bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_8221_orf1
Length=92
Score E
Sequences producing significant alignments: (Bits) Value
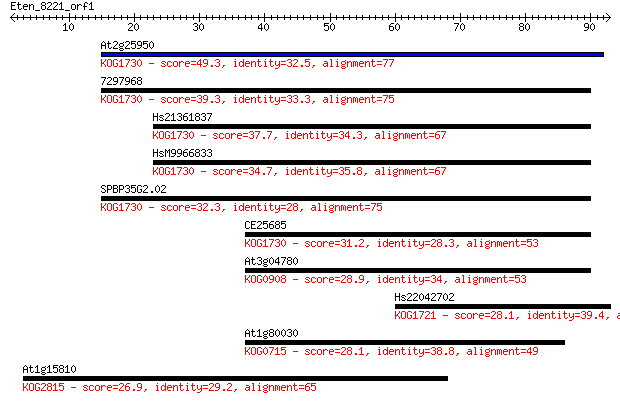
At2g25950 49.3 2e-06
7297968 39.3 0.002
Hs21361837 37.7 0.005
HsM9966833 34.7 0.038
SPBP35G2.02 32.3 0.20
CE25685 31.2 0.44
At3g04780 28.9 2.1
Hs22042702 28.1 3.9
At1g80030 28.1 4.3
At1g15810 26.9 9.6
> At2g25950
Length=192
Score = 49.3 bits (116), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 25/78 (32%), Positives = 44/78 (56%), Gaps = 4/78 (5%)
Query 15 MPGAHGQGCGCKPENEFNLGEFLFGYIDVDGVRGINEQETGSARRTIRPYDERLNDS-VS 73
M H C ++E + L+ +ID+ V +NE +GSA+ + +++RL+ S
Sbjct 1 MSCTHDHNC---EDHECSSDWSLYKHIDLSKVSALNESVSGSAKSVFKAWEQRLHSSGEH 57
Query 74 CKSEEGDAELIIYIPFIS 91
+S EGD EL++++PF S
Sbjct 58 LESNEGDPELLVFVPFTS 75
> 7297968
Length=211
Score = 39.3 bits (90), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 25/83 (30%), Positives = 40/83 (48%), Gaps = 9/83 (10%)
Query 15 MPGAHGQ---GCGCKPEN-----EFNLGEFLFGYIDVDGVRGINEQETGSARRTIRPYDE 66
MP H GC + + E + L+ ID+D V +NE+ G + +PY++
Sbjct 1 MPHGHSHDHGGCSHEASDVDHALEMGIEYSLYTKIDLDNVECLNEETDGQGKSVFKPYEK 60
Query 67 RLNDSVSCKSEEGDAELIIYIPF 89
R + S +S + D EL+ IPF
Sbjct 61 RQDLSKYVES-DADEELLFNIPF 82
> Hs21361837
Length=211
Score = 37.7 bits (86), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 23/72 (31%), Positives = 38/72 (52%), Gaps = 6/72 (8%)
Query 23 CGC-----KPENEFNLGEFLFGYIDVDGVRGINEQETGSARRTIRPYDERLNDSVSCKSE 77
C C +P + L L+ ID++ ++ +NE GS R +P++ER + S +S
Sbjct 12 CRCAAEREEPPEQRGLAYGLYLRIDLERLQCLNESREGSGRGVFKPWEERTDRSKFVES- 70
Query 78 EGDAELIIYIPF 89
+ D EL+ IPF
Sbjct 71 DADEELLFNIPF 82
> HsM9966833
Length=210
Score = 34.7 bits (78), Expect = 0.038, Method: Compositional matrix adjust.
Identities = 24/72 (33%), Positives = 37/72 (51%), Gaps = 7/72 (9%)
Query 23 CGCKPENE-----FNLGEFLFGYIDVDGVRGINEQETGSARRTIRPYDERLNDSVSCKSE 77
C C E E L L+ ID++ ++ +NE GS R +P++ER + S +S
Sbjct 12 CRCAAEREEPPEQRGLAYGLYLRIDLERLQCLNESREGSG-RVFKPWEERTDRSKFVES- 69
Query 78 EGDAELIIYIPF 89
+ D EL+ IPF
Sbjct 70 DADEELLFNIPF 81
> SPBP35G2.02
Length=207
Score = 32.3 bits (72), Expect = 0.20, Method: Compositional matrix adjust.
Identities = 21/77 (27%), Positives = 37/77 (48%), Gaps = 4/77 (5%)
Query 15 MPGAHGQGCGCKPENEFNLG--EFLFGYIDVDGVRGINEQETGSARRTIRPYDERLNDSV 72
M G H C E+ F G + L+ I + + +NE S + +P+D R +D+
Sbjct 1 MSGPHHCSADCD-EHPFESGPNDTLYSCIRKESIVTLNEAVPDSGKLVFKPWDLRYDDTD 59
Query 73 SCKSEEGDAELIIYIPF 89
+S + D +L+ +PF
Sbjct 60 IVES-DADDQLLFQVPF 75
> CE25685
Length=208
Score = 31.2 bits (69), Expect = 0.44, Method: Compositional matrix adjust.
Identities = 15/53 (28%), Positives = 29/53 (54%), Gaps = 1/53 (1%)
Query 37 LFGYIDVDGVRGINEQETGSARRTIRPYDERLNDSVSCKSEEGDAELIIYIPF 89
+ YID++ V +NE G+ ++ + ++R +D + + D EL+ IPF
Sbjct 28 MVSYIDMEKVTTLNESVDGAGKKVFKVMEKR-DDRLEYVESDCDHELLFNIPF 79
> At3g04780
Length=176
Score = 28.9 bits (63), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 18/56 (32%), Positives = 27/56 (48%), Gaps = 8/56 (14%)
Query 37 LFGYIDVDGVRGINEQETGSARRTIRP---YDERLNDSVSCKSEEGDAELIIYIPF 89
L +ID GV +N+ + S ++ DE LN + D +L+IYIPF
Sbjct 16 LLDFIDWSGVECLNQSSSHSLPNALKQGYREDEGLN-----LESDADEQLLIYIPF 66
> Hs22042702
Length=551
Score = 28.1 bits (61), Expect = 3.9, Method: Composition-based stats.
Identities = 13/33 (39%), Positives = 19/33 (57%), Gaps = 1/33 (3%)
Query 60 TIRPYDERLNDSVSCKSEEGDAELIIYIPFISP 92
++ P D+RLN S SC+ G + +P ISP
Sbjct 68 SLEPVDKRLN-SASCEFIRGHQSCCVVVPVISP 99
> At1g80030
Length=435
Score = 28.1 bits (61), Expect = 4.3, Method: Compositional matrix adjust.
Identities = 19/50 (38%), Positives = 25/50 (50%), Gaps = 1/50 (2%)
Query 37 LFGYIDVDGVRGINEQETG-SARRTIRPYDERLNDSVSCKSEEGDAELII 85
L+ Y+DV+ VRGI + +I D L V K+ EGD EL I
Sbjct 326 LYVYLDVEDVRGIERDGINLLSTLSISYLDAILGAVVKVKTVEGDTELQI 375
> At1g15810
Length=419
Score = 26.9 bits (58), Expect = 9.6, Method: Composition-based stats.
Identities = 19/65 (29%), Positives = 27/65 (41%), Gaps = 9/65 (13%)
Query 3 EKTIGAANSTQAMPGAHGQGCGCKPENEFNLGEFLFGYIDVDGVRGINEQETGSARRTIR 62
+ T+G A + G + E E + EFL Y NE+E G RT+R
Sbjct 183 KSTVGEALPKYLFETQMDESKGSEGETEEAMTEFLMFY---------NEEELGEKLRTLR 233
Query 63 PYDER 67
P E+
Sbjct 234 PEGEK 238
Lambda K H
0.314 0.137 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1171209254
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40