bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_7313_orf1
Length=74
Score E
Sequences producing significant alignments: (Bits) Value
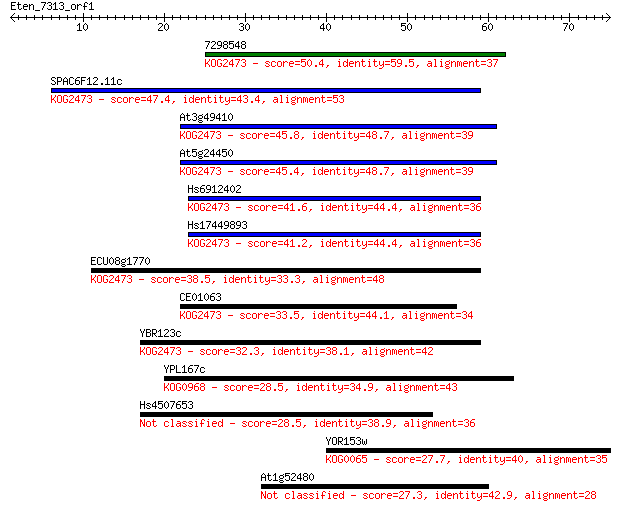
7298548 50.4 7e-07
SPAC6F12.11c 47.4 6e-06
At3g49410 45.8 2e-05
At5g24450 45.4 2e-05
Hs6912402 41.6 3e-04
Hs17449893 41.2 4e-04
ECU08g1770 38.5 0.003
CE01063 33.5 0.11
YBR123c 32.3 0.22
YPL167c 28.5 3.2
Hs4507653 28.5 3.3
YOR153w 27.7 5.1
At1g52480 27.3 8.0
> 7298548
Length=480
Score = 50.4 bits (119), Expect = 7e-07, Method: Composition-based stats.
Identities = 22/37 (59%), Positives = 26/37 (70%), Gaps = 0/37 (0%)
Query 25 FLFADGPFRGCLCRLGYDPRKDPESRHYQTIDFRDPF 61
F F++GP+R R GYDPRKD SR+YQT DFR F
Sbjct 270 FYFSNGPWRTLYVRFGYDPRKDFNSRYYQTFDFRLRF 306
> SPAC6F12.11c
Length=456
Score = 47.4 bits (111), Expect = 6e-06, Method: Composition-based stats.
Identities = 23/53 (43%), Positives = 30/53 (56%), Gaps = 1/53 (1%)
Query 6 HLPPTYTAWRKKPAYAKTCFLFADGPFRGCLCRLGYDPRKDPESRHYQTIDFR 58
HL P+YT + K A +L+ GPFR R GYDPRKD + YQ + F+
Sbjct 243 HLDPSYTHYLKF-ALPYLSYLWTSGPFRDTYTRFGYDPRKDSNAAAYQALFFK 294
> At3g49410
Length=566
Score = 45.8 bits (107), Expect = 2e-05, Method: Composition-based stats.
Identities = 19/39 (48%), Positives = 25/39 (64%), Gaps = 0/39 (0%)
Query 22 KTCFLFADGPFRGCLCRLGYDPRKDPESRHYQTIDFRDP 60
+ + F+ GPF + GYDPR DPESR YQ ++FR P
Sbjct 270 RAAYYFSSGPFLRFWIKRGYDPRNDPESRVYQRMEFRVP 308
> At5g24450
Length=545
Score = 45.4 bits (106), Expect = 2e-05, Method: Composition-based stats.
Identities = 19/39 (48%), Positives = 26/39 (66%), Gaps = 0/39 (0%)
Query 22 KTCFLFADGPFRGCLCRLGYDPRKDPESRHYQTIDFRDP 60
+ + F+ GPF + GYDPRKDPESR +Q ++FR P
Sbjct 278 RAAYYFSGGPFLRFWIKRGYDPRKDPESRVFQRMEFRVP 316
> Hs6912402
Length=519
Score = 41.6 bits (96), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 16/36 (44%), Positives = 23/36 (63%), Gaps = 0/36 (0%)
Query 23 TCFLFADGPFRGCLCRLGYDPRKDPESRHYQTIDFR 58
+ GP+R R GYDPRK+P+++ YQ +DFR
Sbjct 287 IAYYMITGPWRSLWIRFGYDPRKNPDAKIYQVLDFR 322
> Hs17449893
Length=526
Score = 41.2 bits (95), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 16/36 (44%), Positives = 23/36 (63%), Gaps = 0/36 (0%)
Query 23 TCFLFADGPFRGCLCRLGYDPRKDPESRHYQTIDFR 58
+ GP+R R GYDPRK+P+++ YQ +DFR
Sbjct 322 IAYYMITGPWRSLWIRFGYDPRKNPDAKIYQVLDFR 357
> ECU08g1770
Length=338
Score = 38.5 bits (88), Expect = 0.003, Method: Composition-based stats.
Identities = 16/48 (33%), Positives = 24/48 (50%), Gaps = 0/48 (0%)
Query 11 YTAWRKKPAYAKTCFLFADGPFRGCLCRLGYDPRKDPESRHYQTIDFR 58
++ W+ K + GP+RGC R G DP++D + YQ D R
Sbjct 222 FSDWKIKKFLPLHAYFIDSGPWRGCWARFGVDPKRDQGNFRYQIYDSR 269
> CE01063
Length=433
Score = 33.5 bits (75), Expect = 0.11, Method: Composition-based stats.
Identities = 15/34 (44%), Positives = 17/34 (50%), Gaps = 0/34 (0%)
Query 22 KTCFLFADGPFRGCLCRLGYDPRKDPESRHYQTI 55
K F GP+ C+ GYDPR D YQTI
Sbjct 275 KYAFYILSGPWGRLWCKFGYDPRTDRNGGLYQTI 308
> YBR123c
Length=649
Score = 32.3 bits (72), Expect = 0.22, Method: Composition-based stats.
Identities = 16/42 (38%), Positives = 20/42 (47%), Gaps = 0/42 (0%)
Query 17 KPAYAKTCFLFADGPFRGCLCRLGYDPRKDPESRHYQTIDFR 58
K A A + F GP+R + G DPR E YQT F+
Sbjct 350 KIALALISYRFTMGPWRNTYIKFGIDPRSSVEYAQYQTEYFK 391
> YPL167c
Length=1504
Score = 28.5 bits (62), Expect = 3.2, Method: Composition-based stats.
Identities = 15/44 (34%), Positives = 21/44 (47%), Gaps = 8/44 (18%)
Query 20 YAKTCFLFADGPFRGCLCRLGY-DPRKDPESRHYQTIDFRDPFF 62
Y K F++ + PF GY D E + ID++DPFF
Sbjct 516 YGKRAFVYGEPPF-------GYQDILNKLEDEGFPKIDYKDPFF 552
> Hs4507653
Length=245
Score = 28.5 bits (62), Expect = 3.3, Method: Composition-based stats.
Identities = 14/36 (38%), Positives = 16/36 (44%), Gaps = 0/36 (0%)
Query 17 KPAYAKTCFLFADGPFRGCLCRLGYDPRKDPESRHY 52
+ YA T F F+ LC L YDP K P Y
Sbjct 163 RKCYADTMFSLLGKKFQYLLCVLSYDPTKHPGPPFY 198
> YOR153w
Length=1511
Score = 27.7 bits (60), Expect = 5.1, Method: Composition-based stats.
Identities = 14/49 (28%), Positives = 20/49 (40%), Gaps = 14/49 (28%)
Query 40 GYDPRKDPESRHYQTI--------------DFRDPFFRSTNWKAAAAGG 74
GYDP+ DP S ++ + DF P+ WK +A G
Sbjct 90 GYDPKLDPNSENFSSAAWVKNMAHLSAADPDFYKPYSLGCAWKNLSASG 138
> At1g52480
Length=201
Score = 27.3 bits (59), Expect = 8.0, Method: Compositional matrix adjust.
Identities = 12/28 (42%), Positives = 13/28 (46%), Gaps = 0/28 (0%)
Query 32 FRGCLCRLGYDPRKDPESRHYQTIDFRD 59
FR C C GYDP D DFR+
Sbjct 111 FRKCNCLFGYDPVMDQYKVLSMVFDFRE 138
Lambda K H
0.325 0.140 0.489
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1184572648
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40