDKEINE 3152
Query 334 EKEKERESEKEGEKEKEGEKEEEEGREREKEEEREKEKERESEEEREKGKERE-SEKERE 392
+ K+ E E E + E+ +KE +EG ++ EE+ EK+ E + ++G E+E +E +
Sbjct 3153 GEYKKFEEETEKQIEEGNDKEIKEGDHKKFEEKIEKQVEEGMNKVIKEGNEKEINEGNDK 3212
Query 393 EGNEGE 398
E EGE
Sbjct 3213 EIKEGE 3218
> ath:AT5G16780 DOT2; DOT2 (DEFECTIVELY ORGANIZED TRIBUTARIES
2); K11984 U4/U6.U5 tri-snRNP-associated protein 1
Length=820
Score = 48.5 bits (114), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 41/97 (42%), Positives = 61/97 (62%), Gaps = 4/97 (4%)
Query 331 REKEKEKERESEKEGEKEKEGEKEEEEGREREKEEEREKEKERESEEEREKGKER----E 386
R K+KEK+ + EK +K+ +KE+E R+R ++E+ EKE R ++EREK K R E
Sbjct 38 RSKDKEKDYDREKIRDKDHRRDKEKERDRKRSRDEDTEKEISRGRDKEREKDKSRDRVKE 97
Query 387 SEKEREEGNEGEGEGEREEEKERESDRERVGEKIKKK 423
+KE+E + E ER+ EKE++ DR RV E+ KK
Sbjct 98 KDKEKERNRHKDRENERDNEKEKDKDRARVKERASKK 134
Score = 43.9 bits (102), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 45/123 (36%), Positives = 73/123 (59%), Gaps = 12/123 (9%)
Query 283 KDREEGAGREGEAETEVEREREKEREEGEKQREKEREGGEDERERGGEREKEKEKERESE 342
KD+E+ RE + + R++EKER +++R ++ E E E RG ++E+EK+K R+
Sbjct 40 KDKEKDYDREKIRDKDHRRDKEKER---DRKRSRD-EDTEKEISRGRDKEREKDKSRDRV 95
Query 343 KEGEKEKEGEKEEEEGREREKEEEREKEKERESEEEREKGKERESEKEREEGNEGEGEGE 402
KE +KEKE R R K+ E E++ E+E +++R + KER S+K E+ +E E
Sbjct 96 KEKDKEKE--------RNRHKDRENERDNEKEKDKDRARVKERASKKSHEDDDETHKAAE 147
Query 403 REE 405
R E
Sbjct 148 RYE 150
Score = 38.5 bits (88), Expect = 0.047, Method: Compositional matrix adjust.
Identities = 27/75 (36%), Positives = 53/75 (70%), Gaps = 0/75 (0%)
Query 349 KEGEKEEEEGREREKEEEREKEKERESEEEREKGKERESEKEREEGNEGEGEGEREEEKE 408
++G ++E++ R ++KE++ ++EK R+ + R+K KER+ ++ R+E E E R++E+E
Sbjct 28 RDGRRKEKDHRSKDKEKDYDREKIRDKDHRRDKEKERDRKRSRDEDTEKEISRGRDKERE 87
Query 409 RESDRERVGEKIKKK 423
++ R+RV EK K+K
Sbjct 88 KDKSRDRVKEKDKEK 102
Score = 36.2 bits (82), Expect = 0.26, Method: Compositional matrix adjust.
Identities = 39/115 (33%), Positives = 60/115 (52%), Gaps = 16/115 (13%)
Query 283 KDREEGAGREGEAETEVEREREKEREEGEKQREKEREGGEDERERGGEREKEKEKERESE 342
K+R+ R+ + E E+ R R+KERE+ ++ R +EK+KEKER
Sbjct 62 KERDRKRSRDEDTEKEISRGRDKEREK--------------DKSRDRVKEKDKEKERNRH 107
Query 343 KEGEKEKEGEKEEEEGREREKEEEREKEKERESEEEREKGKERESEKEREEGNEG 397
K+ E E++ EKE+++ R R K ER +K E ++E K ER + NEG
Sbjct 108 KDRENERDNEKEKDKDRARVK--ERASKKSHEDDDETHKAAERYEHSDNRGLNEG 160
> dre:553240 rbmx2, im:7141316, zgc:113884; RNA binding motif
protein, X-linked 2; K13107 RNA-binding motif protein, X-linked
2
Length=434
Score = 46.6 bits (109), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 52/142 (36%), Positives = 94/142 (66%), Gaps = 18/142 (12%)
Query 283 KDREEGAGREGEAETEVEREREKEREEGEKQREKEREGG-----EDERERGG--EREKEK 335
K+RE GR+ + E + ER+ ++R+ GEK RE+ER+GG E +RER G +R+ E+
Sbjct 265 KERERDRGRQRDGERQRERDGGRQRD-GEKDRERERDGGRQRDGERQRERDGGRQRDGER 323
Query 336 EKERESEKEGEKEKEGEKEEEEGREREKEEEREKEKERESEEEREKGKERESEKEREEGN 395
++ER E++G ++++G+++ RE+E++ R+++ ER+ E ER+ G++R+ E++RE
Sbjct 324 DRER--ERDGGRQRDGDRD----REQERDGGRQRDVERDQERERDGGRQRDGERDRER-- 375
Query 396 EGEGEGEREEEKERESDRERVG 417
E +G R+ + R+ RER G
Sbjct 376 --ERDGGRQRDGVRDRKRERDG 395
Score = 45.8 bits (107), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 49/139 (35%), Positives = 94/139 (67%), Gaps = 24/139 (17%)
Query 300 EREREKERE-EGEKQREKEREGGEDERERGGEREKEKEKERESEKEGEKEKEGEKEEEE- 357
ERER++ R+ +GE+QRE R+GG +R+ EK++ER E++G ++++GE++ E
Sbjct 266 ERERDRGRQRDGERQRE--RDGGR-------QRDGEKDRER--ERDGGRQRDGERQRERD 314
Query 358 -GREREKEEEREKEK--------ERESEEEREKGKERESEKEREEGNEG--EGEGEREEE 406
GR+R+ E +RE+E+ +R+ E+ER+ G++R+ E+++E +G + +GER+ E
Sbjct 315 GGRQRDGERDRERERDGGRQRDGDRDREQERDGGRQRDVERDQERERDGGRQRDGERDRE 374
Query 407 KERESDRERVGEKIKKKKK 425
+ER+ R+R G + +K+++
Sbjct 375 RERDGGRQRDGVRDRKRER 393
Score = 42.4 bits (98), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 42/121 (34%), Positives = 80/121 (66%), Gaps = 14/121 (11%)
Query 272 EREGESEEAGDKDREE--GAGREGEAETEVEREREKEREEGEKQREKEREGG-------E 322
ER+G + G+KDRE GR+ + E + ER+ ++R+ GE+ RE+ER+GG +
Sbjct 282 ERDGGRQRDGEKDRERERDGGRQRDGERQRERDGGRQRD-GERDRERERDGGRQRDGDRD 340
Query 323 DERERGGEREKEKEKERESEKEGEKEKEGEKEEEEGREREKEEEREKEKERESEEEREKG 382
E+ER G R+++ E+++E E++G ++++GE++ RERE++ R+++ R+ + ER+ G
Sbjct 341 REQERDGGRQRDVERDQERERDGGRQRDGERD----RERERDGGRQRDGVRDRKRERDGG 396
Query 383 K 383
+
Sbjct 397 R 397
> pfa:PF14_0419 conserved Plasmodium protein, unknown function
Length=7231
Score = 42.7 bits (99), Expect = 0.002, Method: Composition-based stats.
Identities = 28/108 (25%), Positives = 50/108 (46%), Gaps = 3/108 (2%)
Query 292 EGEAETEVEREREKEREEGEKQREKEREGGEDERERGGEREKEKEKERESEKEGEKEKEG 351
E + E ++ EKE +K EK G E E+ E+ EK ++ +EK EK E
Sbjct 3398 EIDLEGTIKTYNEKEGNTNDKLNEK---GNEKSNEKSNEKSNEKINDKSNEKINEKSNEK 3454
Query 352 EKEEEEGREREKEEEREKEKERESEEEREKGKERESEKEREEGNEGEG 399
E+ + +K + + +S+++ E KE+ KE + + +G
Sbjct 3455 SNEKSNDKRVDKYRGDKGNEYIKSDQQTENQKEKMILKEEKISGKSKG 3502
Score = 33.9 bits (76), Expect = 1.3, Method: Composition-based stats.
Identities = 22/70 (31%), Positives = 35/70 (50%), Gaps = 3/70 (4%)
Query 353 KEEEEGREREKEEEREKEKERESEEE-REKGKERESEKEREEGNE--GEGEGEREEEKER 409
+EE E + E + EKE + ++ EKG E+ +EK E+ NE + E+ EK
Sbjct 3393 REETEEIDLEGTIKTYNEKEGNTNDKLNEKGNEKSNEKSNEKSNEKINDKSNEKINEKSN 3452
Query 410 ESDRERVGEK 419
E E+ +K
Sbjct 3453 EKSNEKSNDK 3462
Score = 31.6 bits (70), Expect = 5.8, Method: Composition-based stats.
Identities = 23/81 (28%), Positives = 39/81 (48%), Gaps = 12/81 (14%)
Query 348 EKEGEKE---------EEEGREREKEEEREKEKERESEEEREKGKERESEKEREEGNEGE 398
E+ E + E+EG +K EK E+ +E+ EK E+ ++K E+ NE
Sbjct 3394 EETEEIDLEGTIKTYNEKEGNTNDKLN--EKGNEKSNEKSNEKSNEKINDKSNEKINEKS 3451
Query 399 GEGEREEEKERESDRERVGEK 419
E E+ ++ D+ R G+K
Sbjct 3452 NEKSNEKSNDKRVDKYR-GDK 3471
> dre:393572 traf3ip1, Elipsa, MGC63522, zgc:63522; TNF receptor-associated
factor 3 interacting protein 1
Length=629
Score = 40.8 bits (94), Expect = 0.011, Method: Compositional matrix adjust.
Identities = 46/132 (34%), Positives = 82/132 (62%), Gaps = 12/132 (9%)
Query 252 GDGVGDEDAKAEKSEAEDKGEREGESEEAGDKDREE------GAGREGEAETEVEREREK 305
GD + D+ K S ++DK REG +DREE +G + + + +++E
Sbjct 131 GDKL-DQKGKPSTSRSQDKENREGREHH---RDREERKGIKESSGSREQKDPDQPKDQES 186
Query 306 EREEGEKQREKEREGGEDERERGGEREKEKEKERESEKEGEKEKEGEKEEEEGRE--REK 363
+R++ +++R+ ER ERER +R+++K+K R+ EK+ +EKE E+E++ RE RE+
Sbjct 187 KRDDKDRRRDAERSDKGRERERTKDRDRDKDKSRDREKDKTREKEREREKDRNREKERER 246
Query 364 EEEREKEKERES 375
+++R+K+KERES
Sbjct 247 DKDRDKKKERES 258
> mmu:214111 Slc24a1, MGC27617; solute carrier family 24 (sodium/potassium/calcium
exchanger), member 1; K13749 solute carrier
family 24 (sodium/potassium/calcium exchanger), member
1
Length=1130
Score = 40.0 bits (92), Expect = 0.016, Method: Compositional matrix adjust.
Identities = 47/119 (39%), Positives = 66/119 (55%), Gaps = 2/119 (1%)
Query 265 SEAEDK-GEREGESEEAGDKDREEGAGREGEAETEVEREREKEREEGEKQREKEREGGED 323
+EAE K E+EGE+E G KD +EG E E + E E E E E +E E++ E E EG E+
Sbjct 768 TEAEGKEDEQEGETEAEGKKDEQEGE-TEAEGKEEQEGETEAEGKEDEQEGETEAEGKEE 826
Query 324 ERERGGEREKEKEKERESEKEGEKEKEGEKEEEEGREREKEEEREKEKERESEEEREKG 382
+ KE E+ERE+E EG+ + EG+ E + K+ E E E E + E +G
Sbjct 827 QEGETEAESKEVEQERETEAEGKDKHEGQGETQPDDTEVKDGEGETEANAEDQCEATQG 885
Score = 34.7 bits (78), Expect = 0.69, Method: Compositional matrix adjust.
Identities = 69/213 (32%), Positives = 101/213 (47%), Gaps = 25/213 (11%)
Query 229 PQEDSPGKDPQKRPGEKSEGDRDGDGVGDEDAKAEKSEAEDKGEREGESEEAGDKDREEG 288
P ED P + P D GD D + + EAE +GE GE E + +
Sbjct 678 PDEDEPAELPAVTVTPAPAPDAKGDQEDDPGCQEDVDEAERRGEMTGEEGEKETETEGKK 737
Query 289 AGREGEAETEVEREREKEREEGEKQR---EKEREGGEDERERGGERE---KEKEKERESE 342
+EGE E E + + ++E E E + E E EG EDE+E GE E K+ E+E E+E
Sbjct 738 DEQEGETEAERKEDEQEEETEAEGKEQEGETEAEGKEDEQE--GETEAEGKKDEQEGETE 795
Query 343 KEGEK-------------EKEGEKEEE---EGREREKEEEREKEKERESEEEREKGKERE 386
EG++ E+EGE E E E + E +E E+ERE+E E + E +
Sbjct 796 AEGKEEQEGETEAEGKEDEQEGETEAEGKEEQEGETEAESKEVEQERETEAEGKDKHEGQ 855
Query 387 SEKEREEGNEGEGEGEREEEKERESDRERVGEK 419
E + ++ +GEGE E E + + + GEK
Sbjct 856 GETQPDDTEVKDGEGETEANAEDQCEATQ-GEK 887
> pfa:MAL7P1.129 conserved Plasmodium protein, unknown function
Length=1003
Score = 38.5 bits (88), Expect = 0.045, Method: Compositional matrix adjust.
Identities = 39/90 (43%), Positives = 63/90 (70%), Gaps = 6/90 (6%)
Query 298 EVEREREKERE-EGEKQREKEREGGEDERERGGEREKEKEKERESEKEGEKEKEGEKEEE 356
++ER++EK+ E + EKQ E++RE + ER +REK+ EK+RE E + ++E+ E+ E
Sbjct 608 QIERDKEKQMERDKEKQMERDREK---QMER--DREKQMEKDREKEWDRDRERNRERHME 662
Query 357 EGREREKEEEREKEKERESEEEREKGKERE 386
+ REREKE +RE++ +R E+ER +ERE
Sbjct 663 KNREREKEMDRERDMDRNREKERNMDRERE 692
Score = 33.1 bits (74), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 33/88 (37%), Positives = 53/88 (60%), Gaps = 14/88 (15%)
Query 336 EKERESEKEGEKEKEGEKEEEEGREREKEEEREKEKERE--------SEEEREKGKERES 387
E+++E + E +KEK+ E++ E+ ER++E++ EK++E+E E EK +ERE
Sbjct 610 ERDKEKQMERDKEKQMERDREKQMERDREKQMEKDREKEWDRDRERNRERHMEKNREREK 669
Query 388 EKEREEGNEGEGEGEREEEKERESDRER 415
E +RE + +R EKER DRER
Sbjct 670 EMDRER------DMDRNREKERNMDRER 691
> pfa:PF11_0486 MAEBL, putative
Length=2055
Score = 35.0 bits (79), Expect = 0.49, Method: Composition-based stats.
Identities = 53/210 (25%), Positives = 89/210 (42%), Gaps = 8/210 (3%)
Query 189 DHNNPYFTELE--QLQSALETATSDWRQMKSQDGASGAPAGDPQEDSPGKDPQKRPGEK- 245
+ N P + E E Q++ +E T + + A G +E+S K+ KR +
Sbjct 1054 ERNKPSYKENEYDQMEKNVEDETYS-EEFGLFEEARKTETGRIEEESKKKEAMKRAEDAR 1112
Query 246 --SEGDRDGDGVGDEDAK-AEKSEAEDKGEREGESEEAGDKDREEGAGREGEAETEVERE 302
E R D E+A+ AE + + R ++ R E A R A +E
Sbjct 1113 RIEEARRAEDARRIEEARRAEDARRVEIARRVEDARRIEISRRAEDAKRIEAARRAIEVR 1172
Query 303 REKEREEGEKQREKEREGGEDERERGGEREKEKEKERESEKEGEK-EKEGEKEEEEGRER 361
R + R+ + +R + E+ER R E EK E+ K E+ K+ E+ + +ER
Sbjct 1173 RAELRKAEDARRIEAARRYENERRIEEARRYEDEKRIEAVKRAEEVRKDEEEAKRAEKER 1232
Query 362 EKEEEREKEKERESEEEREKGKERESEKER 391
EE R+ E+ R + R + + EK +
Sbjct 1233 NNEEIRKFEEARMAHFARRQAAIKAEEKRK 1262
Score = 32.0 bits (71), Expect = 4.4, Method: Composition-based stats.
Identities = 28/87 (32%), Positives = 38/87 (43%), Gaps = 2/87 (2%)
Query 330 EREKEKEKERESEKEGEKEKEGEKEEEEGREREKEEEREKEKERESEEEREKGKERESEK 389
ER K KE E ++ EK E E EE E+ + E + E +++E K E +
Sbjct 1054 ERNKPSYKENEYDQM-EKNVEDETYSEEFGLFEEARKTETGRIEEESKKKEAMKRAEDAR 1112
Query 390 EREEGNEGEGEGEREEEKERESDRERV 416
EE E + R EE R D RV
Sbjct 1113 RIEEARRAE-DARRIEEARRAEDARRV 1138
> bbo:BBOV_I002740 19.m02242; 200 kDa antigen p200
Length=1023
Score = 32.3 bits (72), Expect = 3.7, Method: Compositional matrix adjust.
Identities = 54/145 (37%), Positives = 88/145 (60%), Gaps = 10/145 (6%)
Query 262 AEKSEAEDKGEREGESEEAGDKDREEGAGREGEAETEVEREREKEREEGEKQREKEREGG 321
AE+ E + ER+ + EA K +E A R+ + E E ER+R+ EE E +R K +E
Sbjct 575 AERKRQEAEAERKRQEAEAERKRQEAEAERKRQEEAEAERKRQ---EEAEAER-KRQEEA 630
Query 322 EDERERGGEREKEKEKERESEKEGEKEKEGEKEEEEGREREKEEEREKEKERESEEEREK 381
E ER+R E E E++++ E+E E ++++E E ER+++EE E E++R+ E E E+
Sbjct 631 EAERKRQEEAEAERKRQEEAEAERKRQEEAEA------ERKRQEEAEAERKRQEEAEAER 684
Query 382 GKERESEKEREEGNEGEGEGEREEE 406
++ E+E ER+ E E E +R+EE
Sbjct 685 KRQEEAEAERKRQEEAEAERKRQEE 709
Score = 30.8 bits (68), Expect = 9.5, Method: Compositional matrix adjust.
Identities = 45/125 (36%), Positives = 80/125 (64%), Gaps = 9/125 (7%)
Query 299 VEREREKEREEGEKQR-----EKEREGGEDERERGGEREKEKEKERESEKEGEKEKEGE- 352
+E ER+++ E E++R E++R+ E ER+R E E E++++ E+E E ++++E E
Sbjct 573 LEAERKRQEAEAERKRQEAEAERKRQEAEAERKRQEEAEAERKRQEEAEAERKRQEEAEA 632
Query 353 ---KEEEEGREREKEEEREKEKERESEEEREKGKERESEKEREEGNEGEGEGEREEEKER 409
++EE ER+++EE E E++R+ E E E+ ++ E+E ER+ E E E +R+EE E
Sbjct 633 ERKRQEEAEAERKRQEEAEAERKRQEEAEAERKRQEEAEAERKRQEEAEAERKRQEEAEA 692
Query 410 ESDRE 414
E R+
Sbjct 693 ERKRQ 697
Lambda K H
0.303 0.127 0.342
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 19577847880
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40