bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_6757_orf2
Length=122
Score E
Sequences producing significant alignments: (Bits) Value
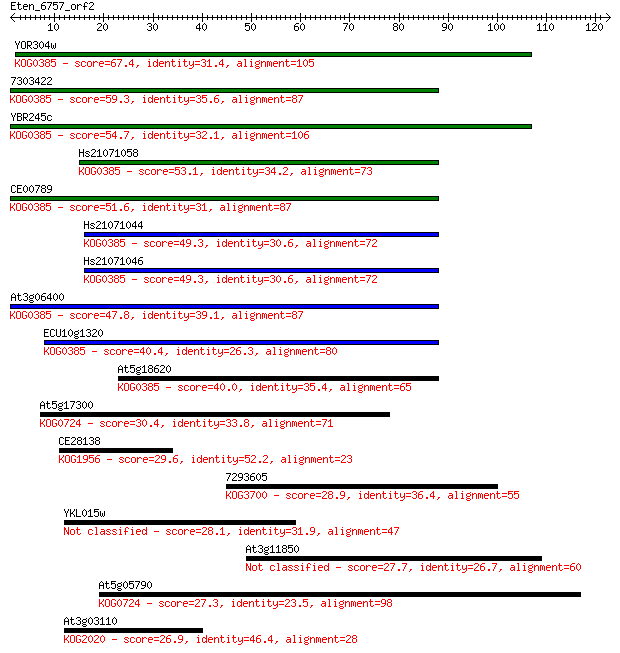
YOR304w 67.4 6e-12
7303422 59.3 2e-09
YBR245c 54.7 4e-08
Hs21071058 53.1 1e-07
CE00789 51.6 3e-07
Hs21071044 49.3 2e-06
Hs21071046 49.3 2e-06
At3g06400 47.8 5e-06
ECU10g1320 40.4 9e-04
At5g18620 40.0 0.001
At5g17300 30.4 0.83
CE28138 29.6 1.5
7293605 28.9 2.3
YKL015w 28.1 3.9
At3g11850 27.7 5.9
At5g05790 27.3 7.1
At3g03110 26.9 8.6
> YOR304w
Length=1120
Score = 67.4 bits (163), Expect = 6e-12, Method: Compositional matrix adjust.
Identities = 33/105 (31%), Positives = 62/105 (59%), Gaps = 3/105 (2%)
Query 2 FTAEMKLQKEQLLSEGFLNWNRTEFTKFVSGLIQFGRDRLEEAWQMHFSNSNKTVEDLRK 61
FT E +L+K++L+S+ F NWN+ +F F++ ++GRD +E + S +KT E++
Sbjct 872 FTQEDELRKQELISKAFTNWNKRDFMAFINACAKYGRDDME---NIKKSIDSKTPEEVEV 928
Query 62 YAAVFFQRYSEIEGGDRMMQRIERAEELRGALDLQRRAIQATVEE 106
YA +F++R EI G ++ + +E E+ L Q ++ +E+
Sbjct 929 YAKIFWERLKEINGWEKYLHNVELGEKKNEKLKFQETLLRQKIEQ 973
> 7303422
Length=1027
Score = 59.3 bits (142), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 31/87 (35%), Positives = 53/87 (60%), Gaps = 3/87 (3%)
Query 1 PFTAEMKLQKEQLLSEGFLNWNRTEFTKFVSGLIQFGRDRLEEAWQMHFSNSNKTVEDLR 60
P T E +KE LLS+GF W + +F +F+ ++GRD ++ + KT E++
Sbjct 780 PLTEEEIQEKENLLSQGFTAWTKRDFNQFIKANEKYGRDDID---NIAKDVEGKTPEEVI 836
Query 61 KYAAVFFQRYSEIEGGDRMMQRIERAE 87
+Y AVF++R +E++ +R+M +IER E
Sbjct 837 EYNAVFWERCTELQDIERIMGQIERGE 863
> YBR245c
Length=1129
Score = 54.7 bits (130), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 34/106 (32%), Positives = 58/106 (54%), Gaps = 2/106 (1%)
Query 1 PFTAEMKLQKEQLLSEGFLNWNRTEFTKFVSGLIQFGRDRLEEAWQMHFSNSNKTVEDLR 60
P T E + K SEGF NWN+ EF KF++ ++GR+ + +A + KT+E++R
Sbjct 867 PLTEEEEKMKADWESEGFTNWNKLEFRKFITVSGKYGRNSI-QAIARELA-PGKTLEEVR 924
Query 61 KYAAVFFQRYSEIEGGDRMMQRIERAEELRGALDLQRRAIQATVEE 106
YA F+ IE ++ ++ IE EE + +Q+ A++ + E
Sbjct 925 AYAKAFWSNIERIEDYEKYLKIIENEEEKIKRVKMQQEALRRKLSE 970
> Hs21071058
Length=1052
Score = 53.1 bits (126), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 25/73 (34%), Positives = 48/73 (65%), Gaps = 3/73 (4%)
Query 15 SEGFLNWNRTEFTKFVSGLIQFGRDRLEEAWQMHFSNSNKTVEDLRKYAAVFFQRYSEIE 74
++GF NWN+ +F +F+ ++GRD +E + KT E++ +Y+AVF++R +E++
Sbjct 839 TQGFTNWNKRDFNQFIKANEKWGRDDIE---NIAREVEGKTPEEVIEYSAVFWERCNELQ 895
Query 75 GGDRMMQRIERAE 87
+++M +IER E
Sbjct 896 DIEKIMAQIERGE 908
> CE00789
Length=971
Score = 51.6 bits (122), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 27/88 (30%), Positives = 55/88 (62%), Gaps = 6/88 (6%)
Query 1 PFTAEMKLQKEQLLSEGFLNWNRTEFTKFVSGLIQFGRDRLEE-AWQMHFSNSNKTVEDL 59
P T + + +K +LL++ +W + EF +FV G ++GR+ LE A +M + +E++
Sbjct 744 PLTDKEQEEKAELLTQSVTDWTKREFQQFVRGNEKYGREDLESIAKEME-----RPLEEI 798
Query 60 RKYAAVFFQRYSEIEGGDRMMQRIERAE 87
+ YA VF++R E++ ++++ +IE+ E
Sbjct 799 QSYAKVFWERIEELQDSEKVLSQIEKGE 826
> Hs21071044
Length=1054
Score = 49.3 bits (116), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 22/72 (30%), Positives = 46/72 (63%), Gaps = 3/72 (4%)
Query 16 EGFLNWNRTEFTKFVSGLIQFGRDRLEEAWQMHFSNSNKTVEDLRKYAAVFFQRYSEIEG 75
+GF NW + +F +F+ ++GRD ++ + K+ E++ +Y+AVF++R +E++
Sbjct 855 QGFTNWTKRDFNQFIKANEKYGRDDID---NIAREVEGKSPEEVMEYSAVFWERCNELQD 911
Query 76 GDRMMQRIERAE 87
+++M +IER E
Sbjct 912 IEKIMAQIERGE 923
> Hs21071046
Length=1033
Score = 49.3 bits (116), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 22/72 (30%), Positives = 46/72 (63%), Gaps = 3/72 (4%)
Query 16 EGFLNWNRTEFTKFVSGLIQFGRDRLEEAWQMHFSNSNKTVEDLRKYAAVFFQRYSEIEG 75
+GF NW + +F +F+ ++GRD ++ + K+ E++ +Y+AVF++R +E++
Sbjct 834 QGFTNWTKRDFNQFIKANEKYGRDDID---NIAREVEGKSPEEVMEYSAVFWERCNELQD 890
Query 76 GDRMMQRIERAE 87
+++M +IER E
Sbjct 891 IEKIMAQIERGE 902
> At3g06400
Length=1057
Score = 47.8 bits (112), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 34/88 (38%), Positives = 52/88 (59%), Gaps = 5/88 (5%)
Query 1 PFTAEMKLQKEQLLSEGFLNWNRTEFTKFVSGLIQFGRDRLEE-AWQMHFSNSNKTVEDL 59
P T E +KE LL EGF W+R +F F+ ++GR+ ++ A +M KT E++
Sbjct 826 PLTTEEVEEKEGLLEEGFSTWSRRDFNTFLRACEKYGRNDIKSIASEME----GKTEEEV 881
Query 60 RKYAAVFFQRYSEIEGGDRMMQRIERAE 87
+YA VF +RY E+ DR+++ IER E
Sbjct 882 ERYAKVFKERYKELNDYDRIIKNIERGE 909
> ECU10g1320
Length=823
Score = 40.4 bits (93), Expect = 9e-04, Method: Compositional matrix adjust.
Identities = 21/80 (26%), Positives = 47/80 (58%), Gaps = 6/80 (7%)
Query 8 LQKEQLLSEGFLNWNRTEFTKFVSGLIQFGRDRLEEAWQMHFSNSNKTVEDLRKYAAVFF 67
L+K++L S+GF +W + +F ++ + + GRD +E+ + K +D+ Y VF+
Sbjct 645 LRKKELQSQGF-DWTKKDFHLYIKAVEKVGRDDVEKIAMLL-----KHKDDVEAYHRVFW 698
Query 68 QRYSEIEGGDRMMQRIERAE 87
+R +E+ ++++ I R++
Sbjct 699 ERVNELSDAEKIIGSINRSQ 718
> At5g18620
Length=1063
Score = 40.0 bits (92), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 23/66 (34%), Positives = 38/66 (57%), Gaps = 5/66 (7%)
Query 23 RTEFTKFVSGLIQFGRDRLEE-AWQMHFSNSNKTVEDLRKYAAVFFQRYSEIEGGDRMMQ 81
R +F F+ ++GR+ ++ A +M KT E++ +YA VF RY E+ DR+++
Sbjct 843 RRDFNAFIRACEKYGRNDIKSIASEME----GKTEEEVERYAQVFQVRYKELNDYDRIIK 898
Query 82 RIERAE 87
IER E
Sbjct 899 NIERGE 904
> At5g17300
Length=385
Score = 30.4 bits (67), Expect = 0.83, Method: Compositional matrix adjust.
Identities = 24/74 (32%), Positives = 36/74 (48%), Gaps = 9/74 (12%)
Query 7 KLQKEQLLSEGFLNWNRTEFTKFVSGLIQFGRD--RLEEAWQMHFSNSNKTVEDLRKYAA 64
K++K +++ W E KFV L +GR R+EE H +KT +R +A
Sbjct 42 KVRKPYTITKERERWTDEEHKKFVEALKLYGRAWRRIEE----HV--GSKTAVQIRSHAQ 95
Query 65 VFFQRYS-EIEGGD 77
FF + + E GGD
Sbjct 96 KFFSKVAREATGGD 109
> CE28138
Length=759
Score = 29.6 bits (65), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 12/23 (52%), Positives = 18/23 (78%), Gaps = 0/23 (0%)
Query 11 EQLLSEGFLNWNRTEFTKFVSGL 33
E+L S+GF+++ RTE KF +GL
Sbjct 324 EKLYSKGFISYPRTETNKFPAGL 346
> 7293605
Length=700
Score = 28.9 bits (63), Expect = 2.3, Method: Composition-based stats.
Identities = 20/58 (34%), Positives = 26/58 (44%), Gaps = 6/58 (10%)
Query 45 WQMHFSNSNKTVEDLRKYAAVFFQRYSEIEGGDRMMQ---RIERAEELRGALDLQRRA 99
WQ S S T+ LR+ A FF +Y I D R+ ++LR DLQ A
Sbjct 41 WQ---SQSYHTISRLRRLAVEFFAQYQNITISDLYRDKEIRLPTTDDLRCLADLQTLA 95
> YKL015w
Length=979
Score = 28.1 bits (61), Expect = 3.9, Method: Composition-based stats.
Identities = 15/49 (30%), Positives = 30/49 (61%), Gaps = 3/49 (6%)
Query 12 QLLSEGFLNWNRTEFTKFVSGLIQFGRDR--LEEAWQMHFSNSNKTVED 58
+LL + F+N+N F F GL++ G ++ LE W +++ N+ K +++
Sbjct 341 KLLIDCFINYNDGCFYFFNEGLVKCGINKLYLENKW-LYYDNTKKALDN 388
> At3g11850
Length=504
Score = 27.7 bits (60), Expect = 5.9, Method: Compositional matrix adjust.
Identities = 16/60 (26%), Positives = 32/60 (53%), Gaps = 2/60 (3%)
Query 49 FSNSNKTVEDLRKYAAVFFQRYSEIEGGDRMMQRIERAEELRGALDLQRRAIQATVEEQL 108
SN +T++DL Y + +R + D ++ I+ E + +DL+ + +Q +V+E L
Sbjct 69 VSNQQQTIQDL--YHELEQERIASSTAADETVKMIQTLEREKAKIDLELKQLQRSVDETL 126
> At5g05790
Length=277
Score = 27.3 bits (59), Expect = 7.1, Method: Compositional matrix adjust.
Identities = 23/100 (23%), Positives = 41/100 (41%), Gaps = 24/100 (24%)
Query 19 LNWNRTEFTKFVSGLIQFGRDRLEEAWQMHFSN--SNKTVEDLRKYAAVFFQRYSEIEGG 76
+ W E +F+ GL+++G+ W+ N +KT + +A ++QR
Sbjct 131 VPWTEEEHRRFLLGLLKYGKG----DWRNISRNFVGSKTPTQVASHAQKYYQR------- 179
Query 77 DRMMQRIERAEELRGALDLQRRAIQATVEEQLLSGTVSSP 116
+L GA D +R +I LL+ +S P
Sbjct 180 -----------QLSGAKDKRRPSIHDITTVNLLNANLSRP 208
> At3g03110
Length=1022
Score = 26.9 bits (58), Expect = 8.6, Method: Compositional matrix adjust.
Identities = 13/28 (46%), Positives = 17/28 (60%), Gaps = 0/28 (0%)
Query 12 QLLSEGFLNWNRTEFTKFVSGLIQFGRD 39
+LLS F N TE T+FV+GL + D
Sbjct 934 KLLSSSFPNMTTTEVTQFVNGLYESRND 961
Lambda K H
0.320 0.133 0.382
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1194805952
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40