bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_7764_orf1
Length=99
Score E
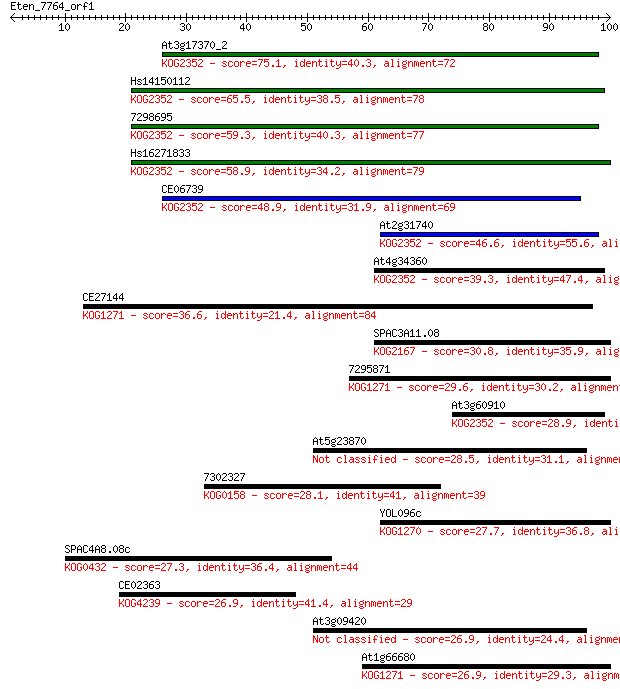
Sequences producing significant alignments: (Bits) Value
At3g17370_2 75.1 3e-14
Hs14150112 65.5 3e-11
7298695 59.3 2e-09
Hs16271833 58.9 2e-09
CE06739 48.9 2e-06
At2g31740 46.6 1e-05
At4g34360 39.3 0.002
CE27144 36.6 0.013
SPAC3A11.08 30.8 0.68
7295871 29.6 1.4
At3g60910 28.9 2.1
At5g23870 28.5 3.0
7302327 28.1 4.3
YOL096c 27.7 5.8
SPAC4A8.08c 27.3 7.3
CE02363 26.9 8.3
At3g09420 26.9 8.7
At1g66680 26.9 8.8
> At3g17370_2
Length=151
Score = 75.1 bits (183), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 29/72 (40%), Positives = 47/72 (65%), Gaps = 0/72 (0%)
Query 26 HWEERYKKDPEPFDWFNKYAALKPFLLSAGLEESHEILMLGCGTSTMSESLWDDGFRNIT 85
+W++RYK + EPFDW+ KY+ L P + + + +L++GCG S SE + DDG+ ++
Sbjct 6 YWDDRYKNESEPFDWYQKYSPLAPLINLYVPQRNQRVLVIGCGNSAFSEGMVDDGYEDVV 65
Query 86 NIDFSSQCINIM 97
+ID SS I+ M
Sbjct 66 SIDISSVVIDTM 77
> Hs14150112
Length=255
Score = 65.5 bits (158), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 30/80 (37%), Positives = 49/80 (61%), Gaps = 3/80 (3%)
Query 21 YASLDHWEERYK--KDPEPFDWFNKYAALKPFLLSAGLEESHEILMLGCGTSTMSESLWD 78
Y +++W++RY+ D P+DWF +++ + LL L IL+LGCG S +S L+
Sbjct 20 YREVEYWDQRYQGAADSAPYDWFGDFSSFRA-LLEPELRPEDRILVLGCGNSALSYELFL 78
Query 79 DGFRNITNIDFSSQCINIMQ 98
GF N+T++D+SS + MQ
Sbjct 79 GGFPNVTSVDYSSVVVAAMQ 98
> 7298695
Length=673
Score = 59.3 bits (142), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 31/78 (39%), Positives = 46/78 (58%), Gaps = 2/78 (2%)
Query 21 YASLDHWEERYKKDPE-PFDWFNKYAALKPFLLSAGLEESHEILMLGCGTSTMSESLWDD 79
+A D+W E +KK E F+W+ +Y L + ++ + ILMLGCG S +S ++D
Sbjct 11 FAQTDYWNEFFKKRGEKAFEWYGEYLELCD-QIHKYIKPADRILMLGCGNSKLSMDMYDT 69
Query 80 GFRNITNIDFSSQCINIM 97
GFR+ITNID S + M
Sbjct 70 GFRDITNIDISPIAVKKM 87
> Hs16271833
Length=543
Score = 58.9 bits (141), Expect = 2e-09, Method: Composition-based stats.
Identities = 27/80 (33%), Positives = 50/80 (62%), Gaps = 2/80 (2%)
Query 21 YASLDHWEERYK-KDPEPFDWFNKYAALKPFLLSAGLEESHEILMLGCGTSTMSESLWDD 79
+ S+D+WE+ ++ + + F+W+ Y L +L ++ ++L++GCG S +SE L+D
Sbjct 11 FGSVDYWEKFFQQRGKKAFEWYGTYLELCG-VLHKYIKPREKVLVIGCGNSELSEQLYDV 69
Query 80 GFRNITNIDFSSQCINIMQQ 99
G+R+I NID S I M++
Sbjct 70 GYRDIVNIDISEVVIKQMKE 89
> CE06739
Length=656
Score = 48.9 bits (115), Expect = 2e-06, Method: Composition-based stats.
Identities = 22/69 (31%), Positives = 36/69 (52%), Gaps = 1/69 (1%)
Query 26 HWEERYKKDPEPFDWFNKYAALKPFLLSAGLEESHEILMLGCGTSTMSESLWDDGFRNIT 85
+W+ + K PF+W+ Y +L ++ L+ L LGCG S ++ L+D+GF I
Sbjct 15 YWKNFFAKRKSPFEWYGDYNSLSN-VIDKYLKPKDTFLQLGCGNSELATQLYDNGFHCIH 73
Query 86 NIDFSSQCI 94
+ID I
Sbjct 74 SIDVEPSVI 82
> At2g31740
Length=690
Score = 46.6 bits (109), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 20/36 (55%), Positives = 27/36 (75%), Gaps = 0/36 (0%)
Query 62 ILMLGCGTSTMSESLWDDGFRNITNIDFSSQCINIM 97
IL+ GCG S ++E L+D GFR+ITN+DFS I+ M
Sbjct 72 ILVPGCGNSRLTEHLYDAGFRDITNVDFSKVVISDM 107
> At4g34360
Length=197
Score = 39.3 bits (90), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 18/38 (47%), Positives = 24/38 (63%), Gaps = 0/38 (0%)
Query 61 EILMLGCGTSTMSESLWDDGFRNITNIDFSSQCINIMQ 98
++L LGCG S + E L+ DG +IT ID SS + MQ
Sbjct 2 KVLELGCGNSQLCEELYKDGIVDITCIDLSSVAVEKMQ 39
> CE27144
Length=236
Score = 36.6 bits (83), Expect = 0.013, Method: Compositional matrix adjust.
Identities = 18/94 (19%), Positives = 46/94 (48%), Gaps = 10/94 (10%)
Query 13 TCALKMTRYASLDHWEERYKKDPEPFD--------WFNKYAALK--PFLLSAGLEESHEI 62
+ + ++ + D W++RY+ + + F WF + + +L+ + + +I
Sbjct 8 SVEIASSQLGTKDFWDQRYELELKNFKQHGDEGEVWFGTSSETRIVKYLIDSKTGKDAKI 67
Query 63 LMLGCGTSTMSESLWDDGFRNITNIDFSSQCINI 96
L LGCG ++ L GF+++ +D+ + +++
Sbjct 68 LDLGCGNGSVLRKLRSKGFQSLKGVDYCQKAVDL 101
> SPAC3A11.08
Length=734
Score = 30.8 bits (68), Expect = 0.68, Method: Composition-based stats.
Identities = 14/39 (35%), Positives = 23/39 (58%), Gaps = 1/39 (2%)
Query 61 EILMLGCGTSTMSESLWDDGFRNITNIDFSSQCINIMQQ 99
+++M G G T + L+ G N+T D +S+C NI+Q
Sbjct 38 QVVMAGLGLKTGYQELYS-GVENLTRADQASRCFNILQH 75
> 7295871
Length=219
Score = 29.6 bits (65), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 13/44 (29%), Positives = 25/44 (56%), Gaps = 1/44 (2%)
Query 57 EESHEILMLGCGTSTMSESLWDDGFR-NITNIDFSSQCINIMQQ 99
+E+ +L LGCG L ++GF ++T +D+S + + + Q
Sbjct 58 KEASRVLDLGCGNGMFLVGLANEGFTGDLTGVDYSPKAVELAQN 101
> At3g60910
Length=248
Score = 28.9 bits (63), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 11/25 (44%), Positives = 16/25 (64%), Gaps = 0/25 (0%)
Query 74 ESLWDDGFRNITNIDFSSQCINIMQ 98
E + DG+ +I N+D SS I +MQ
Sbjct 61 EDMVKDGYEDIMNVDISSVAIEMMQ 85
> At5g23870
Length=415
Score = 28.5 bits (62), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 14/45 (31%), Positives = 24/45 (53%), Gaps = 0/45 (0%)
Query 51 LLSAGLEESHEILMLGCGTSTMSESLWDDGFRNITNIDFSSQCIN 95
LL GL ++H+ L+ GC +S L D F + + S +C++
Sbjct 165 LLPKGLAKAHKALLTGCSAGGLSTFLHCDNFTSYLPKNASVKCMS 209
> 7302327
Length=485
Score = 28.1 bits (61), Expect = 4.3, Method: Composition-based stats.
Identities = 16/39 (41%), Positives = 21/39 (53%), Gaps = 2/39 (5%)
Query 33 KDPEPFDWFNKYAALKPFLLSAGLEESHEILMLGCGTST 71
K PF F YA LKPF+L+ L+ H+I+ G T
Sbjct 65 KGRAPFVGF--YACLKPFILALDLKLVHQIIFTDAGHFT 101
> YOL096c
Length=316
Score = 27.7 bits (60), Expect = 5.8, Method: Compositional matrix adjust.
Identities = 14/42 (33%), Positives = 23/42 (54%), Gaps = 7/42 (16%)
Query 62 ILMLGCGTSTMSESL----WDDGFRNITNIDFSSQCINIMQQ 99
+L +GCG +SESL W +N+ ID + CI + ++
Sbjct 130 VLDVGCGGGILSESLARLKW---VKNVQGIDLTRDCIMVAKE 168
> SPAC4A8.08c
Length=950
Score = 27.3 bits (59), Expect = 7.3, Method: Composition-based stats.
Identities = 16/44 (36%), Positives = 21/44 (47%), Gaps = 0/44 (0%)
Query 10 RAATCALKMTRYASLDHWEERYKKDPEPFDWFNKYAALKPFLLS 53
R LKM Y+ L H E K+ E +FN +K FLL+
Sbjct 671 RQRISLLKMATYSKLHHAVEGVKESFEQRKFFNAADIMKNFLLN 714
> CE02363
Length=615
Score = 26.9 bits (58), Expect = 8.3, Method: Compositional matrix adjust.
Identities = 12/29 (41%), Positives = 19/29 (65%), Gaps = 1/29 (3%)
Query 19 TRYASLDHWEERYKKDPEPFDWFNKYAAL 47
+RYA+L ERY K+ PF+W ++Y +
Sbjct 305 SRYATL-RVVERYVKEDTPFEWTDEYKEM 332
> At3g09420
Length=427
Score = 26.9 bits (58), Expect = 8.7, Method: Compositional matrix adjust.
Identities = 11/45 (24%), Positives = 25/45 (55%), Gaps = 0/45 (0%)
Query 51 LLSAGLEESHEILMLGCGTSTMSESLWDDGFRNITNIDFSSQCIN 95
LLS G+ ++ + ++ GC ++ + D FR+ D + +C++
Sbjct 192 LLSMGMSDAKQAILTGCSAGGLASLIHCDYFRDHLPKDAAVKCVS 236
> At1g66680
Length=358
Score = 26.9 bits (58), Expect = 8.8, Method: Composition-based stats.
Identities = 12/41 (29%), Positives = 20/41 (48%), Gaps = 0/41 (0%)
Query 59 SHEILMLGCGTSTMSESLWDDGFRNITNIDFSSQCINIMQQ 99
S +L LG G + L +GF ++T D+S + + Q
Sbjct 170 SWNVLDLGTGNGLLLHQLAKEGFSDLTGTDYSDGAVELAQH 210
Lambda K H
0.323 0.136 0.441
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1191270180
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40