bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_0014_orf1
Length=154
Score E
Sequences producing significant alignments: (Bits) Value
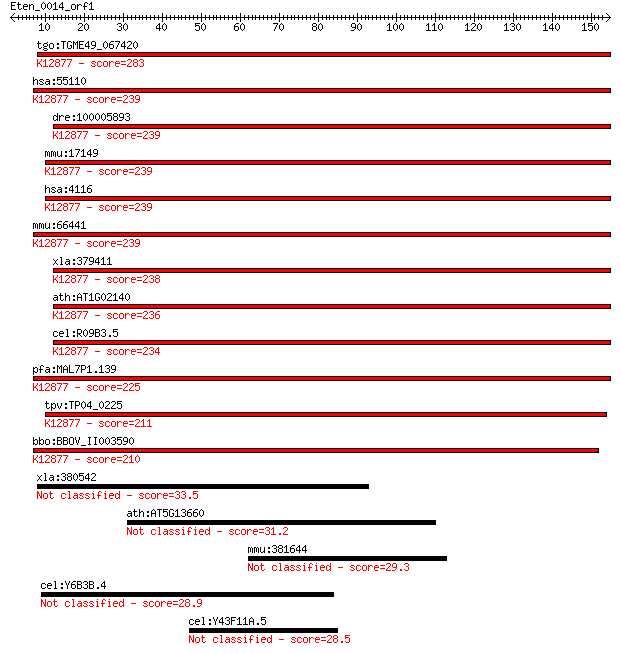
tgo:TGME49_067420 mago nashi protein, putative ; K12877 protei... 283 2e-76
hsa:55110 MAGOHB, FLJ10292, MGN2, mago, magoh; mago-nashi homo... 239 2e-63
dre:100005893 magoh, MGC112220, zgc:112220; mago-nashi homolog... 239 3e-63
mmu:17149 Magoh, Mago-m, Mos2; mago-nashi homolog, proliferati... 239 3e-63
hsa:4116 MAGOH, MAGOH1, MAGOHA; mago-nashi homolog, proliferat... 239 3e-63
mmu:66441 Magohb, 2010012C16Rik, Mago, Magoh, Magoh-rs1; mago-... 239 3e-63
xla:379411 magoh, MGC64274, mago; mago-nashi homolog, prolifer... 238 6e-63
ath:AT1G02140 MAGO; MAGO (MAGO NASHI); protein binding; K12877... 236 2e-62
cel:R09B3.5 mag-1; Drosophila MAGonashi homolog family member ... 234 1e-61
pfa:MAL7P1.139 mago nashi protein homolog, putative; K12877 pr... 225 4e-59
tpv:TP04_0225 mago nashi protein; K12877 protein mago nashi 211 5e-55
bbo:BBOV_II003590 18.m09966; mago nashi protein; K12877 protei... 210 2e-54
xla:380542 tmem168, MGC53845, flj13576; transmembrane protein 168 33.5
ath:AT5G13660 hypothetical protein 31.2 1.5
mmu:381644 Cep135, BC062951, Cep4, Gm1044, MGC86039; centrosom... 29.3 5.2
cel:Y6B3B.4 hypothetical protein 28.9 5.9
cel:Y43F11A.5 set-24; SET (trithorax/polycomb) domain containi... 28.5 9.9
> tgo:TGME49_067420 mago nashi protein, putative ; K12877 protein
mago nashi
Length=150
Score = 283 bits (723), Expect = 2e-76, Method: Compositional matrix adjust.
Identities = 130/147 (88%), Positives = 143/147 (97%), Gaps = 0/147 (0%)
Query 8 GDDDDFYLRYYTGHKGKFGHEFLEFEFLAEGRMRYANNSNYKNDTMIRKEAYVSQAVIKE 67
G+++DFYLRYY GHKGKFGHEFLEFEF EGR+RYANNSNYKNDTMIRKEAYVSQAV+KE
Sbjct 4 GEEEDFYLRYYVGHKGKFGHEFLEFEFRPEGRLRYANNSNYKNDTMIRKEAYVSQAVMKE 63
Query 68 LRRIIEEAEILREDDNNWPAPDRVGRQELEIILGKDHISFTTSKIGSMADVQRCKDQEGL 127
LRRI+EE+EI++EDDNNWPAPDRVGRQELEI+LGKDHISFTTSKIGSMADVQR KDQ+GL
Sbjct 64 LRRIVEESEIIKEDDNNWPAPDRVGRQELEIVLGKDHISFTTSKIGSMADVQRSKDQDGL 123
Query 128 RVFYYLVQDLKCFVYSLIGLHFRIKPV 154
RVFYYLVQDLKCF++SLIGLHFRIKPV
Sbjct 124 RVFYYLVQDLKCFIFSLIGLHFRIKPV 150
> hsa:55110 MAGOHB, FLJ10292, MGN2, mago, magoh; mago-nashi homolog
B (Drosophila); K12877 protein mago nashi
Length=148
Score = 239 bits (611), Expect = 2e-63, Method: Compositional matrix adjust.
Identities = 109/148 (73%), Positives = 129/148 (87%), Gaps = 0/148 (0%)
Query 7 MGDDDDFYLRYYTGHKGKFGHEFLEFEFLAEGRMRYANNSNYKNDTMIRKEAYVSQAVIK 66
M DFYLRYY GHKGKFGHEFLEFEF +G++RYANNSNYKND MIRKEAYV ++V++
Sbjct 1 MAVASDFYLRYYVGHKGKFGHEFLEFEFRPDGKLRYANNSNYKNDVMIRKEAYVHKSVME 60
Query 67 ELRRIIEEAEILREDDNNWPAPDRVGRQELEIILGKDHISFTTSKIGSMADVQRCKDQEG 126
EL+RII+++EI +EDD WP PDRVGRQELEI++G +HISFTTSKIGS+ DV + KD EG
Sbjct 61 ELKRIIDDSEITKEDDALWPPPDRVGRQELEIVIGDEHISFTTSKIGSLIDVNQSKDPEG 120
Query 127 LRVFYYLVQDLKCFVYSLIGLHFRIKPV 154
LRVFYYLVQDLKC V+SLIGLHF+IKP+
Sbjct 121 LRVFYYLVQDLKCLVFSLIGLHFKIKPI 148
> dre:100005893 magoh, MGC112220, zgc:112220; mago-nashi homolog,
proliferation-associated (Drosophila); K12877 protein mago
nashi
Length=147
Score = 239 bits (609), Expect = 3e-63, Method: Compositional matrix adjust.
Identities = 108/143 (75%), Positives = 128/143 (89%), Gaps = 0/143 (0%)
Query 12 DFYLRYYTGHKGKFGHEFLEFEFLAEGRMRYANNSNYKNDTMIRKEAYVSQAVIKELRRI 71
DFYLRYY GHKGKFGHEFLEFEF +G++RYANNSNYKND MIRKEAYV ++V++EL+RI
Sbjct 5 DFYLRYYVGHKGKFGHEFLEFEFRPDGKLRYANNSNYKNDVMIRKEAYVHKSVMEELKRI 64
Query 72 IEEAEILREDDNNWPAPDRVGRQELEIILGKDHISFTTSKIGSMADVQRCKDQEGLRVFY 131
I+++EI +EDD WP PDRVGRQELEI++G +HISFTTSKIGS+ DV + KD EGLRVFY
Sbjct 65 IDDSEITKEDDALWPPPDRVGRQELEIVIGDEHISFTTSKIGSLIDVNQSKDPEGLRVFY 124
Query 132 YLVQDLKCFVYSLIGLHFRIKPV 154
YLVQDLKC V+SLIGLHF+IKP+
Sbjct 125 YLVQDLKCLVFSLIGLHFKIKPI 147
> mmu:17149 Magoh, Mago-m, Mos2; mago-nashi homolog, proliferation-associated
(Drosophila); K12877 protein mago nashi
Length=146
Score = 239 bits (609), Expect = 3e-63, Method: Compositional matrix adjust.
Identities = 108/145 (74%), Positives = 129/145 (88%), Gaps = 0/145 (0%)
Query 10 DDDFYLRYYTGHKGKFGHEFLEFEFLAEGRMRYANNSNYKNDTMIRKEAYVSQAVIKELR 69
+ DFYLRYY GHKGKFGHEFLEFEF +G++RYANNSNYKND MIRKEAYV ++V++EL+
Sbjct 2 ESDFYLRYYVGHKGKFGHEFLEFEFRPDGKLRYANNSNYKNDVMIRKEAYVHKSVMEELK 61
Query 70 RIIEEAEILREDDNNWPAPDRVGRQELEIILGKDHISFTTSKIGSMADVQRCKDQEGLRV 129
RII+++EI +EDD WP PDRVGRQELEI++G +HISFTTSKIGS+ DV + KD EGLRV
Sbjct 62 RIIDDSEITKEDDALWPPPDRVGRQELEIVIGDEHISFTTSKIGSLIDVNQSKDPEGLRV 121
Query 130 FYYLVQDLKCFVYSLIGLHFRIKPV 154
FYYLVQDLKC V+SLIGLHF+IKP+
Sbjct 122 FYYLVQDLKCLVFSLIGLHFKIKPI 146
> hsa:4116 MAGOH, MAGOH1, MAGOHA; mago-nashi homolog, proliferation-associated
(Drosophila); K12877 protein mago nashi
Length=146
Score = 239 bits (609), Expect = 3e-63, Method: Compositional matrix adjust.
Identities = 108/145 (74%), Positives = 129/145 (88%), Gaps = 0/145 (0%)
Query 10 DDDFYLRYYTGHKGKFGHEFLEFEFLAEGRMRYANNSNYKNDTMIRKEAYVSQAVIKELR 69
+ DFYLRYY GHKGKFGHEFLEFEF +G++RYANNSNYKND MIRKEAYV ++V++EL+
Sbjct 2 ESDFYLRYYVGHKGKFGHEFLEFEFRPDGKLRYANNSNYKNDVMIRKEAYVHKSVMEELK 61
Query 70 RIIEEAEILREDDNNWPAPDRVGRQELEIILGKDHISFTTSKIGSMADVQRCKDQEGLRV 129
RII+++EI +EDD WP PDRVGRQELEI++G +HISFTTSKIGS+ DV + KD EGLRV
Sbjct 62 RIIDDSEITKEDDALWPPPDRVGRQELEIVIGDEHISFTTSKIGSLIDVNQSKDPEGLRV 121
Query 130 FYYLVQDLKCFVYSLIGLHFRIKPV 154
FYYLVQDLKC V+SLIGLHF+IKP+
Sbjct 122 FYYLVQDLKCLVFSLIGLHFKIKPI 146
> mmu:66441 Magohb, 2010012C16Rik, Mago, Magoh, Magoh-rs1; mago-nashi
homolog B (Drosophila); K12877 protein mago nashi
Length=148
Score = 239 bits (609), Expect = 3e-63, Method: Compositional matrix adjust.
Identities = 109/148 (73%), Positives = 129/148 (87%), Gaps = 0/148 (0%)
Query 7 MGDDDDFYLRYYTGHKGKFGHEFLEFEFLAEGRMRYANNSNYKNDTMIRKEAYVSQAVIK 66
M DFYLRYY GHKGKFGHEFLEFEF +G++RYANNSNYKND MIRKEAYV ++V++
Sbjct 1 MSMGSDFYLRYYVGHKGKFGHEFLEFEFRPDGKLRYANNSNYKNDVMIRKEAYVHKSVME 60
Query 67 ELRRIIEEAEILREDDNNWPAPDRVGRQELEIILGKDHISFTTSKIGSMADVQRCKDQEG 126
EL+RII+++EI +EDD WP PDRVGRQELEI++G +HISFTTSKIGS+ DV + KD EG
Sbjct 61 ELKRIIDDSEITKEDDALWPPPDRVGRQELEIVIGDEHISFTTSKIGSLIDVNQSKDPEG 120
Query 127 LRVFYYLVQDLKCFVYSLIGLHFRIKPV 154
LRVFYYLVQDLKC V+SLIGLHF+IKP+
Sbjct 121 LRVFYYLVQDLKCLVFSLIGLHFKIKPI 148
> xla:379411 magoh, MGC64274, mago; mago-nashi homolog, proliferation-associated;
K12877 protein mago nashi
Length=146
Score = 238 bits (607), Expect = 6e-63, Method: Compositional matrix adjust.
Identities = 107/143 (74%), Positives = 128/143 (89%), Gaps = 0/143 (0%)
Query 12 DFYLRYYTGHKGKFGHEFLEFEFLAEGRMRYANNSNYKNDTMIRKEAYVSQAVIKELRRI 71
DFYLRYY GHKGKFGHEFLEFEF +G++RYANNSNYKND MIRKEAYV ++V++EL+RI
Sbjct 4 DFYLRYYVGHKGKFGHEFLEFEFRPDGKLRYANNSNYKNDVMIRKEAYVHKSVMEELKRI 63
Query 72 IEEAEILREDDNNWPAPDRVGRQELEIILGKDHISFTTSKIGSMADVQRCKDQEGLRVFY 131
I+++E+ +EDD WP PDRVGRQELEI++G +HISFTTSKIGS+ DV + KD EGLRVFY
Sbjct 64 IDDSEVTKEDDALWPPPDRVGRQELEIVIGDEHISFTTSKIGSLIDVNQSKDPEGLRVFY 123
Query 132 YLVQDLKCFVYSLIGLHFRIKPV 154
YLVQDLKC V+SLIGLHF+IKP+
Sbjct 124 YLVQDLKCLVFSLIGLHFKIKPI 146
> ath:AT1G02140 MAGO; MAGO (MAGO NASHI); protein binding; K12877
protein mago nashi
Length=150
Score = 236 bits (602), Expect = 2e-62, Method: Compositional matrix adjust.
Identities = 104/143 (72%), Positives = 125/143 (87%), Gaps = 0/143 (0%)
Query 12 DFYLRYYTGHKGKFGHEFLEFEFLAEGRMRYANNSNYKNDTMIRKEAYVSQAVIKELRRI 71
+FYLRYY GHKGKFGHEFLEFEF +G++RYANNSNYKNDT+IRKE +++ AV+KE +RI
Sbjct 8 EFYLRYYVGHKGKFGHEFLEFEFREDGKLRYANNSNYKNDTIIRKEVFLTPAVLKECKRI 67
Query 72 IEEAEILREDDNNWPAPDRVGRQELEIILGKDHISFTTSKIGSMADVQRCKDQEGLRVFY 131
+ E+EIL+EDDNNWP PDRVG+QELEI+LG +HISF TSKIGS+ D Q D EGLR+FY
Sbjct 68 VSESEILKEDDNNWPEPDRVGKQELEIVLGNEHISFATSKIGSLVDCQSSNDPEGLRIFY 127
Query 132 YLVQDLKCFVYSLIGLHFRIKPV 154
YLVQDLKC V+SLI LHF+IKP+
Sbjct 128 YLVQDLKCLVFSLISLHFKIKPI 150
> cel:R09B3.5 mag-1; Drosophila MAGonashi homolog family member
(mag-1); K12877 protein mago nashi
Length=152
Score = 234 bits (596), Expect = 1e-61, Method: Compositional matrix adjust.
Identities = 104/143 (72%), Positives = 125/143 (87%), Gaps = 0/143 (0%)
Query 12 DFYLRYYTGHKGKFGHEFLEFEFLAEGRMRYANNSNYKNDTMIRKEAYVSQAVIKELRRI 71
DFY+RYY GHKGKFGHEFLEFEF G +RYANNSNYKNDTMIRKEA VS++V+ EL+RI
Sbjct 10 DFYVRYYVGHKGKFGHEFLEFEFRPNGSLRYANNSNYKNDTMIRKEATVSESVLSELKRI 69
Query 72 IEEAEILREDDNNWPAPDRVGRQELEIILGKDHISFTTSKIGSMADVQRCKDQEGLRVFY 131
IE++EI++EDD+NWP PD++GRQELEI+ +HISFTT KIG++ADV KD +GLR FY
Sbjct 70 IEDSEIMQEDDDNWPEPDKIGRQELEILYKNEHISFTTGKIGALADVNNSKDPDGLRSFY 129
Query 132 YLVQDLKCFVYSLIGLHFRIKPV 154
YLVQDLKC V+SLIGLHF+IKP+
Sbjct 130 YLVQDLKCLVFSLIGLHFKIKPI 152
> pfa:MAL7P1.139 mago nashi protein homolog, putative; K12877
protein mago nashi
Length=148
Score = 225 bits (573), Expect = 4e-59, Method: Compositional matrix adjust.
Identities = 104/148 (70%), Positives = 124/148 (83%), Gaps = 0/148 (0%)
Query 7 MGDDDDFYLRYYTGHKGKFGHEFLEFEFLAEGRMRYANNSNYKNDTMIRKEAYVSQAVIK 66
M D F RYY GH+GKFGHEFLEFEF ++GR+RYANNSNYKND +IRKEAYVS++V+K
Sbjct 1 MSKKDKFSFRYYVGHEGKFGHEFLEFEFNSKGRLRYANNSNYKNDKIIRKEAYVSKSVLK 60
Query 67 ELRRIIEEAEILREDDNNWPAPDRVGRQELEIILGKDHISFTTSKIGSMADVQRCKDQEG 126
EL+RIIEE+EI +E D WP PD++G+QELEI L FTTSKIGS++DV++C D EG
Sbjct 61 ELKRIIEESEICKESDKEWPMPDKIGKQELEIYLDGVEYYFTTSKIGSLSDVKQCSDPEG 120
Query 127 LRVFYYLVQDLKCFVYSLIGLHFRIKPV 154
LRVFYYLVQDLKCF++SLI LHFRIKPV
Sbjct 121 LRVFYYLVQDLKCFLFSLICLHFRIKPV 148
> tpv:TP04_0225 mago nashi protein; K12877 protein mago nashi
Length=153
Score = 211 bits (538), Expect = 5e-55, Method: Compositional matrix adjust.
Identities = 93/144 (64%), Positives = 118/144 (81%), Gaps = 0/144 (0%)
Query 10 DDDFYLRYYTGHKGKFGHEFLEFEFLAEGRMRYANNSNYKNDTMIRKEAYVSQAVIKELR 69
++DFYLRYY GH+GKFGHEFLEFE + GR+RYANNSNYK DTMI+KE Y+ V++E +
Sbjct 3 EEDFYLRYYVGHEGKFGHEFLEFELDSNGRLRYANNSNYKRDTMIKKEVYLLNEVVEEFK 62
Query 70 RIIEEAEILREDDNNWPAPDRVGRQELEIILGKDHISFTTSKIGSMADVQRCKDQEGLRV 129
RII E+ I+ EDDN WP PDR+G+QELEI L ++ +FTTSKIGS+ ++Q KD GLR+
Sbjct 63 RIIVESGIVHEDDNEWPHPDRIGKQELEIKLNGNYYTFTTSKIGSLQEIQSSKDPHGLRI 122
Query 130 FYYLVQDLKCFVYSLIGLHFRIKP 153
FYYL+QDLKCFV+S+I LHFR+ P
Sbjct 123 FYYLIQDLKCFVFSMISLHFRVSP 146
> bbo:BBOV_II003590 18.m09966; mago nashi protein; K12877 protein
mago nashi
Length=218
Score = 210 bits (534), Expect = 2e-54, Method: Compositional matrix adjust.
Identities = 97/145 (66%), Positives = 122/145 (84%), Gaps = 1/145 (0%)
Query 7 MGDDDDFYLRYYTGHKGKFGHEFLEFEFLAEGRMRYANNSNYKNDTMIRKEAYVSQAVIK 66
MGDD F+LRYY GH+GKFGHEFLEFE +G++RY NNSNY+ D+ I+KEA+V++AV+
Sbjct 1 MGDDK-FFLRYYVGHEGKFGHEFLEFELNDDGKLRYTNNSNYRKDSKIKKEAHVTRAVVA 59
Query 67 ELRRIIEEAEILREDDNNWPAPDRVGRQELEIILGKDHISFTTSKIGSMADVQRCKDQEG 126
ELRRIIEE+EI ED ++WP PDR+GRQELE+ LG H +F+TSKIGS+++VQ KD +G
Sbjct 60 ELRRIIEESEITSEDHSDWPIPDRIGRQELELRLGGKHYTFSTSKIGSLSEVQNSKDPDG 119
Query 127 LRVFYYLVQDLKCFVYSLIGLHFRI 151
LRVFYYLVQDLKCFV+SLI L FR+
Sbjct 120 LRVFYYLVQDLKCFVFSLINLCFRV 144
> xla:380542 tmem168, MGC53845, flj13576; transmembrane protein
168
Length=700
Score = 33.5 bits (75), Expect = 0.27, Method: Composition-based stats.
Identities = 29/97 (29%), Positives = 43/97 (44%), Gaps = 14/97 (14%)
Query 8 GDDDDFYLRYYTGHKGKFGH------EFLEFEFLAEGRMRYANNSNYKNDTMIRKEAYVS 61
G D Y+ YYTGH G E L FE L E N ++ + +I + S
Sbjct 495 GPRHDTYIIYYTGHSHSTGEWALAGGETLRFETLLE--WWREKNGSFCSRLIIVLDTESS 552
Query 62 QAVIKELRRI------IEEAEILREDDNNWPAPDRVG 92
Q +KE+RR+ ++ AE+ R D P ++G
Sbjct 553 QPWVKEVRRVGDQYVAVQGAEMARVVDIEEADPPQLG 589
> ath:AT5G13660 hypothetical protein
Length=537
Score = 31.2 bits (69), Expect = 1.5, Method: Composition-based stats.
Identities = 25/90 (27%), Positives = 41/90 (45%), Gaps = 17/90 (18%)
Query 31 EFEFLAEGRMRYANNSNYKNDTMIRKEAYVSQAVIKELRRIIEE-----------AEILR 79
EFE A + ++ NN Y + K Y V++EL+ +E ++I
Sbjct 454 EFEQHANCKTKHPNNHIYFENG---KTIY---GVVQELKNTPQEKLFDAIQNVTGSDINH 507
Query 80 EDDNNWPAPDRVGRQELEIILGKDHISFTT 109
++ N W A V R EL+ I GKD ++ +
Sbjct 508 KNFNTWKASYHVARLELQRIYGKDDVTLAS 537
> mmu:381644 Cep135, BC062951, Cep4, Gm1044, MGC86039; centrosomal
protein 135
Length=1140
Score = 29.3 bits (64), Expect = 5.2, Method: Composition-based stats.
Identities = 16/51 (31%), Positives = 28/51 (54%), Gaps = 2/51 (3%)
Query 62 QAVIKELRRIIEEAEILREDDNNWPAPDRVGRQELEIILGKDHISFTTSKI 112
+ + ELRRI E E LRE N + VG+ +LE + +H+++ ++
Sbjct 572 ERALSELRRITAEKEALREKLKNIQERNAVGKSDLEKTI--EHLTYINHQL 620
> cel:Y6B3B.4 hypothetical protein
Length=965
Score = 28.9 bits (63), Expect = 5.9, Method: Composition-based stats.
Identities = 23/75 (30%), Positives = 34/75 (45%), Gaps = 9/75 (12%)
Query 9 DDDDFYLRYYTGHKGKFGHEFLEFEFLAEGRMRYANNSNYKNDTMIRKEAYVSQAVIKEL 68
DDDDF R+ K K E + +++ A S K T +E + Q +KE
Sbjct 208 DDDDFGKRHEM--KQKHAEELRQ-------QLQEAKASKLKELTSRVEEVRMKQEALKER 258
Query 69 RRIIEEAEILREDDN 83
+R + EA + R DN
Sbjct 259 KRQLLEARMQRASDN 273
> cel:Y43F11A.5 set-24; SET (trithorax/polycomb) domain containing
family member (set-24)
Length=868
Score = 28.5 bits (62), Expect = 9.9, Method: Compositional matrix adjust.
Identities = 11/38 (28%), Positives = 22/38 (57%), Gaps = 0/38 (0%)
Query 47 NYKNDTMIRKEAYVSQAVIKELRRIIEEAEILREDDNN 84
+Y DT IR + S A+ +I++++ ++ DD+N
Sbjct 584 DYDTDTKIRMLVFTSSAITDSFLKILQQSAVVEVDDSN 621
Lambda K H
0.324 0.142 0.425
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3321543300
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40