bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_0016_orf2
Length=121
Score E
Sequences producing significant alignments: (Bits) Value
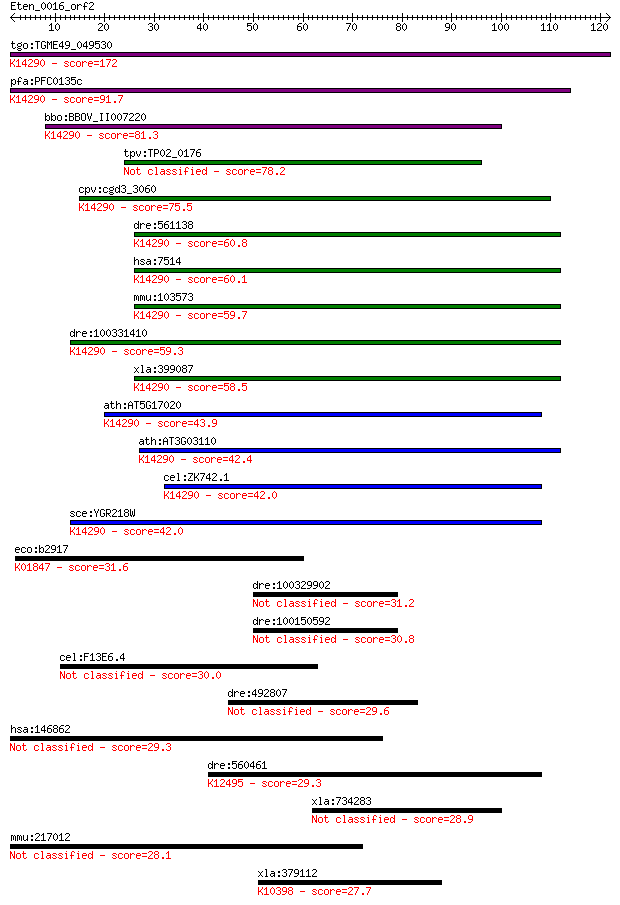
tgo:TGME49_049530 exportin, putative ; K14290 exportin-1 172 2e-43
pfa:PFC0135c exportin 1, putative; K14290 exportin-1 91.7 5e-19
bbo:BBOV_II007220 18.m06600; exportin 1; K14290 exportin-1 81.3 8e-16
tpv:TP02_0176 hypothetical protein 78.2 6e-15
cpv:cgd3_3060 exportin 1 ; K14290 exportin-1 75.5 4e-14
dre:561138 xpo1b, xpo1; exportin 1 (CRM1 homolog, yeast) b; K1... 60.8 1e-09
hsa:7514 XPO1, CRM1, DKFZp686B1823, emb; exportin 1 (CRM1 homo... 60.1 2e-09
mmu:103573 Xpo1, AA420417, Crm1, Exp1; exportin 1, CRM1 homolo... 59.7 2e-09
dre:100331410 exportin 1-like; K14290 exportin-1 59.3 3e-09
xla:399087 xpo1, crm1, exportin-1; exportin 1 (CRM1 homolog); ... 58.5 4e-09
ath:AT5G17020 XPO1A; XPO1A; protein binding / protein transpor... 43.9 1e-04
ath:AT3G03110 XPO1B; XPO1B; binding / protein transporter; K14... 42.4 4e-04
cel:ZK742.1 xpo-1; eXPOrtin (nuclear export receptor) family m... 42.0 4e-04
sce:YGR218W CRM1, KAP124, XPO1; Crm1p; K14290 exportin-1 42.0 5e-04
eco:b2917 scpA, ECK2913, JW2884, mcm, sbm, yliK; methylmalonyl... 31.6 0.64
dre:100329902 transposase domain-containing protein-like 31.2 0.91
dre:100150592 transposase domain-containing protein-like 30.8 0.96
cel:F13E6.4 hypothetical protein 30.0 2.1
dre:492807 cdadc1, zgc:101622; cytidine and dCMP deaminase dom... 29.6 2.7
hsa:146862 UNC45B, CMYA4, FLJ38610, MGC119540, MGC119541, SMUN... 29.3 3.0
dre:560461 cytohesin 2-like; K12495 IQ motif and SEC7 domain-c... 29.3 3.5
xla:734283 cep57l1, MGC84957, XCep57, c6orf182, cep57, cep57r;... 28.9 3.7
mmu:217012 Unc45b, AA445617, Cmya4, D230041A13Rik, MGC91090, U... 28.1 7.1
xla:379112 kif11-a, Cos2, Costal2, MGC52588, XLEg5K2, eg5, hks... 27.7 10.0
> tgo:TGME49_049530 exportin, putative ; K14290 exportin-1
Length=1125
Score = 172 bits (437), Expect = 2e-43, Method: Compositional matrix adjust.
Identities = 81/121 (66%), Positives = 102/121 (84%), Gaps = 0/121 (0%)
Query 1 RIVAFRLVENEQQGLTQENVTKSLIDLLCRSFQTLNPKQVEAFVLDLFNFCVDSNPSRFQ 60
RIV F +V+N GLT+ENV +SLIDLL RSFQT+N KQVEAFV+DLFN+C D P+RFQ
Sbjct 1004 RIVEFGVVQNPGNGLTRENVMQSLIDLLSRSFQTVNQKQVEAFVVDLFNYCRDPKPTRFQ 1063
Query 61 EHMRDFLISLKEFAGDNEALFEAERKEALARAAELEKQKRGMVPGLLPQYESMVSIRNME 120
+HMRDFLISLKEFAGDN+ LFEAER+EALARA EL++Q+R VPG++ QY++ V++R +
Sbjct 1064 QHMRDFLISLKEFAGDNDPLFEAEREEALARARELDRQRRMQVPGMIEQYDTTVTVRGGD 1123
Query 121 D 121
D
Sbjct 1124 D 1124
> pfa:PFC0135c exportin 1, putative; K14290 exportin-1
Length=1254
Score = 91.7 bits (226), Expect = 5e-19, Method: Compositional matrix adjust.
Identities = 48/117 (41%), Positives = 77/117 (65%), Gaps = 5/117 (4%)
Query 1 RIVAFRLVENEQQGLTQENVTKSLIDLLCRSFQTLNPKQVEAFVLDLFNFCVDSNPSRFQ 60
R++ F +V + +T+ ++ K + + L +SF+ LN KQ+E F +D+FNFCV+S PS F+
Sbjct 1129 RLLEFEVVNIPEVEITKPHIIKHVQNFLTQSFENLNQKQIETFSVDMFNFCVES-PSAFR 1187
Query 61 EHMRDFLISLKEFAGDNEALFEAERKEALARAAELEKQK----RGMVPGLLPQYESM 113
+RD LISLKEFA + + L+EA+R+EAL RA E K RG++ +P + ++
Sbjct 1188 SFVRDLLISLKEFATNQDELYEADRQEALQRAKMAEDNKLIKLRGLMKEDVPSFSAI 1244
> bbo:BBOV_II007220 18.m06600; exportin 1; K14290 exportin-1
Length=1186
Score = 81.3 bits (199), Expect = 8e-16, Method: Compositional matrix adjust.
Identities = 48/113 (42%), Positives = 65/113 (57%), Gaps = 21/113 (18%)
Query 8 VENEQQGLTQENVTKSLIDLLCRSFQTLNPKQVEAFVLDLFNFC---------------- 51
V + LT+ +V K L++LL SF TLN KQVEAFV+DLFNF
Sbjct 1057 VNDPSSELTRVHVMKFLVELLGNSFITLNVKQVEAFVVDLFNFAGETIAEQNESMMSSGL 1116
Query 52 --VDSNPSRFQEHMRDFLISLKEFAGDNEA---LFEAERKEALARAAELEKQK 99
P RFQ H++DFL+SLKEFAG + +FE +R+ A+ RA +E++K
Sbjct 1117 SITSGQPMRFQTHVKDFLLSLKEFAGSGDEFDRIFEQDRQNAIERARAIEQRK 1169
> tpv:TP02_0176 hypothetical protein
Length=139
Score = 78.2 bits (191), Expect = 6e-15, Method: Compositional matrix adjust.
Identities = 49/119 (41%), Positives = 62/119 (52%), Gaps = 47/119 (39%)
Query 24 LIDLLCRSFQTLNPKQVEAFVLDLFNFCVDS-----------------------NPS--- 57
L+DLL RSFQ+LN KQ+EAFV+DLFN+ VD+ NP+
Sbjct 4 LVDLLTRSFQSLNNKQIEAFVVDLFNYSVDTTAPSGMGATQFFESDYNYVNVYENPTISG 63
Query 58 ------------------RFQEHMRDFLISLKEFAGDNE---ALFEAERKEALARAAEL 95
RFQ H+RDFL+SLKEFAG E A+FE +R EA+ RA +L
Sbjct 64 VLNTNVKEEKEKDKDKDIRFQTHVRDFLLSLKEFAGCTEEFQAIFEKDRDEAIERARQL 122
> cpv:cgd3_3060 exportin 1 ; K14290 exportin-1
Length=1266
Score = 75.5 bits (184), Expect = 4e-14, Method: Composition-based stats.
Identities = 41/96 (42%), Positives = 61/96 (63%), Gaps = 1/96 (1%)
Query 15 LTQENVTKSLIDLLCRSFQTLNPKQVEAFVLDLFNFCVDSNPSRFQEHMRDFLISLKEFA 74
+++ V + + DLL +SF T+ +QVE FVL+LFN S FQ + DFLI +KEF
Sbjct 1148 ISKVGVMEYIADLLIKSFITVQKEQVEVFVLELFNSVHSKTISDFQRLVHDFLIQIKEFT 1207
Query 75 G-DNEALFEAERKEALARAAELEKQKRGMVPGLLPQ 109
+++ +FE E+ AL RA E+E K+ M+PGL+ Q
Sbjct 1208 NEESKQMFEIEKSIALKRAIEIENSKQWMIPGLINQ 1243
> dre:561138 xpo1b, xpo1; exportin 1 (CRM1 homolog, yeast) b;
K14290 exportin-1
Length=1071
Score = 60.8 bits (146), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 36/87 (41%), Positives = 55/87 (63%), Gaps = 3/87 (3%)
Query 26 DLLCRSFQTLNPKQVEAFVLDLFNFCVDSNPSRFQEHMRDFLISLKEFAGDNEALFEAER 85
+LL +F L QV+ FV LF+ ++ + + F+EH+RDFL+ +KEFAG++ + E
Sbjct 980 NLLKSAFPHLQDAQVKVFVTGLFS--LNQDIAAFKEHLRDFLVQIKEFAGEDTSDLFLEE 1037
Query 86 KEALARAAELEKQKRGM-VPGLLPQYE 111
+EA R A+ EK K M VPG+L +E
Sbjct 1038 REASLRQAQEEKHKLQMSVPGILNPHE 1064
> hsa:7514 XPO1, CRM1, DKFZp686B1823, emb; exportin 1 (CRM1 homolog,
yeast); K14290 exportin-1
Length=1071
Score = 60.1 bits (144), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 40/88 (45%), Positives = 55/88 (62%), Gaps = 5/88 (5%)
Query 26 DLLCRSFQTLNPKQVEAFVLDLFNFCVDSNPSRFQEHMRDFLISLKEFAG-DNEALFEAE 84
+LL +F L QV+ FV LF+ ++ + F+EH+RDFL+ +KEFAG D LF E
Sbjct 980 NLLKSAFPHLQDAQVKLFVTGLFS--LNQDIPAFKEHLRDFLVQIKEFAGEDTSDLFLEE 1037
Query 85 RKEALARAAELEKQKRGM-VPGLLPQYE 111
R+ AL R A+ EK KR M VPG+ +E
Sbjct 1038 REIAL-RQADEEKHKRQMSVPGIFNPHE 1064
> mmu:103573 Xpo1, AA420417, Crm1, Exp1; exportin 1, CRM1 homolog
(yeast); K14290 exportin-1
Length=1071
Score = 59.7 bits (143), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 40/88 (45%), Positives = 55/88 (62%), Gaps = 5/88 (5%)
Query 26 DLLCRSFQTLNPKQVEAFVLDLFNFCVDSNPSRFQEHMRDFLISLKEFAG-DNEALFEAE 84
+LL +F L QV+ FV LF+ ++ + F+EH+RDFL+ +KEFAG D LF E
Sbjct 980 NLLKSAFPHLQDAQVKLFVTGLFS--LNQDIPAFKEHLRDFLVQIKEFAGEDTSDLFLEE 1037
Query 85 RKEALARAAELEKQKRGM-VPGLLPQYE 111
R+ AL +A E EK K M VPG+L +E
Sbjct 1038 RETALRQAQE-EKHKLQMSVPGILNPHE 1064
> dre:100331410 exportin 1-like; K14290 exportin-1
Length=131
Score = 59.3 bits (142), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 39/100 (39%), Positives = 58/100 (58%), Gaps = 7/100 (7%)
Query 13 QGLTQENVTKSLIDLLCRSFQTLNPKQVEAFVLDLFNFCVDSNPSRFQEHMRDFLISLKE 72
QG Q+ V +LL +F L QV+ FV LF+ ++ + F+EH+RDFL+ +KE
Sbjct 31 QGYVQDYVA----NLLKTAFPHLQDAQVKVFVTGLFS--LNQDIPAFKEHLRDFLVQIKE 84
Query 73 FAGDNEALFEAERKEALARAAELEKQKRGM-VPGLLPQYE 111
FAG++ E +EA R A+ EK K + VPG+L +E
Sbjct 85 FAGEDSTDLFLEEREASLRQAQEEKHKIQLSVPGILNPHE 124
> xla:399087 xpo1, crm1, exportin-1; exportin 1 (CRM1 homolog);
K14290 exportin-1
Length=1071
Score = 58.5 bits (140), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 38/88 (43%), Positives = 56/88 (63%), Gaps = 5/88 (5%)
Query 26 DLLCRSFQTLNPKQVEAFVLDLFNFCVDSNPSRFQEHMRDFLISLKEFAG-DNEALFEAE 84
+LL +F L QV+ FV LF+ ++ + + F+EH+RDFL+ +KE+AG D LF E
Sbjct 980 NLLKSAFPHLQDAQVKLFVTGLFS--LNQDIAAFKEHLRDFLVQIKEYAGEDTSDLFLEE 1037
Query 85 RKEALARAAELEKQKRGM-VPGLLPQYE 111
R+ +L +A E EK K M VPG+L +E
Sbjct 1038 RESSLRQAQE-EKHKLQMSVPGILNPHE 1064
> ath:AT5G17020 XPO1A; XPO1A; protein binding / protein transporter/
receptor; K14290 exportin-1
Length=1075
Score = 43.9 bits (102), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 32/89 (35%), Positives = 48/89 (53%), Gaps = 7/89 (7%)
Query 20 VTKSLIDLLCRSFQTLNPKQVEAFVLDLFNFCVDSNPSRFQEHMRDFLISLKEF-AGDNE 78
V + I LL SF + +V FV L+ ++PS F+ ++RDFL+ KEF A DN+
Sbjct 981 VREYTIKLLSSSFPNMTAAEVTQFVNGLYE--SRNDPSGFKNNIRDFLVQSKEFSAQDNK 1038
Query 79 ALFEAERKEALARAAELEKQKRGMVPGLL 107
L+ E A E E+Q+ +PGL+
Sbjct 1039 DLYAEEA----AAQRERERQRMLSIPGLI 1063
> ath:AT3G03110 XPO1B; XPO1B; binding / protein transporter; K14290
exportin-1
Length=1076
Score = 42.4 bits (98), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 31/88 (35%), Positives = 50/88 (56%), Gaps = 11/88 (12%)
Query 27 LLCRSFQTLNPKQVEAFVLDLFNFCVDSNPSRFQEHMRDFLISLKEF-AGDNEALFEAER 85
LL SF + +V FV L+ ++ RF++++RDFLI KEF A DN+ L+ E
Sbjct 989 LLSSSFPNMTTTEVTQFVNGLYE--SRNDVGRFKDNIRDFLIQSKEFSAQDNKDLYAEE- 1045
Query 86 KEALARAAELEKQKRGM--VPGLLPQYE 111
AA++E++++ M +PGL+ E
Sbjct 1046 -----AAAQMERERQRMLSIPGLIAPSE 1068
> cel:ZK742.1 xpo-1; eXPOrtin (nuclear export receptor) family
member (xpo-1); K14290 exportin-1
Length=1080
Score = 42.0 bits (97), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 24/76 (31%), Positives = 42/76 (55%), Gaps = 5/76 (6%)
Query 32 FQTLNPKQVEAFVLDLFNFCVDSNPSRFQEHMRDFLISLKEFAGDNEALFEAERKEALAR 91
F +N Q+ + F+F ++ S + H+RDFLI +KE G++ + E +EA +
Sbjct 999 FDNMNQDQIRIIIKGFFSF--NTEISSMRNHLRDFLIQIKEHNGEDTSDLYLEEREAEIQ 1056
Query 92 AAELEKQKRGMVPGLL 107
A+ Q++ VPG+L
Sbjct 1057 QAQ---QRKRDVPGIL 1069
> sce:YGR218W CRM1, KAP124, XPO1; Crm1p; K14290 exportin-1
Length=1084
Score = 42.0 bits (97), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 30/97 (30%), Positives = 52/97 (53%), Gaps = 4/97 (4%)
Query 13 QGLT-QENVTKSLIDLLCRSFQTLNPKQVEAFVLDLFNFCVDSNPSRFQEHMRDFLISLK 71
QG + Q +++ L ++L +F L +Q+ +F+ L D F+ +RDFL+ +K
Sbjct 983 QGTSNQVYLSQYLANMLSNAFPHLTSEQIASFLSALTKQYKDL--VVFKGTLRDFLVQIK 1040
Query 72 EFAGD-NEALFEAERKEALARAAELEKQKRGMVPGLL 107
E GD + LF +++ AL LE++K + GLL
Sbjct 1041 EVGGDPTDYLFAEDKENALMEQNRLEREKAAKIGGLL 1077
> eco:b2917 scpA, ECK2913, JW2884, mcm, sbm, yliK; methylmalonyl-CoA
mutase (EC:5.4.99.2); K01847 methylmalonyl-CoA mutase
[EC:5.4.99.2]
Length=714
Score = 31.6 bits (70), Expect = 0.64, Method: Composition-based stats.
Identities = 19/63 (30%), Positives = 33/63 (52%), Gaps = 6/63 (9%)
Query 2 IVAFRLVENEQQGLTQENVTKS-----LIDLLCRSFQTLNPKQVEAFVLDLFNFCVDSNP 56
++AF +V E+QG+T + +T + L + LCR+ PK + D+ +C + P
Sbjct 163 VLAFYIVAAEEQGVTPDKLTGTIQNDILKEYLCRNTYIYPPKPSMRIIADIIAWCSGNMP 222
Query 57 SRF 59
RF
Sbjct 223 -RF 224
> dre:100329902 transposase domain-containing protein-like
Length=695
Score = 31.2 bits (69), Expect = 0.91, Method: Composition-based stats.
Identities = 12/29 (41%), Positives = 17/29 (58%), Gaps = 0/29 (0%)
Query 50 FCVDSNPSRFQEHMRDFLISLKEFAGDNE 78
FC DS PS E+++DF+ +KE E
Sbjct 218 FCGDSKPSSLDEYLKDFISEIKELGNGFE 246
> dre:100150592 transposase domain-containing protein-like
Length=811
Score = 30.8 bits (68), Expect = 0.96, Method: Composition-based stats.
Identities = 12/29 (41%), Positives = 17/29 (58%), Gaps = 0/29 (0%)
Query 50 FCVDSNPSRFQEHMRDFLISLKEFAGDNE 78
FC DS PS E+++DF+ +KE E
Sbjct 218 FCGDSKPSSLDEYLKDFISEIKELGNGFE 246
> cel:F13E6.4 hypothetical protein
Length=442
Score = 30.0 bits (66), Expect = 2.1, Method: Composition-based stats.
Identities = 21/55 (38%), Positives = 32/55 (58%), Gaps = 5/55 (9%)
Query 11 EQQGLTQENVTKSL-IDLLCRSFQTLNPKQVEAFVLDL-FNFCVDSNPSR-FQEH 62
+QQG+ Q+N KSL +D + R F T P+ VE + + C DS+ R F++H
Sbjct 172 QQQGMMQQNREKSLSLDPMRRPFMT--PQDVEQLPMPQGWEMCYDSDGVRYFKDH 224
> dre:492807 cdadc1, zgc:101622; cytidine and dCMP deaminase domain
containing 1
Length=471
Score = 29.6 bits (65), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 14/38 (36%), Positives = 21/38 (55%), Gaps = 0/38 (0%)
Query 45 LDLFNFCVDSNPSRFQEHMRDFLISLKEFAGDNEALFE 82
+ L NFC++ N S ++HMR+ + L A AL E
Sbjct 255 MGLENFCMEPNFSNLRQHMRNLIRILASVASSVPALLE 292
> hsa:146862 UNC45B, CMYA4, FLJ38610, MGC119540, MGC119541, SMUNC45,
UNC45; unc-45 homolog B (C. elegans)
Length=929
Score = 29.3 bits (64), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 22/75 (29%), Positives = 37/75 (49%), Gaps = 6/75 (8%)
Query 1 RIVAFRLVENEQQGLTQENVTKSLIDLLCRSFQTLNPKQVEAFVLDLFNFCVDSNPSRFQ 60
RI + VENE+ L N+ +++ID L + + + EA VLD + +
Sbjct 225 RICSLMAVENEEMSLAVCNLLQAIIDSLSGEDKREHRGKEEALVLD-----TKKDLKQIT 279
Query 61 EHMRDFLISLKEFAG 75
H+ D L+S K+ +G
Sbjct 280 SHLLDMLVS-KKVSG 293
> dre:560461 cytohesin 2-like; K12495 IQ motif and SEC7 domain-containing
protein
Length=1431
Score = 29.3 bits (64), Expect = 3.5, Method: Composition-based stats.
Identities = 22/67 (32%), Positives = 30/67 (44%), Gaps = 13/67 (19%)
Query 41 EAFVLDLFNFCVDSNPSRFQEHMRDFLISLKEFAGDNEALFEAERKEALARAAELEKQKR 100
E VL +FN + +RF +R E++ E + E +ELEKQK
Sbjct 913 ERKVLIVFNAPSQQDRTRFTSDLR-------------ESIAEVQDMEKYRVESELEKQKG 959
Query 101 GMVPGLL 107
M PGLL
Sbjct 960 VMRPGLL 966
> xla:734283 cep57l1, MGC84957, XCep57, c6orf182, cep57, cep57r;
centrosomal protein 57kDa-like 1
Length=488
Score = 28.9 bits (63), Expect = 3.7, Method: Compositional matrix adjust.
Identities = 14/38 (36%), Positives = 22/38 (57%), Gaps = 0/38 (0%)
Query 62 HMRDFLISLKEFAGDNEALFEAERKEALARAAELEKQK 99
H RD L +L AG+++ + E+E++ A A E QK
Sbjct 77 HARDRLTNLSRAAGEHKKVLESEKRSAEWAAQEATSQK 114
> mmu:217012 Unc45b, AA445617, Cmya4, D230041A13Rik, MGC91090,
Unc45; unc-45 homolog B (C. elegans)
Length=929
Score = 28.1 bits (61), Expect = 7.1, Method: Compositional matrix adjust.
Identities = 20/71 (28%), Positives = 34/71 (47%), Gaps = 5/71 (7%)
Query 1 RIVAFRLVENEQQGLTQENVTKSLIDLLCRSFQTLNPKQVEAFVLDLFNFCVDSNPSRFQ 60
RI + +ENE+ L N+ +++ID L + + + EA VLD + +
Sbjct 225 RICSLMALENEEMSLAVCNLLQAIIDSLSGEDKREHRGKEEALVLD-----TKKDLKQIT 279
Query 61 EHMRDFLISLK 71
H+ D L+S K
Sbjct 280 SHLLDMLVSKK 290
> xla:379112 kif11-a, Cos2, Costal2, MGC52588, XLEg5K2, eg5, hksp,
kif11, kif11a, knsl1, trip5; kinesin family member 11;
K10398 kinesin family member 11
Length=1067
Score = 27.7 bits (60), Expect = 10.0, Method: Compositional matrix adjust.
Identities = 10/37 (27%), Positives = 20/37 (54%), Gaps = 0/37 (0%)
Query 51 CVDSNPSRFQEHMRDFLISLKEFAGDNEALFEAERKE 87
C+ S+ S+F E + + +++ AG N + E K+
Sbjct 775 CISSHHSKFTEQSQAVAVEIRQLAGSNMSTLEESSKQ 811
Lambda K H
0.320 0.135 0.372
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2013067560
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40