bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_6758_orf3
Length=106
Score E
Sequences producing significant alignments: (Bits) Value
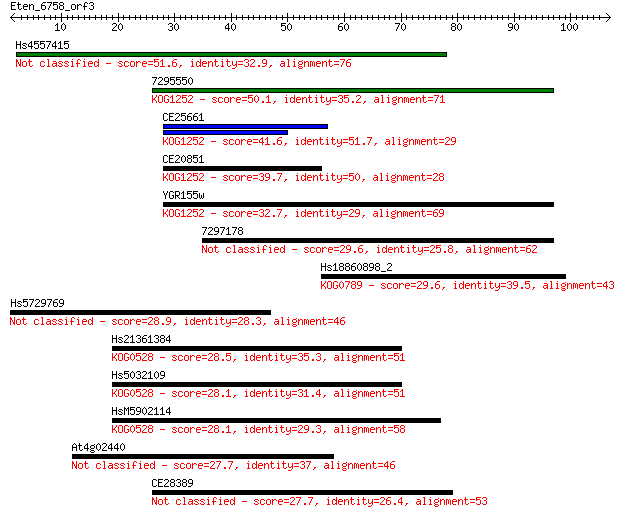
Hs4557415 51.6 3e-07
7295550 50.1 1e-06
CE25661 41.6 3e-04
CE20851 39.7 0.001
YGR155w 32.7 0.15
7297178 29.6 1.4
Hs18860898_2 29.6 1.5
Hs5729769 28.9 2.6
Hs21361384 28.5 2.8
Hs5032109 28.1 3.8
HsM5902114 28.1 3.9
At4g02440 27.7 4.7
CE28389 27.7 5.6
> Hs4557415
Length=551
Score = 51.6 bits (122), Expect = 3e-07, Method: Composition-based stats.
Identities = 25/76 (32%), Positives = 42/76 (55%), Gaps = 3/76 (3%)
Query 2 GTVAATTIGANSRIFRSLWKDDSSRRIAMVLPDGSRNYTSKFVCDEWMAEKGFLERSELS 61
G A +T+ + + L +R ++LPD RNY +KF+ D WM +KGFL+ +L+
Sbjct 347 GGSAGSTVAVAVKAAQEL---QEGQRCVVILPDSVRNYMTKFLSDRWMLQKGFLKEEDLT 403
Query 62 RQYEQYSNMSVQSLQL 77
+ + ++ VQ L L
Sbjct 404 EKKPWWWHLRVQELGL 419
> 7295550
Length=522
Score = 50.1 bits (118), Expect = 1e-06, Method: Composition-based stats.
Identities = 25/72 (34%), Positives = 43/72 (59%), Gaps = 3/72 (4%)
Query 26 RRIAMVLPDGSRNYTSKFVCDEWMAEKGFLERSELSRQYEQYSNMSVQSLQLPRLTFV-S 84
+R ++LPDG RNY +KFV D WM + F E ++ + ++++ L+LP +
Sbjct 337 QRCVVILPDGIRNYMTKFVSDNWMEARNFKE--PVNEHGHWWWSLAIAELELPAPPVILK 394
Query 85 SDSSLKEAAALL 96
SD+++ EA AL+
Sbjct 395 SDATVGEAIALM 406
> CE25661
Length=700
Score = 41.6 bits (96), Expect = 3e-04, Method: Composition-based stats.
Identities = 15/29 (51%), Positives = 23/29 (79%), Gaps = 0/29 (0%)
Query 28 IAMVLPDGSRNYTSKFVCDEWMAEKGFLE 56
+ ++LPDG RNY +KF+ D+WM E+ FL+
Sbjct 671 VVVLLPDGIRNYITKFLDDDWMNERHFLD 699
Score = 28.1 bits (61), Expect = 4.0, Method: Composition-based stats.
Identities = 11/22 (50%), Positives = 15/22 (68%), Gaps = 0/22 (0%)
Query 28 IAMVLPDGSRNYTSKFVCDEWM 49
I +VL DG RNY F+ D+W+
Sbjct 291 IVVVLMDGIRNYLRHFLDDDWI 312
> CE20851
Length=755
Score = 39.7 bits (91), Expect = 0.001, Method: Composition-based stats.
Identities = 14/28 (50%), Positives = 21/28 (75%), Gaps = 0/28 (0%)
Query 28 IAMVLPDGSRNYTSKFVCDEWMAEKGFL 55
+ ++LPDG RNY +KF+ D+WM +GF
Sbjct 635 VVVLLPDGIRNYLTKFLDDDWMKARGFF 662
> YGR155w
Length=507
Score = 32.7 bits (73), Expect = 0.15, Method: Composition-based stats.
Identities = 20/81 (24%), Positives = 37/81 (45%), Gaps = 12/81 (14%)
Query 28 IAMVLPDGSRNYTSKFVCDEWMAEKGFLERSELSR-----------QY-EQYSNMSVQSL 75
I + PD R+Y +KFV DEW+ + + L+R +Y + + N +V+ L
Sbjct 313 IVAIFPDSIRSYLTKFVDDEWLKKNNLWDDDVLARFDSSKLEASTTKYADVFGNATVKDL 372
Query 76 QLPRLTFVSSDSSLKEAAALL 96
L + V + + + +L
Sbjct 373 HLKPVVSVKETAKVTDVIKIL 393
> 7297178
Length=238
Score = 29.6 bits (65), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 16/62 (25%), Positives = 35/62 (56%), Gaps = 1/62 (1%)
Query 35 GSRNYTSKFVCDEWMAEKGFLERSELSRQYEQYSNMSVQSLQLPRLTFVSSDSSLKEAAA 94
G Y + FV + ++ + R LS ++ + ++ Q + +PR++F++ S +EAA+
Sbjct 103 GISTYDALFVHEIAFPDELYFNRDFLSTIFDVFGFVA-QHMHMPRVSFIALSSVDQEAAS 161
Query 95 LL 96
L+
Sbjct 162 LI 163
> Hs18860898_2
Length=745
Score = 29.6 bits (65), Expect = 1.5, Method: Composition-based stats.
Identities = 17/45 (37%), Positives = 23/45 (51%), Gaps = 2/45 (4%)
Query 56 ERSELSRQYEQYSNMSVQSLQLPRLTFV--SSDSSLKEAAALLVQ 98
ER + Y Q+ +M V LP LTFV SS + + E +LV
Sbjct 315 ERVVIQYHYTQWPDMGVPEYALPVLTFVRRSSAARMPETGPVLVH 359
> Hs5729769
Length=943
Score = 28.9 bits (63), Expect = 2.6, Method: Composition-based stats.
Identities = 13/46 (28%), Positives = 20/46 (43%), Gaps = 0/46 (0%)
Query 1 KGTVAATTIGANSRIFRSLWKDDSSRRIAMVLPDGSRNYTSKFVCD 46
K TV N +F W+ I + PDG + YT+ F+ +
Sbjct 511 KNTVTVDNTVGNDTMFLVTWQASGPPEIILFDPDGRKYYTNNFITN 556
> Hs21361384
Length=763
Score = 28.5 bits (62), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 18/58 (31%), Positives = 29/58 (50%), Gaps = 7/58 (12%)
Query 19 LWKDDSSRRIAMVLPDGSRNYTSKFVCDEWMA----EKG--FLERSELSRQY-EQYSN 69
+W D R+I PD + SK + W A EK + E++ LS+Q+ E+Y +
Sbjct 565 VWAKDERRKILQAFPDMHNSNISKILGSRWKAMTNLEKQPYYEEQARLSKQHLEKYPD 622
> Hs5032109
Length=622
Score = 28.1 bits (61), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 16/58 (27%), Positives = 28/58 (48%), Gaps = 7/58 (12%)
Query 19 LWKDDSSRRIAMVLPDGSRNYTSKFVCDEWMA------EKGFLERSELSRQY-EQYSN 69
+W D R+I PD + SK + W + + + E++ LSRQ+ E+Y +
Sbjct 433 VWAKDERRKILQAFPDMHNSSISKILGSRWKSMTNQEKQPYYEEQARLSRQHLEKYPD 490
> HsM5902114
Length=347
Score = 28.1 bits (61), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 17/60 (28%), Positives = 27/60 (45%), Gaps = 8/60 (13%)
Query 19 LWKDDSSRRIAMVLPDGSRNYTSKFVCDEWMAEKGFLERSELSRQ--YEQYSNMSVQSLQ 76
+W D R+I PD + SK + W A + L +Q YE+ + +S Q L+
Sbjct 149 VWAKDERRKILQAFPDMHNSNISKILGSRWKA------MTNLEKQPYYEEQARLSKQHLE 202
> At4g02440
Length=336
Score = 27.7 bits (60), Expect = 4.7, Method: Composition-based stats.
Identities = 17/52 (32%), Positives = 25/52 (48%), Gaps = 6/52 (11%)
Query 12 NSRIFRSLWKDDSSRRIAMVLPDGSRNYTSKF-----VCDE-WMAEKGFLER 57
N +FR ++KD R+ + DG+R+ TS +CDE W F R
Sbjct 251 NYMLFRGVFKDFKRSRVWRTINDGNRSKTSGLKCAFCLCDETWDLHSSFCLR 302
> CE28389
Length=563
Score = 27.7 bits (60), Expect = 5.6, Method: Composition-based stats.
Identities = 14/53 (26%), Positives = 27/53 (50%), Gaps = 0/53 (0%)
Query 26 RRIAMVLPDGSRNYTSKFVCDEWMAEKGFLERSELSRQYEQYSNMSVQSLQLP 78
R++A+V + Y S+ D +E+ FLER+ L +Y++ + + P
Sbjct 510 RKVAVVQRESESAYESEQGSDGETSEESFLERASLFDEYDRMIEILAEEYDTP 562
Lambda K H
0.316 0.127 0.355
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1167556980
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40