bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_7790_orf1
Length=64
Score E
Sequences producing significant alignments: (Bits) Value
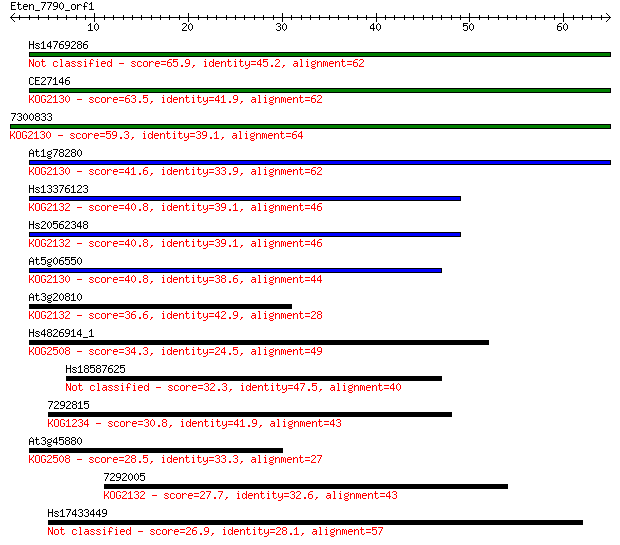
Hs14769286 65.9 2e-11
CE27146 63.5 9e-11
7300833 59.3 2e-09
At1g78280 41.6 4e-04
Hs13376123 40.8 6e-04
Hs20562348 40.8 6e-04
At5g06550 40.8 6e-04
At3g20810 36.6 0.012
Hs4826914_1 34.3 0.061
Hs18587625 32.3 0.21
7292815 30.8 0.64
At3g45880 28.5 3.2
7292005 27.7 5.2
Hs17433449 26.9 8.5
> Hs14769286
Length=414
Score = 65.9 bits (159), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 28/62 (45%), Positives = 41/62 (66%), Gaps = 0/62 (0%)
Query 3 FIRNYEVPGRPVLLCGWMEDWPAMRRWDIESLRRAYRTAKFKVGEKDNGDKIKLKIKYFI 62
F+ YE P +PV+L E W A +W +E L+R YR KFK GE ++G +K+K+KY+I
Sbjct 59 FVERYERPYKPVVLLNAQEGWSAQEKWTLERLKRKYRNQKFKCGEDNDGYSVKMKMKYYI 118
Query 63 DY 64
+Y
Sbjct 119 EY 120
> CE27146
Length=400
Score = 63.5 bits (153), Expect = 9e-11, Method: Compositional matrix adjust.
Identities = 26/62 (41%), Positives = 41/62 (66%), Gaps = 0/62 (0%)
Query 3 FIRNYEVPGRPVLLCGWMEDWPAMRRWDIESLRRAYRTAKFKVGEKDNGDKIKLKIKYFI 62
F R++E P PV++ G ++W A +W +E L + YR FK GE DNG+ +++K+KY+
Sbjct 64 FRRDFERPRIPVIITGLTDNWAAKDKWTVERLSKKYRNQNFKCGEDDNGNSVRMKMKYYH 123
Query 63 DY 64
DY
Sbjct 124 DY 125
> 7300833
Length=408
Score = 59.3 bits (142), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 25/64 (39%), Positives = 41/64 (64%), Gaps = 0/64 (0%)
Query 1 STFIRNYEVPGRPVLLCGWMEDWPAMRRWDIESLRRAYRTAKFKVGEKDNGDKIKLKIKY 60
S FI +E P +PV++ G + W A+ +W + L + YR KFK GE + G +K+K+KY
Sbjct 62 SEFIERFERPYKPVVIRGCTDGWLALEKWTLARLAKKYRNQKFKCGEDNEGYSVKMKMKY 121
Query 61 FIDY 64
+++Y
Sbjct 122 YVEY 125
> At1g78280
Length=889
Score = 41.6 bits (96), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 21/62 (33%), Positives = 34/62 (54%), Gaps = 2/62 (3%)
Query 3 FIRNYEVPGRPVLLCGWMEDWPAMRRWDIESLRRAYRTAKFKVGEKDNGDKIKLKIKYFI 62
F + Y+ +PVLL G + WPA W I+ L Y F++ ++ + +KI +K K +I
Sbjct 111 FSKEYDA-KKPVLLSGLADSWPASNTWTIDQLSEKYGEVPFRISQR-SPNKISMKFKDYI 168
Query 63 DY 64
Y
Sbjct 169 AY 170
> Hs13376123
Length=416
Score = 40.8 bits (94), Expect = 6e-04, Method: Composition-based stats.
Identities = 18/48 (37%), Positives = 28/48 (58%), Gaps = 2/48 (4%)
Query 3 FIRNYEVPGRPVLLCGWMEDWPAMRRWDIESLRR--AYRTAKFKVGEK 48
F + VPGRPV+L G + WP M++W +E ++ RT +VG +
Sbjct 195 FREQFLVPGRPVILKGVADHWPCMQKWSLEYIQEIAGCRTVPVEVGSR 242
> Hs20562348
Length=416
Score = 40.8 bits (94), Expect = 6e-04, Method: Composition-based stats.
Identities = 18/48 (37%), Positives = 28/48 (58%), Gaps = 2/48 (4%)
Query 3 FIRNYEVPGRPVLLCGWMEDWPAMRRWDIESLRR--AYRTAKFKVGEK 48
F + VPGRPV+L G + WP M++W +E ++ RT +VG +
Sbjct 195 FREQFLVPGRPVILKGVADHWPCMQKWSLEYIQEIAGCRTVPVEVGSR 242
> At5g06550
Length=425
Score = 40.8 bits (94), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 17/44 (38%), Positives = 26/44 (59%), Gaps = 0/44 (0%)
Query 3 FIRNYEVPGRPVLLCGWMEDWPAMRRWDIESLRRAYRTAKFKVG 46
FI +E P +PVLL G ++ WPA+ +W + L + +F VG
Sbjct 205 FITKFEEPNKPVLLEGCLDGWPAIEKWSRDYLTKVVGDVEFAVG 248
> At3g20810
Length=418
Score = 36.6 bits (83), Expect = 0.012, Method: Composition-based stats.
Identities = 12/28 (42%), Positives = 19/28 (67%), Gaps = 0/28 (0%)
Query 3 FIRNYEVPGRPVLLCGWMEDWPAMRRWD 30
F+R+Y +PG PV++ M WPA +W+
Sbjct 188 FLRDYYLPGTPVVITNSMAHWPARTKWN 215
> Hs4826914_1
Length=234
Score = 34.3 bits (77), Expect = 0.061, Method: Composition-based stats.
Identities = 12/49 (24%), Positives = 25/49 (51%), Gaps = 0/49 (0%)
Query 3 FIRNYEVPGRPVLLCGWMEDWPAMRRWDIESLRRAYRTAKFKVGEKDNG 51
F R++ P RP ++ ++ WPA+++W + R + + V +G
Sbjct 41 FYRDWVCPNRPCIIRNALQHWPALQKWSLPYFRATVGSTEVSVAVTPDG 89
> Hs18587625
Length=475
Score = 32.3 bits (72), Expect = 0.21, Method: Composition-based stats.
Identities = 19/51 (37%), Positives = 25/51 (49%), Gaps = 11/51 (21%)
Query 7 YEVPGRPVLLC----GWMEDWPAMRR-------WDIESLRRAYRTAKFKVG 46
Y+V G VL+C W ED P+ +R D+E RR + KF VG
Sbjct 223 YQVVGSSVLMCQWDLTWSEDLPSCQRVTSCHDPGDVEHSRRLISSPKFPVG 273
> 7292815
Length=999
Score = 30.8 bits (68), Expect = 0.64, Method: Composition-based stats.
Identities = 18/43 (41%), Positives = 20/43 (46%), Gaps = 12/43 (27%)
Query 5 RNYEVPGRPVLLCGWMEDWPAMRRWDIESLRRAYRTAKFKVGE 47
R E PGRPV W + W RR+YRT KF GE
Sbjct 939 RAEEHPGRPV--------WVKAKAWQ----RRSYRTVKFSRGE 969
> At3g45880
Length=431
Score = 28.5 bits (62), Expect = 3.2, Method: Composition-based stats.
Identities = 9/27 (33%), Positives = 16/27 (59%), Gaps = 0/27 (0%)
Query 3 FIRNYEVPGRPVLLCGWMEDWPAMRRW 29
F+RNY +P ++ + WPA++ W
Sbjct 33 FLRNYVSQSKPCVISKAITHWPALKLW 59
> 7292005
Length=394
Score = 27.7 bits (60), Expect = 5.2, Method: Composition-based stats.
Identities = 14/46 (30%), Positives = 24/46 (52%), Gaps = 3/46 (6%)
Query 11 GRPVLLCGWMEDWPAMRRW-DIESLRR--AYRTAKFKVGEKDNGDK 53
G+P LL ++ WPA+ +W D+ L + RT ++G D+
Sbjct 175 GQPTLLLNTIQHWPALHKWLDLNYLLQVAGNRTVPIEIGSNYASDE 220
> Hs17433449
Length=220
Score = 26.9 bits (58), Expect = 8.5, Method: Composition-based stats.
Identities = 16/57 (28%), Positives = 27/57 (47%), Gaps = 0/57 (0%)
Query 5 RNYEVPGRPVLLCGWMEDWPAMRRWDIESLRRAYRTAKFKVGEKDNGDKIKLKIKYF 61
RN++ V C +WP M ++E++ + A G+ N DK K K+K +
Sbjct 125 RNHDKGKSTVEQCSHTSNWPLMGADEVETVSQRPFVAIGLSGKGRNHDKGKAKVKQY 181
Lambda K H
0.325 0.143 0.475
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1169706042
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40