bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
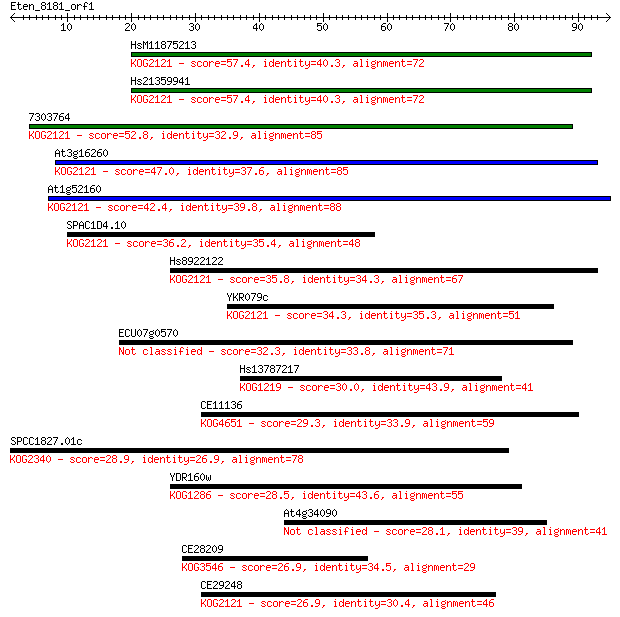
Query= Eten_8181_orf1
Length=94
Score E
Sequences producing significant alignments: (Bits) Value
HsM11875213 57.4 6e-09
Hs21359941 57.4 6e-09
7303764 52.8 1e-07
At3g16260 47.0 9e-06
At1g52160 42.4 2e-04
SPAC1D4.10 36.2 0.014
Hs8922122 35.8 0.019
YKR079c 34.3 0.061
ECU07g0570 32.3 0.21
Hs13787217 30.0 1.0
CE11136 29.3 1.7
SPCC1827.01c 28.9 2.1
YDR160w 28.5 3.4
At4g34090 28.1 3.6
CE28209 26.9 9.4
CE29248 26.9 9.7
> HsM11875213
Length=826
Score = 57.4 bits (137), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 29/72 (40%), Positives = 43/72 (59%), Gaps = 0/72 (0%)
Query 20 SLHIFYEGSRLLINAGENVQRYHLEKKLHIHRISDVFLTSLSPAAAAGLIGLLLSLEGTG 79
+L++F E +R L N GE VQR E KL + R+ ++FLT + + GL G++L+L+ TG
Sbjct 73 ALYVFSEFNRYLFNCGEGVQRLMQEHKLKVARLDNIFLTRMHWSNVGGLSGMILTLKETG 132
Query 80 SAAVTLWGPHGL 91
L GP L
Sbjct 133 LPKCVLSGPPQL 144
> Hs21359941
Length=826
Score = 57.4 bits (137), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 29/72 (40%), Positives = 43/72 (59%), Gaps = 0/72 (0%)
Query 20 SLHIFYEGSRLLINAGENVQRYHLEKKLHIHRISDVFLTSLSPAAAAGLIGLLLSLEGTG 79
+L++F E +R L N GE VQR E KL + R+ ++FLT + + GL G++L+L+ TG
Sbjct 73 ALYVFSEFNRYLFNCGEGVQRLMQEHKLKVARLDNIFLTRMHWSNVGGLSGMILTLKETG 132
Query 80 SAAVTLWGPHGL 91
L GP L
Sbjct 133 LPKCVLSGPPQL 144
> 7303764
Length=743
Score = 52.8 bits (125), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 28/85 (32%), Positives = 46/85 (54%), Gaps = 0/85 (0%)
Query 4 SITAQLLGSHRLSVAPSLHIFYEGSRLLINAGENVQRYHLEKKLHIHRISDVFLTSLSPA 63
++ Q+LGS ++++F + +R L N GE QR E K + R+ +FLT + A
Sbjct 35 TVNLQVLGSGANGAPAAVYLFTDQARYLFNCGEGTQRLAHEHKTRLSRLEQIFLTQNTWA 94
Query 64 AAAGLIGLLLSLEGTGSAAVTLWGP 88
+ GL GL L+++ G + L GP
Sbjct 95 SCGGLPGLTLTIQDAGVRDIGLHGP 119
> At3g16260
Length=937
Score = 47.0 bits (110), Expect = 9e-06, Method: Compositional matrix adjust.
Identities = 32/89 (35%), Positives = 53/89 (59%), Gaps = 4/89 (4%)
Query 8 QLLGS--HRLSVAPSLHIFYEGSRLLINAGENVQRYHLEKKLHIHRISDVFLTSLSPAAA 65
Q+LG+ +PS+ +F++ R + NAGE +QR+ E K+ + ++ +FL+ + A
Sbjct 127 QILGTGMDTQDTSPSVLLFFDKQRFIFNAGEGLQRFCTEHKIKLSKVDHIFLSRVCSETA 186
Query 66 AGLIGLLLSLEGTGSA--AVTLWGPHGLR 92
GL GLLL+L G G +V +WGP L+
Sbjct 187 GGLPGLLLTLAGIGEQGLSVNVWGPSDLK 215
> At1g52160
Length=837
Score = 42.4 bits (98), Expect = 2e-04, Method: Composition-based stats.
Identities = 35/92 (38%), Positives = 53/92 (57%), Gaps = 4/92 (4%)
Query 7 AQLLGS--HRLSVAPSLHIFYEGSRLLINAGENVQRYHLEKKLHIHRISDVFLTSLSPAA 64
AQ+LG+ + S+ +F++ R + NAGE +QR+ E K+ + +I VFL+ +
Sbjct 53 AQILGTGMDTQDTSSSVLLFFDKQRFIFNAGEGLQRFCTEHKIKLSKIDHVFLSRVCSET 112
Query 65 AAGLIGLLLSLEGTGSA--AVTLWGPHGLRPL 94
A GL GLLL+L G G +V +WGP L L
Sbjct 113 AGGLPGLLLTLAGIGEEGLSVNVWGPSDLNYL 144
> SPAC1D4.10
Length=809
Score = 36.2 bits (82), Expect = 0.014, Method: Composition-based stats.
Identities = 17/49 (34%), Positives = 28/49 (57%), Gaps = 1/49 (2%)
Query 10 LGSHRLSVAPSLHIFYEGSRLLINA-GENVQRYHLEKKLHIHRISDVFL 57
+ S S P +H+F++ R + + GE QR L ++L + +I DVFL
Sbjct 20 VSSRDTSCIPCIHLFFDSKRYVFGSVGEGCQRAILSQQLRLSKIKDVFL 68
> Hs8922122
Length=363
Score = 35.8 bits (81), Expect = 0.019, Method: Composition-based stats.
Identities = 23/72 (31%), Positives = 36/72 (50%), Gaps = 5/72 (6%)
Query 26 EGSRLLINAGENVQRYHLEKKLHIHRISDVFLTSLSPAAAAGLIGLLLSLEGTGSAAVT- 84
EG L + GE Q ++ +L RI+ +F+T L GL GLL ++ + V+
Sbjct 29 EGECWLFDCGEGTQTQLMKSQLKAGRITKIFITHLHGDHFFGLPGLLCTISLQSGSMVSK 88
Query 85 ----LWGPHGLR 92
++GP GLR
Sbjct 89 QPIEIYGPVGLR 100
> YKR079c
Length=838
Score = 34.3 bits (77), Expect = 0.061, Method: Composition-based stats.
Identities = 18/52 (34%), Positives = 30/52 (57%), Gaps = 1/52 (1%)
Query 35 GENVQRYHLEKKLHIHRISDVFLT-SLSPAAAAGLIGLLLSLEGTGSAAVTL 85
GE QR E K+ I ++ D+FLT L+ + GL G++L++ G + + L
Sbjct 35 GEGSQRSLTENKIRISKLKDIFLTGELNWSDIGGLPGMILTIADQGKSNLVL 86
> ECU07g0570
Length=512
Score = 32.3 bits (72), Expect = 0.21, Method: Compositional matrix adjust.
Identities = 24/87 (27%), Positives = 39/87 (44%), Gaps = 16/87 (18%)
Query 18 APSLHIFYEGSRLLINAGENVQRYH--LEK--------------KLHIHRISDVFLTSLS 61
+ ++H+ + SRL + G+ + LEK + I R S V+L S+S
Sbjct 384 SSTIHVIFSPSRLPLLTGDGSDEFEMRLEKLVFSGKVVVPIDLTAVWIRRKSAVYLKSIS 443
Query 62 PAAAAGLIGLLLSLEGTGSAAVTLWGP 88
P G + LL+L G A ++ GP
Sbjct 444 PPNKEGKVFFLLTLSNLGGFASSILGP 470
> Hs13787217
Length=4349
Score = 30.0 bits (66), Expect = 1.0, Method: Composition-based stats.
Identities = 18/41 (43%), Positives = 25/41 (60%), Gaps = 2/41 (4%)
Query 37 NVQRYHLEKKLHIHRISDVFLTSLSPAAAAGLIGLLLSLEG 77
N+QR+ L KL I R +++ L SL PA A + +LL EG
Sbjct 3650 NLQRF-LSHKLDIKR-ANIHLASLQPAEAVAGVDVLLVFEG 3688
> CE11136
Length=304
Score = 29.3 bits (64), Expect = 1.7, Method: Composition-based stats.
Identities = 20/64 (31%), Positives = 30/64 (46%), Gaps = 5/64 (7%)
Query 31 LINAGENVQRYH-----LEKKLHIHRISDVFLTSLSPAAAAGLIGLLLSLEGTGSAAVTL 85
LI G + Q H L L+ +R+SD +T +S + + + L L + SA TL
Sbjct 186 LIKIGFDYQSRHAAITQLTDILYTNRVSDTLITKISNESMSKIFTLYLQWQCLVSAGETL 245
Query 86 WGPH 89
G H
Sbjct 246 HGTH 249
> SPCC1827.01c
Length=652
Score = 28.9 bits (63), Expect = 2.1, Method: Composition-based stats.
Identities = 21/78 (26%), Positives = 36/78 (46%), Gaps = 0/78 (0%)
Query 1 STMSITAQLLGSHRLSVAPSLHIFYEGSRLLINAGENVQRYHLEKKLHIHRISDVFLTSL 60
+ MS+ + LG+ L++ H+F R+L N+ Q LE + + V +
Sbjct 206 TNMSMKSHKLGTSLLALHALNHVFKTRDRVLKNSARISQNPELEFRDQGYTRPKVLILLP 265
Query 61 SPAAAAGLIGLLLSLEGT 78
+ +A I LL+S GT
Sbjct 266 TRNSAFEFINLLISYSGT 283
> YDR160w
Length=852
Score = 28.5 bits (62), Expect = 3.4, Method: Composition-based stats.
Identities = 24/71 (33%), Positives = 32/71 (45%), Gaps = 16/71 (22%)
Query 26 EGSRLL------INAGENVQRYHLEKKL---HIHRIS-------DVFLTSLSPAAAAGLI 69
EG LL N ++YH+++KL HI +S +FLTS + AG
Sbjct 254 EGGSLLHDIEKVFNRSRATRKYHIQRKLKVRHIQMLSIGACFSVGLFLTSGKAFSIAGPF 313
Query 70 GLLLSLEGTGS 80
G LL TGS
Sbjct 314 GTLLGFGLTGS 324
> At4g34090
Length=377
Score = 28.1 bits (61), Expect = 3.6, Method: Composition-based stats.
Identities = 16/45 (35%), Positives = 22/45 (48%), Gaps = 4/45 (8%)
Query 44 EKKLHIHRISDVFLTS----LSPAAAAGLIGLLLSLEGTGSAAVT 84
E +H+H + V T +SP A GLI L + L S +VT
Sbjct 111 EPPMHLHHLLKVLQTRGETIISPGAKQGLIPLAIPLSKNSSGSVT 155
> CE28209
Length=864
Score = 26.9 bits (58), Expect = 9.4, Method: Compositional matrix adjust.
Identities = 10/29 (34%), Positives = 20/29 (68%), Gaps = 0/29 (0%)
Query 28 SRLLINAGENVQRYHLEKKLHIHRISDVF 56
++L++ EN+ +YH ++ L +HRI+ F
Sbjct 834 NKLVVLCVENMSKYHGDRILRLHRITSDF 862
> CE29248
Length=850
Score = 26.9 bits (58), Expect = 9.7, Method: Composition-based stats.
Identities = 14/46 (30%), Positives = 25/46 (54%), Gaps = 0/46 (0%)
Query 31 LINAGENVQRYHLEKKLHIHRISDVFLTSLSPAAAAGLIGLLLSLE 76
+ N EN R+ + ++ + D+F+TS + AG+ +LLS E
Sbjct 125 MFNCPENACRFLWQLRIRSSSVVDLFITSANWDNIAGISSILLSKE 170
Lambda K H
0.320 0.136 0.393
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1164659894
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40