bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_7055_orf3
Length=62
Score E
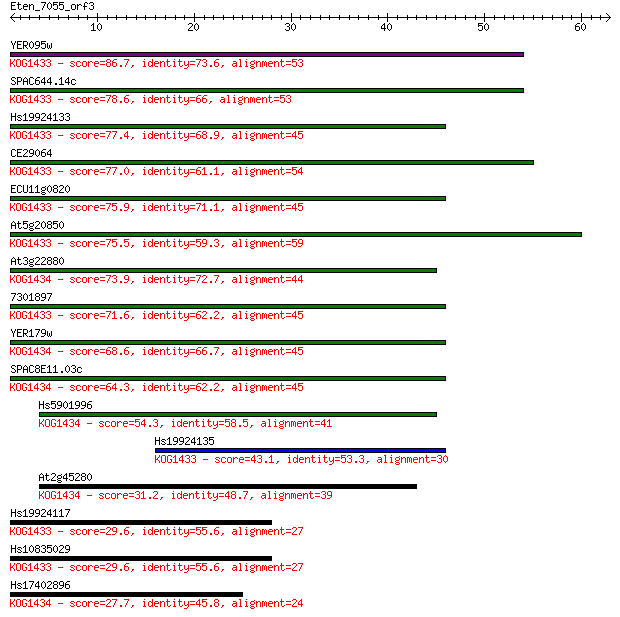
Sequences producing significant alignments: (Bits) Value
YER095w 86.7 9e-18
SPAC644.14c 78.6 2e-15
Hs19924133 77.4 7e-15
CE29064 77.0 7e-15
ECU11g0820 75.9 2e-14
At5g20850 75.5 2e-14
At3g22880 73.9 6e-14
7301897 71.6 3e-13
YER179w 68.6 3e-12
SPAC8E11.03c 64.3 5e-11
Hs5901996 54.3 5e-08
Hs19924135 43.1 1e-04
At2g45280 31.2 0.43
Hs19924117 29.6 1.3
Hs10835029 29.6 1.3
Hs17402896 27.7 5.8
> YER095w
Length=400
Score = 86.7 bits (213), Expect = 9e-18, Method: Composition-based stats.
Identities = 39/53 (73%), Positives = 45/53 (84%), Gaps = 0/53 (0%)
Query 1 GGEGKCLWIDTEGTFRPERIVAIARRFGLDPADCLDNIAYARAYNCGPLLLLL 53
GGEGKCL+IDTEGTFRP R+V+IA+RFGLDP D L+N+AYARAYN L LL
Sbjct 210 GGEGKCLYIDTEGTFRPVRLVSIAQRFGLDPDDALNNVAYARAYNADHQLRLL 262
> SPAC644.14c
Length=365
Score = 78.6 bits (192), Expect = 2e-15, Method: Composition-based stats.
Identities = 35/53 (66%), Positives = 43/53 (81%), Gaps = 0/53 (0%)
Query 1 GGEGKCLWIDTEGTFRPERIVAIARRFGLDPADCLDNIAYARAYNCGPLLLLL 53
GGEGKCL+IDTEGTFRP R++A+A R+GL+ + LDN+AYARAYN L LL
Sbjct 174 GGEGKCLYIDTEGTFRPVRLLAVADRYGLNGEEVLDNVAYARAYNADHQLELL 226
> Hs19924133
Length=339
Score = 77.4 bits (189), Expect = 7e-15, Method: Composition-based stats.
Identities = 31/45 (68%), Positives = 40/45 (88%), Gaps = 0/45 (0%)
Query 1 GGEGKCLWIDTEGTFRPERIVAIARRFGLDPADCLDNIAYARAYN 45
GGEGK ++IDTEGTFRPER++A+A R+GL +D LDN+AYARA+N
Sbjct 152 GGEGKAMYIDTEGTFRPERLLAVAERYGLSGSDVLDNVAYARAFN 196
> CE29064
Length=395
Score = 77.0 bits (188), Expect = 7e-15, Method: Composition-based stats.
Identities = 33/54 (61%), Positives = 43/54 (79%), Gaps = 0/54 (0%)
Query 1 GGEGKCLWIDTEGTFRPERIVAIARRFGLDPADCLDNIAYARAYNCGPLLLLLL 54
GGEGKC++IDT TFRPERI+AIA+R+ +D A L+NIA ARAYN L+ L++
Sbjct 206 GGEGKCMYIDTNATFRPERIIAIAQRYNMDSAHVLENIAVARAYNSEHLMALII 259
> ECU11g0820
Length=334
Score = 75.9 bits (185), Expect = 2e-14, Method: Composition-based stats.
Identities = 32/45 (71%), Positives = 38/45 (84%), Gaps = 0/45 (0%)
Query 1 GGEGKCLWIDTEGTFRPERIVAIARRFGLDPADCLDNIAYARAYN 45
GG GK ++IDTEGTFR ER+V IA RFGLDP + +DNI+YARAYN
Sbjct 146 GGGGKAMYIDTEGTFRSERLVPIAERFGLDPNEVMDNISYARAYN 190
> At5g20850
Length=339
Score = 75.5 bits (184), Expect = 2e-14, Method: Composition-based stats.
Identities = 35/59 (59%), Positives = 44/59 (74%), Gaps = 0/59 (0%)
Query 1 GGEGKCLWIDTEGTFRPERIVAIARRFGLDPADCLDNIAYARAYNCGPLLLLLLFGVSV 59
GGEGK ++ID EGTFRP+R++ IA RFGL+ AD L+N+AYARAYN LLL S+
Sbjct 152 GGEGKAMYIDAEGTFRPQRLLQIADRFGLNGADVLENVAYARAYNTDHQSRLLLEAASM 210
> At3g22880
Length=339
Score = 73.9 bits (180), Expect = 6e-14, Method: Composition-based stats.
Identities = 32/44 (72%), Positives = 35/44 (79%), Gaps = 0/44 (0%)
Query 1 GGEGKCLWIDTEGTFRPERIVAIARRFGLDPADCLDNIAYARAY 44
GG GK +IDTEGTFRP+RIV IA RFG+DP LDNI YARAY
Sbjct 158 GGNGKVAYIDTEGTFRPDRIVPIAERFGMDPGAVLDNIIYARAY 201
> 7301897
Length=336
Score = 71.6 bits (174), Expect = 3e-13, Method: Composition-based stats.
Identities = 28/45 (62%), Positives = 39/45 (86%), Gaps = 0/45 (0%)
Query 1 GGEGKCLWIDTEGTFRPERIVAIARRFGLDPADCLDNIAYARAYN 45
GGEGKC++IDTE TFRPER+ AIA+R+ L+ ++ LDN+A+ RA+N
Sbjct 149 GGEGKCMYIDTENTFRPERLAAIAQRYKLNESEVLDNVAFTRAHN 193
> YER179w
Length=334
Score = 68.6 bits (166), Expect = 3e-12, Method: Composition-based stats.
Identities = 30/45 (66%), Positives = 34/45 (75%), Gaps = 0/45 (0%)
Query 1 GGEGKCLWIDTEGTFRPERIVAIARRFGLDPADCLDNIAYARAYN 45
GGEGK +IDTEGTFRPERI IA + LDP CL N++YARA N
Sbjct 146 GGEGKVAYIDTEGTFRPERIKQIAEGYELDPESCLANVSYARALN 190
> SPAC8E11.03c
Length=332
Score = 64.3 bits (155), Expect = 5e-11, Method: Composition-based stats.
Identities = 28/45 (62%), Positives = 34/45 (75%), Gaps = 0/45 (0%)
Query 1 GGEGKCLWIDTEGTFRPERIVAIARRFGLDPADCLDNIAYARAYN 45
G EGK +IDTEGTFRP+RI AIA RFG+D ++NI +RAYN
Sbjct 145 GAEGKVAFIDTEGTFRPDRIKAIAERFGVDADQAMENIIVSRAYN 189
> Hs5901996
Length=340
Score = 54.3 bits (129), Expect = 5e-08, Method: Composition-based stats.
Identities = 24/41 (58%), Positives = 30/41 (73%), Gaps = 0/41 (0%)
Query 4 GKCLWIDTEGTFRPERIVAIARRFGLDPADCLDNIAYARAY 44
GK ++IDTE TFRP+R+ IA RF +D LDN+ YARAY
Sbjct 154 GKIIFIDTENTFRPDRLRDIADRFNVDHDAVLDNVLYARAY 194
> Hs19924135
Length=242
Score = 43.1 bits (100), Expect = 1e-04, Method: Composition-based stats.
Identities = 16/30 (53%), Positives = 25/30 (83%), Gaps = 0/30 (0%)
Query 16 RPERIVAIARRFGLDPADCLDNIAYARAYN 45
+ ++I+A+A R+GL +D LDN+AYARA+N
Sbjct 70 KADKILAVAERYGLSGSDVLDNVAYARAFN 99
> At2g45280
Length=332
Score = 31.2 bits (69), Expect = 0.43, Method: Composition-based stats.
Identities = 19/59 (32%), Positives = 26/59 (44%), Gaps = 20/59 (33%)
Query 4 GKCLWIDTEGTFRPERIVAIAR--------------------RFGLDPADCLDNIAYAR 42
GK ++IDTEG+F ER + IA + + P D L+NI Y R
Sbjct 128 GKAIYIDTEGSFMVERALQIAEACVEDMEEYTGYMHKHFQANQVQMKPEDILENIFYFR 186
> Hs19924117
Length=350
Score = 29.6 bits (65), Expect = 1.3, Method: Composition-based stats.
Identities = 15/28 (53%), Positives = 18/28 (64%), Gaps = 1/28 (3%)
Query 1 GGEGKCLWIDTEGTFRPERIVAIAR-RF 27
G EG ++IDTE F ER+V IA RF
Sbjct 133 GLEGAVVYIDTESAFSAERLVEIAESRF 160
> Hs10835029
Length=350
Score = 29.6 bits (65), Expect = 1.3, Method: Composition-based stats.
Identities = 15/28 (53%), Positives = 18/28 (64%), Gaps = 1/28 (3%)
Query 1 GGEGKCLWIDTEGTFRPERIVAIAR-RF 27
G EG ++IDTE F ER+V IA RF
Sbjct 133 GLEGAVVYIDTESAFSAERLVEIAESRF 160
> Hs17402896
Length=376
Score = 27.7 bits (60), Expect = 5.8, Method: Composition-based stats.
Identities = 11/24 (45%), Positives = 18/24 (75%), Gaps = 0/24 (0%)
Query 1 GGEGKCLWIDTEGTFRPERIVAIA 24
G G+ ++IDTEG+F +R+V +A
Sbjct 150 GVAGEAVFIDTEGSFMVDRVVDLA 173
Lambda K H
0.326 0.145 0.467
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1175803722
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40