bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_6741_orf1
Length=125
Score E
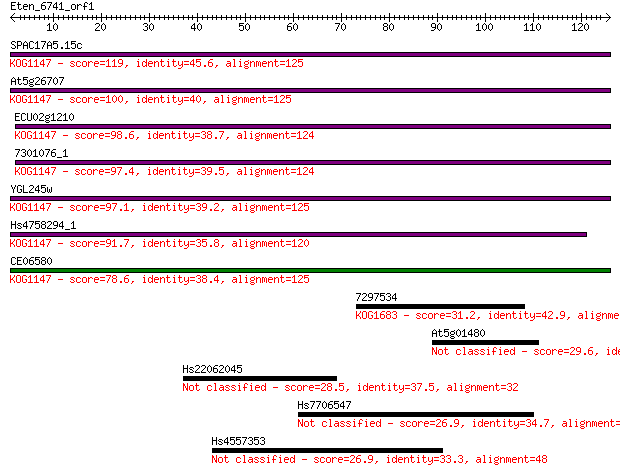
Sequences producing significant alignments: (Bits) Value
SPAC17A5.15c 119 2e-27
At5g26707 100 9e-22
ECU02g1210 98.6 3e-21
7301076_1 97.4 6e-21
YGL245w 97.1 7e-21
Hs4758294_1 91.7 3e-19
CE06580 78.6 2e-15
7297534 31.2 0.43
At5g01480 29.6 1.4
Hs22062045 28.5 3.5
Hs7706547 26.9 8.2
Hs4557353 26.9 8.3
> SPAC17A5.15c
Length=716
Score = 119 bits (297), Expect = 2e-27, Method: Compositional matrix adjust.
Identities = 57/125 (45%), Positives = 74/125 (59%), Gaps = 0/125 (0%)
Query 1 EWDKLWTLNRQVIDPIVPRFMAVSRTDGVKVTISGAPENVATQPRRLHAKNEALGQVPLY 60
+W W N+++IDP+ PR AV D VK TI P + R H KN LG
Sbjct 497 DWTSFWATNKKIIDPVAPRHTAVESGDVVKATIVNGPAAPYAEDRPRHKKNPELGNKKSI 556
Query 61 LHNTILIETDDAQLCQDGEEVTLMHWGNCVFERLERDSTGKVTAISAKLHLEGDFKKTKK 120
N ILIE DAQ + EEVTLM WGN + RD++GKVT++ +LHL+GDFKKT+K
Sbjct 557 FANEILIEQADAQSFKQDEEVTLMDWGNAYVREINRDASGKVTSLKLELHLDGDFKKTEK 616
Query 121 KLNWV 125
K+ W+
Sbjct 617 KVTWL 621
> At5g26707
Length=719
Score = 100 bits (248), Expect = 9e-22, Method: Compositional matrix adjust.
Identities = 50/125 (40%), Positives = 72/125 (57%), Gaps = 0/125 (0%)
Query 1 EWDKLWTLNRQVIDPIVPRFMAVSRTDGVKVTISGAPENVATQPRRLHAKNEALGQVPLY 60
EWDKLW++N+++IDP+ PR AV V T++ P+ + H K E G+
Sbjct 504 EWDKLWSINKRIIDPVCPRHTAVVAERRVLFTLTDGPDEPFVRMIPKHKKFEGAGEKATT 563
Query 61 LHNTILIETDDAQLCQDGEEVTLMHWGNCVFERLERDSTGKVTAISAKLHLEGDFKKTKK 120
+I +E DA GEEVTLM WGN + + + +D G+VTA+S L+L+G K TK
Sbjct 564 FTKSIWLEEADASAISVGEEVTLMDWGNAIVKEITKDEEGRVTALSGVLNLQGSVKTTKL 623
Query 121 KLNWV 125
KL W+
Sbjct 624 KLTWL 628
> ECU02g1210
Length=642
Score = 98.6 bits (244), Expect = 3e-21, Method: Compositional matrix adjust.
Identities = 48/124 (38%), Positives = 73/124 (58%), Gaps = 3/124 (2%)
Query 2 WDKLWTLNRQVIDPIVPRFMAVSRTDGVKVTISGAPENVATQPRRLHAKNEALGQVPLYL 61
WDK+W +NR+ IDP+ R+ V + D V+V+I E P+ H KN LG ++
Sbjct 439 WDKVWAINRKKIDPVSARYFCVQQRDAVEVSIDNTSEYTMDVPK--HKKNGDLGTKEVFY 496
Query 62 HNTILIETDDAQLCQDGEEVTLMHWGNCVFERLERDSTGKVTAISAKLHLEGDFKKTKKK 121
+ IL+ +D ++ QD EE TLM+WGN + + + G VT + L+ +GDFK TK K
Sbjct 497 SSQILLSQEDGRVLQDNEEFTLMNWGNAIV-KSKTVENGTVTKMEVSLNPDGDFKLTKNK 555
Query 122 LNWV 125
++WV
Sbjct 556 MSWV 559
> 7301076_1
Length=1128
Score = 97.4 bits (241), Expect = 6e-21, Method: Compositional matrix adjust.
Identities = 49/125 (39%), Positives = 79/125 (63%), Gaps = 4/125 (3%)
Query 2 WDKLWTLNRQVIDPIVPRFMAVSRTDGVKVTISGAPENVATQPRRLHAKNEALGQVPLYL 61
WDK+W N++VIDPI PR+ A+ + V V ++GA V +H K+E+LG+ + L
Sbjct 495 WDKIWAFNKKVIDPIAPRYTALEKEKRVIVNVAGA--KVERIQVSVHPKDESLGKKTVLL 552
Query 62 HNTILIETDDAQLCQDGEEVTLMHWGNCVFERLERDSTGKVTAISAKLHLEG-DFKKTKK 120
I I+ DA+ ++GE T ++WGN + ++ +D++G +T++ A L+LE DFKKT
Sbjct 553 GPRIYIDYVDAEALKEGENATFINWGNILIRKVNKDASGNITSVDAALNLENKDFKKT-L 611
Query 121 KLNWV 125
KL W+
Sbjct 612 KLTWL 616
> YGL245w
Length=724
Score = 97.1 bits (240), Expect = 7e-21, Method: Compositional matrix adjust.
Identities = 49/125 (39%), Positives = 73/125 (58%), Gaps = 5/125 (4%)
Query 1 EWDKLWTLNRQVIDPIVPRFMAVSRTDGVKVTISGAPENVATQPRRLHAKNEALGQVPLY 60
EW+ +W N++VIDPI PR A+ + + S AP+ + + H KN A+G+ +
Sbjct 509 EWNLIWAFNKKVIDPIAPRHTAIVNPVKIHLEGSEAPQEPKIEMKPKHKKNPAVGEKKVI 568
Query 61 LHNTILIETDDAQLCQDGEEVTLMHWGNCVFERLERDSTGKVTAISAKLHLEGDFKKTKK 120
+ I+++ DDA + EEVTLM WGN + + D + + AKL+LEGDFKKTK
Sbjct 569 YYKDIVVDKDDADVINVDEEVTLMDWGNVIITKKNDDGS-----MVAKLNLEGDFKKTKH 623
Query 121 KLNWV 125
KL W+
Sbjct 624 KLTWL 628
> Hs4758294_1
Length=941
Score = 91.7 bits (226), Expect = 3e-19, Method: Compositional matrix adjust.
Identities = 43/121 (35%), Positives = 74/121 (61%), Gaps = 3/121 (2%)
Query 1 EWDKLWTLNRQVIDPIVPRFMAVSRTDGVKVTISGAPENVATQPRRLHAKNEALGQVPLY 60
EWDK+W N++VIDP+ PR++A+ + + + V + A E + + H KN +G P++
Sbjct 416 EWDKIWAFNKKVIDPVAPRYVALLKKEVIPVNVPEAQEEMKEVAK--HPKNPEVGLKPVW 473
Query 61 LHNTILIETDDAQLCQDGEEVTLMHWGNCVFERLERDSTGKVTAISAKLHLEG-DFKKTK 119
+ IE DA+ +GE VT ++WGN ++ +++ GK+ ++ AK +LE D+KKT
Sbjct 474 YSPKVFIEGADAETFSEGEMVTFINWGNLNITKIHKNADGKIISLDAKFNLENKDYKKTT 533
Query 120 K 120
K
Sbjct 534 K 534
> CE06580
Length=1149
Score = 78.6 bits (192), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 48/131 (36%), Positives = 75/131 (57%), Gaps = 13/131 (9%)
Query 1 EWDKLWTLNRQVIDPIVPRFMAVSRTD---GVKVT--ISGAPENVATQPRRLHAKNEALG 55
EWDK+W N++VIDP+ PR+ A+ T +++T IS NV+ LH KN +G
Sbjct 484 EWDKIWAFNKKVIDPVAPRYTALDSTSPLVSIELTDSISDDTSNVS-----LHPKNAEIG 538
Query 56 QVPLYLHNTILIETDDAQLCQDGEEVTLMHWGNCVFERLERDSTGKVTAISAKLHLEG-D 114
++ +L+E DA ++GE VT ++WGN ++E+ +T ISA L L+ D
Sbjct 539 SKDVHKGKKLLLEQVDAAALKEGEIVTFVNWGNIKIGKIEKKG-AVITKISATLQLDNTD 597
Query 115 FKKTKKKLNWV 125
+KKT K+ W+
Sbjct 598 YKKT-TKVTWL 607
> 7297534
Length=783
Score = 31.2 bits (69), Expect = 0.43, Method: Compositional matrix adjust.
Identities = 15/36 (41%), Positives = 20/36 (55%), Gaps = 1/36 (2%)
Query 73 QLCQDGEEVTLM-HWGNCVFERLERDSTGKVTAISA 107
+ CQ EE TL+ H +F+R+ER V AIS
Sbjct 119 EACQTAEEATLISHGAQVMFDRMERSKKPIVAAISG 154
> At5g01480
Length=413
Score = 29.6 bits (65), Expect = 1.4, Method: Composition-based stats.
Identities = 11/22 (50%), Positives = 15/22 (68%), Gaps = 0/22 (0%)
Query 89 CVFERLERDSTGKVTAISAKLH 110
C ER+++D GKVT IS +H
Sbjct 14 CPVERIQKDKDGKVTTISGGIH 35
> Hs22062045
Length=553
Score = 28.5 bits (62), Expect = 3.5, Method: Composition-based stats.
Identities = 12/32 (37%), Positives = 14/32 (43%), Gaps = 0/32 (0%)
Query 37 PENVATQPRRLHAKNEALGQVPLYLHNTILIE 68
P A QP +H E G +P Y T L E
Sbjct 178 PYQAAFQPSVIHLPEEGFGYLPFYFAGTSLFE 209
> Hs7706547
Length=704
Score = 26.9 bits (58), Expect = 8.2, Method: Composition-based stats.
Identities = 17/49 (34%), Positives = 24/49 (48%), Gaps = 2/49 (4%)
Query 61 LHNTILIETDDAQLCQDGEEVTLMHWGNCVFERLERDSTGKVTAISAKL 109
LHN+ + A CQ E+ L WG+ V+ R+E S G I +L
Sbjct 299 LHNSSSVACGGASGCQ--LELALPPWGHWVYVRVETSSRGPGRTIRFQL 345
> Hs4557353
Length=392
Score = 26.9 bits (58), Expect = 8.3, Method: Composition-based stats.
Identities = 16/48 (33%), Positives = 27/48 (56%), Gaps = 5/48 (10%)
Query 43 QPRRLHAKNEALGQVPLYLHNTILIETDDAQLCQDGEEVTLMHWGNCV 90
+P+ L+ A +VP+ +N I A++ Q+G +VTL+ WG V
Sbjct 239 EPKILY--RAAAEEVPIEPYN---IPLSQAEVIQEGSDVTLVAWGTQV 281
Lambda K H
0.318 0.134 0.412
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1183965632
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40